Abstract
• Premise of the study: Phylogenetic analysis of DNA and amino acid sequences requires the creation of files formatted specifically for each analysis package. Programs currently available cannot simultaneously code inferred insertion/deletion (indel) events in sequence alignments and concatenate data sets.
• Methods and Results: A novel Perl script, 2matrix, was created to concatenate matrices of non-molecular characters and/or aligned sequences and to code indels. 2matrix outputs a variety of formats compatible with popular phylogenetic programs.
• Conclusions: 2matrix efficiently codes indels and concatenates matrices of sequences and non-molecular data. It is available for free download under a GPL (General Public License) open source license (https://github.com/nrsalinas/2matrix/archive/master.zip).
Keywords: 2matrix, file conversion, indel coding, software
To make robust phylogenetic inferences, data from several unlinked sources are often required. Commonly, researchers evaluate DNA (or amino acid) sequences from a number of different regions and/or anatomical, morphological, developmental, biochemical, or behavioral characteristics. To conduct phylogenetic analyses of aligned sequences, a researcher must concatenate sequence files (typically in FASTA format) into a single matrix file formatted specifically for the analysis package used. Binary characters, representing inferred insertion/deletion (indel) events, are often appended to the matrix along with non-molecular data.
Indel events are usually incorporated in phylogenetic matrices using the “simple indel coding” algorithm (Simmons and Ochoterena, 2000). The algorithm creates a character for each unique combination of 5′ and 3′ indel termini in an alignment (5′ termini must be preceded by a nucleotide/amino acid sequence and 3′ termini must be followed by a nucleotide/amino acid sequence). For each character, each sequence is assigned a state based on what is contained between the indel termini: (0) nucleotide/amino acid sequence and/or an indel with termini that do not extend up to or beyond both the 5′ and 3′ indel termini; (1) an indel with the exact same combination of termini; (— [inapplicable]) an indel that extends up to or beyond both the 5′ and 3′ indel termini; or (? [missing]) the sequence begins after the 5′ indel terminus or ends before the 3′ indel terminus. Several software implementations are available: gapcode (part of NEXUS Class Library; Lewis, 2003), GapCoder (Young and Healy, 2003; no longer publicly distributed), 2xread (Little, 2005), and SeqState (Müller, 2006). Although useful, these implementations cannot simultaneously code indel events and concatenate data sets nor can they process sequences along with non-molecular data sets in a straightforward manner. Therefore, we created a program that can code indels, concatenate DNA and amino acid sequences, incorporate non-molecular data, and produce output files compatible with the most widely used analysis programs.
METHODS AND RESULTS
2matrix is an open source Perl (5.10) script that concatenates and translates phylogenetic data sets into a variety of useful file formats. It can be executed on any operating system that has a Perl interpreter (e.g., Linux, Mac OS X, and Windows). Perl interpreters are installed, by default, on Linux and Mac OS X. Users of Windows must install a Perl distribution—available Perl distributions and installation instructions can be found at the Perl Programming Language web site (http://www.perl.org/get.html). Once installed, Perl can be accessed by the user via a terminal window.
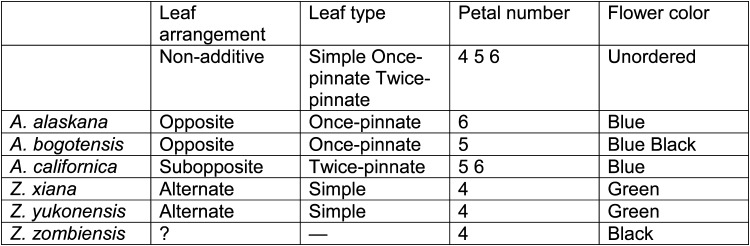
2matrix accepts DNA and amino acid sequence alignments in FASTA format and non-molecular data in xread or comma-separated value (csv) formats. FASTA is the most widely used format for sequence alignments and is output by most alignment programs. Non-molecular data are often compiled using specialized software (e.g., WinClada, Mesquite) that can export xread files or, in some cases, spreadsheet programs that can export csv files. The csv files accepted by 2matrix must be consistently organized: the first row contains character names; the second row describes character state additivity; the first column contains taxon names; the remaining cells contain the scores of a single character for a given taxon (polymorphic entries can be accommodated). Sample files and detailed information on file formats is provided with the program distribution (Fig. 1).
Fig. 1.
Example data matrix in csv format. The first column contains taxon names. The remaining columns are used for individual characters. The first row contains character names, the second row indicates additivity (the order of the additive states must be given; non-additive/unordered characters must be indicated), and remaining rows contain taxon scores. Polymorphic scores are separated by spaces. Missing data are indicated by question marks or dashes.
By default, 2matrix implements the “simple indel coding” algorithm (Simmons and Ochoterena, 2000) to create binary characters that describe indel size and distribution throughout each sequence alignment. Optionally, users can prevent indel coding (“-d”), but still concatenate and/or reformat matrices. Nucleotide and amino acid positions in xread and NEXUS output files can, optionally, be named (“-s”) with a stem phrase (one per partition). This facilitates post-analysis data interpretation—particularly if indels have been coded. All 2matrix command-line options are summarized in Table 1.
Table 1.
