Summary
Description of macrophage activation is currently contentious and confusing. Like the biblical Tower of Babel, macrophage activation encompasses a panoply of descriptors used in different ways. The lack of consensus on how to define macrophage activation in experiments in vitro and in vivo impedes progress in multiple ways, including the fact that many researchers still consider there to be only the two types of activated macrophages often termed M1 and M2. Here we describe a set of standards for the field encompassing three principles: the source of macrophages, definition of the activators, and a consensus collection of markers to describe macrophage activation, with the goal of unifying experimental standards for diverse experimental scenarios. Collectively, we propose a common framework for macrophage activation nomenclature.
Overview
Activation of macrophages has emerged as a key area of immunology, tissue homeostasis, disease pathogenesis, and in resolving and non-resolving inflammation (Biswas and Mantovani, 2010; Gordon and Martinez, 2010; Lawrence and Natoli, 2011; Mantovani et al., 2008; Mantovani et al., 2005; Martinez et al., 2008; Murray and Wynn, 2011b; Nathan and Ding, 2010; Wynn et al., 2013). Over the last several years, diverse terms have been applied to macrophage activation and ‘polarization’ where a stimulus such as cytokines or toll-like receptor (TLR) agonists produces distinct patterns of gene and protein expression. Here we use the term ‘activation’ to mean the perturbation of macrophages with exogenous agents in the same vein as many use ‘polarization’. We also note the ability of macrophages to change their activation states in response to growth factors (e.g., CSF-1 and GM-CSF) and external cues such as cytokines, microbes, microbial products and other modulators including nucleotide derivatives, antibody-Fc receptor stimulation, glucocorticoids, infection, phagocytosis and potentially any other entity capable of being recognized by macrophages. Because macrophage activation is involved in the outcome of many diseases, including metabolic diseases, allergic disorders including airway hyperreactivity, autoimmune diseases, cancer and bacterial, parasitic, fungal, and viral infections we need to establish a common language for describing the properties of the macrophages under investigation.
Background to the problem
We note widespread use of at least four definitions of macrophage activation, combining terms such as M1 and M2, alternative and classical activation, ‘regulatory’ macrophages and subdivisions originating from the parent terms. The origins of these terms originated in the early 1990s when differential effects of IL-4 compared to IFN-γ and/or lipopolysaccharide (LPS) on macrophage gene expression were described (Martinez and Gordon, 2014; Stein et al., 1992). IL- 4 was described to induce ‘alternative activation’ compared to the effects of IFN-γ. It should be noted the term ‘classical’ activation originally referred to macrophages stimulated with IFN-γ is now interchangeably used with IFN-γ and TLR stimulation (Martinez and Gordon, 2014). The second definition came several years later when Mills proposed the M1–M2 terminology (Mills et al., 2000). Mills’ idea originated from the differential metabolism of arginine between macrophages from C57BL/6 and Balb/c mice; an effect he correlated with differences between Th1 and Th2 cell responses in the same strains. Mills and colleagues went further and proposed the M1–M2 dichotomy was an intrinsic property of macrophages associated with transitions from inflammation to healing that would occur in the absence of an adaptive immune response and arose early in evolution (Mills, 2012). Several lines of evidence suggest this theory requires rethinking. First, C57BL/6 mice bear a deletion in the promoter of Slc7a2, the key arginine transporter in macrophages causing large differences in arginine utilization between C57BL/6 and BALB/c mice. This genetic difference between the strains was not known at the time Mills’ hypothesis was published and was therefore not taken into account (Sans-Fons et al., 2013). Second, while Mills’ notion on ‘innate’ shifts in macrophage activation may be true, most immunologists are concerned with immunity in the presence of lymphocytes, which through cytokine secretion, profoundly affect the activation state of macrophages. Third, no molecular definition has yet accounted for an ‘innate’ M1 to M2 transition, although new information from epigenetics and metabolism (see below) may provide a means to dissect intrinsic macrophage activation states.
The third set of nomenclature expanded the M1–M2 definitions to account for different activation scenarios (M2a, M2b etc), balanced by the idea that activation exists on a spectrum and cannot easily be binned into defined groups (Biswas and Mantovani, 2010; Edwards et al., 2006; Mantovani et al., 2005; Martinez and Gordon, 2014; Stout et al., 2005; Stout and Suttles, 2004). The fourth definition refers to macrophages grown in GM-CSF-1 as M1 and CSF-1 as M2 (Joshi et al., 2014). Notably, significant differences have been documented in the transcriptomes of macrophage populations primarily generated with the use of CSF-1 or GM-CSF, without and with exogenous perturbation (Fleetwood et al., 2009) but there is no compelling evidence to assign CSF-1- or GM-CSF-derived macrophages as M1 or M2.
The diversity of terminology and inconsistent use of markers to describe macrophage activation impedes research in several ways. First, researchers entering the field encounter confusion about which terms to use, and which markers are representative of their experimental or human-based system; many researchers may erroneously consider there to be only ‘two types of macrophages.’ Second, established researchers in the field have yet to agree on nomenclature or standards for describing activation. Third, grant and manuscript writers and their reviewers, funding and regulatory agencies, and journal editors can be exasperated at the breadth of terminology in use. Fourth, the lack of experimental standards impedes studies where comparisons are required (e.g., microarray and proteomic datasets) and fifth, deployment of therapeutic macrophage modulators requires translatable standards across disciplines that can be used by pharma and regulatory bodies to draw meaningful comparisons in terms of diagnostic or efficacy metrics. A final issue is the diversity in macrophage activation across species (discussed briefly below).
To address obstacles and pitfalls in describing macrophage activation, and in achieving experimental standards, a small group of macrophage biologists met informally at the International Congress of Immunology in Milan in August 2013. We discussed the issues surrounding terminology and set about providing an initial set of nomenclature and experimental guidelines. A draft letter was then circulated to a broader group of researchers active in this area. We did not attempt to capture everyone who had published on macrophage activation and polarization; rather this perspective is an attempt to attain consensus about the problems within the field and to propose solutions. As such, discussion and revision will be essential for refining the properties and mechanisms of macrophage polarization.
Recommendations
A reproducible experimental standard: We concluded that a starting point was to frame a nomenclature system within a reproducible in vitro experimental standard. CSF-1 cultured macrophages from the murine bone marrow and peripheral blood monocytes from humans remain the predominant in vitro systems used to generate macrophages and therefore will be used as references (Figure 1A). Other commonly used macrophage sources are peritoneal macrophages (resident or elicited) from mice and GM-CSF-cultured macrophages from murine bone marrow (Figure 1A), each of which can be perturbed to generate activated populations of macrophages with overlapping gene expression profiles to CSF-1-generated cells. On this basis, the culture conditions for generating the two paradigmatic in vitro M1 and M2 populations are straightforward, i.e., post-differentiation stimulation with IFN-γ or IL-4. IL-4 and IFN-γ often exert clear-cut antagonistic effects on macrophage polarization mediated to a large extent by the transcription factors STAT6 or STAT1 signaling, respectively, and induce defined and comprehensively investigated macrophage subpopulations (Lawrence and Natoli, 2011; Mills, 2012; Rutschman et al., 2001; Taub and Cox, 1995; Wynn et al., 2013).
Recommendation for minimal reporting standards: Incomplete descriptions of how macrophages are isolated, stimulated, and analyzed are contrary to the value of replication and reproducibility across laboratories. To this end, macrophages isolated from in vitro and in vivo systems require, at a minimum, reporting standards encapsulated in Table 1. Using these standards as a guide, in vitro experiments from different laboratories may be directly compared. Finally, we favor the use of purified endotoxin-free recombinant CSF-1, rather than L cellconditioned media, as the source of CSF-1 to generate bone marrow-derived macrophages (BMDMs) versus L cell-conditioned media, as the latter is not readily defined, and can vary from batch to batch. For example, L cell-conditioned media contains variable amounts of type I interferons, that may cause confounding effects in subsequent activation experiments (Warren and Vogel, 1985).