Command-line options available in 2matrix.
| Option flaga | Description | Required for operation |
| -d | Produce output without coded indels (the default is to code indels). | No |
| -i file-name | Specify input files (aligned FASTA, csv, or xread [cf. Hennig86, NONA, WinClada]). If several files are to be merged, file names should be input with multiple “-i” flags. | Yes |
| -n root-name | Specify the root name for output files. | Yes |
| -o format | Specify the output file format: “-o x” for xread; “-o n” for NEXUS; “-o p” for extended PHYLIP. If NEXUS format is selected, files compatible with both Garli (“root-name.garli.nex”) and MrBayes (“root-name.mrbayes.nex”) will be created. Additionally, a Garli configuration file will be automatically generated (“root-name.conf”). If PHYLIP format is selected, a RAxML partition file will automatically be created (“root-name.part”). | Yes |
| -s stem-name | Specify the stem name to be used for sequence and indel characters in xread and NEXUS files. If characters are to be named, there must be an “-s” flag for each FASTA file (the “-s” and “-i” flags should be in the same order). | No |
Italicized text following option flags should be specified by the user.
The output of 2matrix is compatible with popular phylogenetic programs: Garli (NEXUS sensu Garli; Zwickl, 2006), RAxML (extended PHYLIP; Stamatakis, 2006), TNT (xread; Goloboff et al., 2008), and MrBayes (NEXUS sensu MrBayes; Ronquist et al., 2012). RAxML and Garli require additional configuration files to read partitioned data sets—2matrix outputs these files using default settings. Users should tailor these configuration files to suit their data and analytic needs. NEXUS files formatted specifically for MrBayes and Garli are output by 2matrix when the NEXUS option is selected (“-o n”). The NEXUS file format (Maddison et al., 1997) is not fully or consistently implemented in most programs that use it. As a result, a NEXUS file that can be read correctly by all programs cannot be created. 2matrix outputs NEXUS files compatible with MrBayes and Garli—due to their current popularity. Unfortunately, this comes at the cost of compatibility with other NEXUS-utilizing programs. With slight manual modification, the MrBayes and Garli NEXUS files can be made compatible with sundry NEXUS-utilizing programs.
The 2matrix distribution includes morphological data (csv and xread format) and sequences for three molecular markers (FASTA files) reconstructed from an analysis of basal angiosperms (Doyle and Endress, 2000). To recreate the combined matrix in TNT format, the user should issue the following command from within a terminal window (assuming that all the files are in the user’s current directory; users of Windows should omit the “./” proceeding the command):
./2matrix.pl -i morphology-example.csv -i 18S-example.fasta -i atpB-example.fasta -i rbcL-example.fasta -s 18S -s atpB -s rbcL -o x -n example -d
To output the same matrix in NEXUS format, the user should replace “-o x” with “-o n” in the command. If the user wishes to add coded indels to the matrix using the “simple indel coding” algorithm, the “-d” option should be omitted (indels were not coded in the original analysis).
In addition to the instructions included in the 2matrix distribution’s README file (https://github.com/nrsalinas/2matrix/blob/master/README), a complete description of all available options can be viewed by invoking 2matrix without any of the required options (“./2matrix.pl” on Linux and Mac OS X, “2matrix.pl” on Windows; Table 1).
CONCLUSIONS
2matrix is hosted on GitHub (https://github.com/nrsalinas/2matrix) and available for free download (https://github.com/nrsalinas/2matrix/archive/master.zip; this is a direct link to a download of the complete 2matrix distribution) under the General Public License (GPL). It is capable of coding indel events, concatenating sequences, incorporating non-molecular data into matrices, and producing output formatted specifically for the popular analytic programs Garli, MrBayes, RAxML, and TNT. In addition, 2matrix can be used within shell scripts and analysis pipelines. No matter how one chooses to use 2matrix, it is vastly more efficient that manually coding indels and/or concatenating matrices.
LITERATURE CITED
- Doyle J. A., Endress P. K. 2000. Morphological phylogenetic analysis of basal angiosperms: Comparison and combination with molecular data. International Journal of Plant Sciences 161: S121–S153 [DOI] [PubMed] [Google Scholar]
- Goloboff P. A., Farris J. S., Nixon K. C. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786 [Google Scholar]
- Lewis P. O. 2003. NCL: A C++ class library for interpreting data files in NEXUS format. Bioinformatics (Oxford, England) 19: 2330–2331 [DOI] [PubMed] [Google Scholar]
- Little D. P. 2005. 2xread: A simple indel coding tool. Available at: http://www.nybg.org/files/scientists/2xread.html [accessed 16 December 2013].
- Maddison D. R., Swofford D. L., Maddison W. P. 1997. NEXUS: An extensible file format for systematic information. Systematic Biology 46: 590–621 [DOI] [PubMed] [Google Scholar]
- Müller K. 2006. Incorporating information from length-mutational events into phylogenetic analysis. Molecular Phylogenetics and Evolution 38: 667–676 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., et al. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M. P., Ochoterena H. 2000. Gaps as characters in sequence-based phylogenetic analysis. Systematic Biology 49: 369–381 [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Young N. D., Healy J. 2003. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation, The University of Texas at Austin, Austin, Texas, USA.