Define the activator: In general, as diverse mediators have been used alone or in various combinations to generate polarized macrophage populations, we propose researchers describe stimulation scenarios and adopt a nomenclature linked to the activation standards, i.e., M(IL-4), M(Ig), M(IL-10), M(GC), M(IFN-γ), M(LPS) and so forth (Figure 1B). Such a system avoids the complexity of M2a, M2b etc. where one laboratory may experimentally define activation differently to another, and allows new activation conditions to be compared and contrasted with these core examples. Figure 1 also depicts the concept of a ‘spectrum’ of activation to denote ‘states’ of activation states commonly observed (Mosser and Edwards, 2008; Stout et al., 2005; Stout and Suttles, 2004; Xue et al., 2014). The employment of the spectrum concept is useful where ambiguity exists or researchers are operating outside the in vitro CSF-1 schema described above. In summary, we note standards need to be simple for adoption but at the same time not causing sudden conceptual shifts. Therefore, researchers should consider harnessing the terminology and markers for CSF-1-grown macrophages activated under defined conditions as a reference standard (Xue et al., 2014). Where ambiguity exists, for example in a macrophage population isolated from an in vivo system, researchers should emphasize the marker combinations used, stating who the closest relative(s) is along the spectrum shown in Figure 1 (discussed below).
Terms to avoid: We propose the term ‘regulatory’ macrophages should be avoided, as all macrophages are regulatory in some capacity. The use of macrophages derived from mice with specific targeted mutations that prevent development of an M(IL-4) profile (e.g., through the use of IL-4Rα–, or STAT6-deficient macrophages) is recommended to confirm a specific phenotype. Some researchers often ascribe the subset terminology M1 and M2 to GM-CSF- and CSF-1-generated macrophages, respectively: such terminology should be abandoned. When CSF-1 or GM-CSF is used to generate activated macrophage populations it should be clearly indicated. A further complication is GM-CSF cultures contain substantial numbers of CD11c+ cells with distinct antigen presenting activities that need to be accounted for in gene profiling or functional analyses.
Markers of activation: CD4 defines helper T cells. Within CD4+ cells, Foxp3 defines regulatory T cells. These are just two examples of markers defining cell lineages. By contrast macrophage activation is associated with substantial shifts (hundreds of genes) depending on the specific stimuli, but none define a sub-lineage or activation state of macrophages. To the researcher outside the macrophage sphere, marker use probably appears confusing, as immunologists are used to tight marker-lineage association. An example of problematic marker use is expression of Arginase-1 (Arg1) as a ‘marker’ for M2 or M(IL-4) spectrum macrophages, which has lead to interpretive problems as Arg1 is also induced in M1 spectrum macrophages, is expressed in some resident macrophage populations, and highly induced in mycobacteria-infected macrophages, further emphasizing the need for criteria encompassing multiple markers (El Kasmi et al., 2008). Accordingly, we favor an approach using combinations of markers (or lack of marker expression used) to ascribe activation outcomes as outlined in Figure 1B. Clearly, there is significant scope to expand upon marker assignment such as transcription factor and cell surface marker combinations within the standardized experimental framework proposed here, which should serve as a starting cartography for the field.
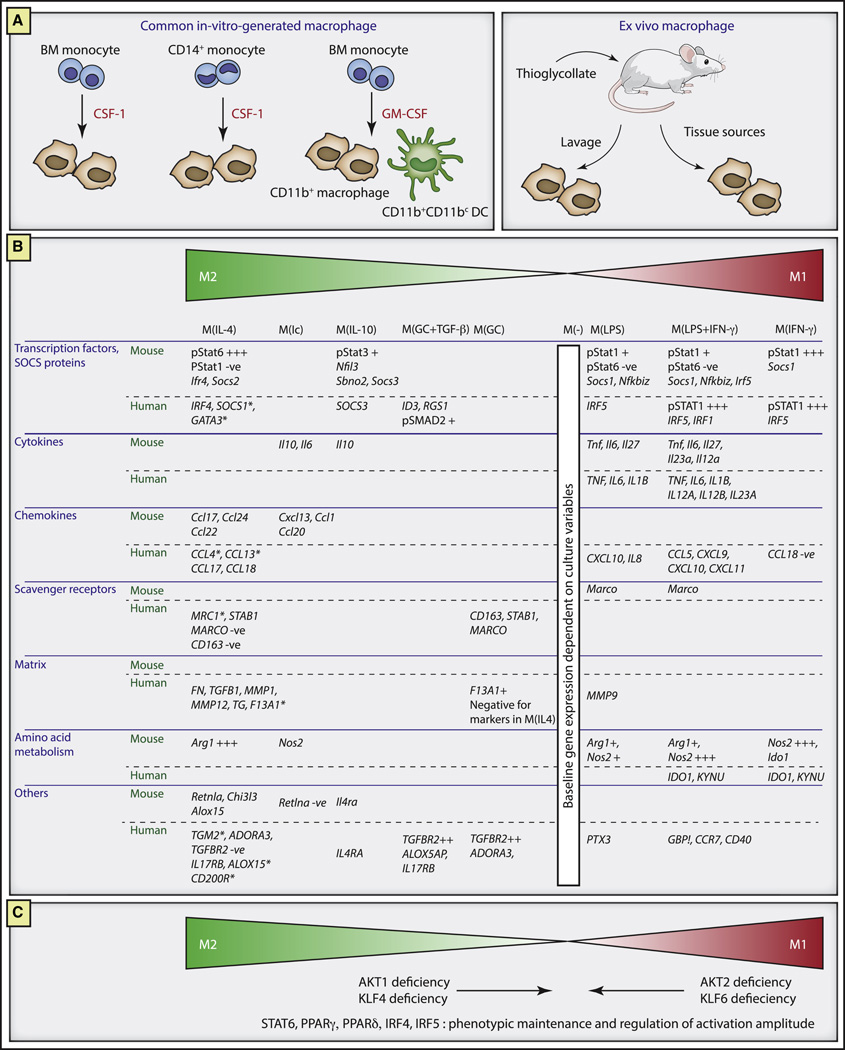
Figure 1. Framework for describing activated macrophages.
A. Examples of widely used macrophage preparations. CSF-1 grown mouse adherent macrophages from the bone marrow or CD14+ monocytes were used as the exemplars for marker evaluation and standardized activation conditions. Macrophages can also be generated with GMCSF, where a CD11c+ DC population is also present depending on the culture conditions. In the mouse, thioglycollate injection followed by peritoneal lavages are used to generate macrophage populations with differing yields and properties, while many organ systems in the mouse and human are sources of tissue infiltrating macrophages.
B. Marker systems for activated macrophages. Shown are functional subdivisions according to stimulation of mouse CSF-1 macrophages or human monocyte-derived CSF-1 macrophages with the existing M1–M2 spectrum concept (Martinez and Gordon, 2014; Mosser and Edwards, 2008; Stout and Suttles, 2004). Stimulation conditions are IL-4, immune complexes (Ic), IL-10, glucocorticoids with TGFβ, glucocorticoids alone, LPS, LPS and IFN-γ and IFN-γ alone. Marker data was drawn from a wide range of published and unpublished data from the authors’ laboratories and represents a starting consensus (Edwards et al., 2006; Fleetwood et al., 2009; Gratchev et al., 2008; Gundra et al., 2014; Krausgruber et al., 2011; Lang et al., 2002; Shirey et al., 2008; Shirey et al., 2014; Shirey et al., 2010; Xue et al., 2014). Asterisk indicates corroboration of human IL-4 genes by deep sequencing (KAS and SNV, unpublished).
C. Use of genetics to aid in macrophage activation studies. Mutations in Akt1 and Klf4 cause a ‘switch’ to M(LPS) and M(IFNγ) associated gene expression while mutations in Akt2 and Klf6 show the reverse phenotype. Mutations in Stat6, Ppard, Pparg, Irf4 and IRF5 depletion are involved in the maintenance and/or amplitude of activation.
Table 1.
Reporting standards for in vitro experiments
| Parameter | Notes |
|---|---|
| Mouse strain | How was the bone marrow isolated and processed? |
| Starting cell number, media and supplements | Media (DMEM versus RPMI) has substantial effects of growth rate, development and activation status |
| Tissue culture conditions | Different types of plastic affect macrophage growth and activation. Tissue culture conditions should be documented for reproducibility |
| Time of culture | What were the precise conditions used? Were cytokines and/or media supplemented during the culture period |
| Source and concentration of differentiation cytokines | The source and concentration of CSF-1 |
| Macrophage yield | The yield relative to the starting number should be recorded |
| Activation conditions | Variables include whether the macrophages were rested prior to activation and how, whether CSF-1 was present in the activation cultures, the source and concentrations of the activating agents, and the time to assay |
| Processing and analysis | How were the cells processed, and what marker readouts used |
Translation to in vivo experiments
When isolating macrophages from tissue and analyzing their activation state, each laboratory will confront a familiar problem: what do I call them? What if there are different populations present? Our recommendation is to acquire sufficient evidence to place a given population within the framework shown in Figure 1. It seems unlikely a particular in vivo scenario will fall exactly within the groups in Figure 1. However, as more macrophage populations are dissected ex vivo, more information will accumulate toward understanding the general and specific nature of in vivo macrophage activation.
Ex vivo characterization of macrophage activation
Each laboratory has individualized macrophage isolation procedures. Because of the breadth of conditions used, we favor describing in detail how macrophages were isolated, from which tissue and pathological or homeostatic condition, and which marker combinations were used to ascertain macrophage activation. All authors stress the need for rapid isolation techniques to preserve the underlying phenotype quickly, and without additional ex vivo culture. Advances in technologies for in situ gene expression within individual tissues and cells will likely advance the understanding of spatial macrophage activation. Regardless of the technology employed, combinations of markers need to be applied to the populations being analyzed and a full description of the isolation techniques provided. For example, the Immgen Consortium has a mandate for isolation and sorting conditions for immune cells and we favor their degree of descriptive rigor for ex vivo macrophages (Gautier et al., 2012). Another complication from ex vivo analysis of macrophage activation is plasticity across different disease stages. For example, in obesity research, adipose tissue resident macrophages are thought to become more proinflammatory as fat accumulates and thus fall toward the M1 end of the activation spectrum (Wynn et al., 2013). In atherosclerosis, resolution of lesions is associated with the reverse: macrophage populations on the M1 spectrum convert to the M2 part of the spectrum without evidence of local STAT6 activation by IL-4 or IL-13 (Moore et al., 2013). One solution to the problem of describing macrophage activation in scenarios in vivo is to begin with an explicit description of the populations under investigation and how they were isolated (as Immgen defines, for example). Markers can then be used to reflect the perturbations they have encountered. For example, Arg1hi, Retnlahi, pSTAT6+, pSTAT1−, could be used to enhance the description of a specific lung macrophage population isolated from a Th2 cell type-driven disease, and thus be reasonably related to the M(IL-4) cells (Figure 1B). Reporting the time points of ex vivo macrophage isolation and analysis are therefore mandatory when describing tissue and disease associated macrophage populations.
Translation to human macrophages
How can we define and categorize activated human macrophages? This question continues to confound researchers in part because human macrophages are generally isolated from blood monocytes as opposed to the bone marrow or tissues commonly used in murine studies. This distinction is particularly important with the new knowledge that many tissue resident populations are not of bone marrow origin (Sieweke and Allen, 2013). Many of the markers used for murine macrophages have not translated to human macrophages. Plausible reasons for these discrepancies have been discussed (Murray and Wynn, 2011a), but it is worth emphasizing no study has systematically compared the responses of blood monocyte-derived macrophages from mice and humans in a side-by-side way. We expect a range of interspecies variability on macrophage activation, reflecting different evolutionary outcomes sculpted by different pathogens, diets, longevity etc. Despite the variables involved, experimental rigor can be used to find information about human (and any other species) macrophage biology by following the principles and practices outlined here. Recently, systematic studies have begun to explore the conservation between macrophages from different species, including the swine where large numbers of different tissue macrophages can be isolated (Fairbairn et al., 2011; Martinez et al., 2013; Schroder et al., 2012; Xue et al., 2014). Therefore, researchers should describe how they generated their macrophages and subsequently stimulated them. When combined with microarray, deep sequencing, and proteomic studies, we anticipate a consensus will emerge about human macrophage activation amenable to new drug discovery.
Genetics to alter activation states: Recent work has identified genetic modifications producing shifts in activation phenotype. For example, deletion of transcription factors IRF4 or KLF6 fail to make M(IL4) macrophages whereas, PPARγ and PPAR∂ are required for the amplitude of the M(IL4) state (Chawla, 2010; Date et al., 2014; Ivashkiv, 2013). Ablation of proteins involved in anabolic growth such AKT2 and PTEN enhance an activation state where gene expression is linked to M(IL4) macrophages, whereas deletion of TSC1, an inhibitor of mTOR, causes the opposite effect (Arranz et al., 2012; Byles et al., 2013; Yue et al., 2014). Other mutations in the mTOR pathway have produced disparate results. However, systematic investigation of mTOR pathway mutants using the principles described here will likely resolve why rapamycin treated macrophages and macrophages from Raptor, Rictor and TSC1 mutants have diverse phenotypes (Ai et al., 2014; Byles et al., 2013; Festuccia et al., 2014; Weichhart et al., 2008). Some of these mutants are summarized in Figure 1C. We contend these, and related, mutants will be increasingly useful to define activation states. Finally, it is important to recognize the effect of timing on altering the activation state: several parameters can effect activation state across time including (i) removal of the stimulus, (ii) enforcement of feedback and feedforward signaling loops including autocrine production of cytokines, and (iii) epigenetic and/or developmental effects built into the life history of a macrophage (Ivashkiv, 2013; Lawrence and Natoli, 2011; Porta et al., 2009). This would go back to Mills’ notion of an activated to healing transition.
Perspectives and conclusions
Understanding macrophage behavior is a keystone of deciphering disease pathogenesis. Macrophages are straightforward to isolate and propagate, facilitating their links to disease. By contrast, nomenclature and standardization issues are stunting progress because a lingua franca has yet to be established and accepted. We hope our attempts are a starting point to resolve some of the immediate issues. We emphasize our goal is to initiate dialog rather than act as arbiters of language and experiment. In doing so, we hope scientists new to macrophage biology, established researchers, pharmaceutical companies, and regulatory agencies can appreciate the history of our field and the need for a common framework open to frequent revision.
Acknowledgements
This work was supported by NIH grants AI062921, Alex’s Lemonade Stand Foundation and the Hartwell Foundation, Cancer Center Core Grant P30 CA21765 and the American Lebanese Syrian Associated Charities (PJM), AI080621 (JPS), HL084312 (EAF), AI18797 (KAS, SNV), The NIH Intramural Program (TAW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Peter J. Murray, Departments of Infectious Diseases & Immunology, St. Jude Children’s Research Hospital, Memphis, TN 38105 USA.
Judith E. Allen, Centre for Immunity, Infection and Evolution, School of Biological Science, University of Edinburgh, UK
Subhra K. Biswas, Singapore Immunology Network (SIgN), A*STAR, 8A Biomedical Grove, #04 138648, Singapore
Edward A. Fisher, The Center for the Prevention of Cardiovascular Disease, New York University School of Medicine, Smilow 7, 522 First Avenue, New York, NY, USA
Derek W. Gilroy, Division of Medicine, Rayne Institute, 5 University Street, University College London WC1 6JJ, UK
Sergij Goerdt, Dept. Dermatology, University Medical Center Mannheim, University of Heidelberg, Mannheim, Germany.
Siamon Gordon, Sir William Dunn School of Pathology, Oxford University, UK.
John A. Hamilton, University of Melbourne, Department of Medicine, Royal Melbourne Hospital, Parkville, Victoria 3050, Australia
Lionel B. Ivashkiv, Hospital for Special Surgery and Weill Medical College of Cornell University, 535 E 70th St, New York, NY 10021
Toby Lawrence, Centre d'Immunologie de Marseille-Luminy, Marseille, France.
Massimo Locati, University of Milan School of Medicine, Humanitas Clinical and Research Center, Via Manzoni 56, I-20089 Rozzano, Italy.
Alberto Mantovani, Humanitas Clinical and Research Center, University of Milan, Via Manzoni 56, 20089 Rozzano, Italy.
Fernando O. Martinez, Botnar Research Centre, Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, University of Oxford, Headington, Oxford OX3 7LD, UK
Jean-Louis Mege, Infectious Diseases, Aix Marseille University, 27 Bd J. Moulin, Marseille, 13285, France.
David M. Mosser, Department of Cell Biology, University of Maryland, College Park, MD 20742
Gioacchino Natoli, Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, Milan, Italy.
Jeroen P. Saeij, Massachusetts Institute of Technology, Department of Biology, Cambridge, Massachusetts, United States of America
Joachim L. Schultze, Genomics & Immunoregulation, LIMES-Institute, University of Bonn, Bonn, 32115, Germany
Kari Ann Shirey, Department of Microbiology and Immunology, Univ. of Maryland, School of Medicine, Baltimore, MD 21201 USA.
Antonio Sica, Humanitas Clinical and Research Center, Via Manzoni 56, 20089 Rozzano, Milan, Italy and Department of Pharmaceutical Sciences, Università del Piemonte Orientale "Amedeo Avogadro", via Bovio 6, Novara, Italy.
Jill Suttles, Microbiology & Immunology, University of Louisville School of Medicine, 319 Abraham Flexner Way, Louisville, KY, 40292, USA.
Irina Udalova, Kennedy Institute of Rheumatology Oxford, UK.
Jo A. van Ginderachter, Laboratory of Cellular and Molecular Immunology, Vrije Universiteit Brussel, & Myeloid Cell Immunology Lab, VIB, Pleinlaan 2, B-1050 Brussels, Belgium
Stefanie N. Vogel, Department of Microbiology and Immunology, Univ. of Maryland, School of Medicine, Baltimore, MD 21201 USA
Thomas A. Wynn, Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892 USA
References
- Ai D, Jiang H, Westerterp M, Murphy AJ, Wang M, Ganda A, Abramowicz S, Welch C, Almazan F, Zhu Y, et al. Disruption of Mammalian target of rapamycin complex 1 in macrophages decreases chemokine gene expression and atherosclerosis. Circ Res. 2014;114:1576–1584. doi: 10.1161/CIRCRESAHA.114.302313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A, Venihaki M, Margioris AN, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Byles V, Covarrubias AJ, Ben-Sahra I, Lamming DW, Sabatini DM, Manning BD, Horng T. The TSC-mTOR pathway regulates macrophage polarization. Nat Commun. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A. Control of macrophage activation and function by PPARs. Circ Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date D, Das R, Narla G, Simon DI, Jain MK, Mahabeleshwar GH. Kruppellike transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem. 2014;289:10318–10329. doi: 10.1074/jbc.M113.526749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J Leukoc Biol. 2006;80:1298–1307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, Konig T, Schleicher U, Koo MS, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn L, Kapetanovic R, Sester DP, Hume DA. The mononuclear phagocyte system of the pig as a model for understanding human innate immunity and disease. J Leukoc Biol. 2011;89:855–871. doi: 10.1189/jlb.1110607. [DOI] [PubMed] [Google Scholar]
- Festuccia WT, Pouliot P, Bakan I, Sabatini DM, Laplante M. Myeloid-specific Rictor deletion induces M1 macrophage polarization and potentiates in vivo pro-inflammatory response to lipopolysaccharide. PLoS One. 2014;9:e95432. doi: 10.1371/journal.pone.0095432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleetwood AJ, Dinh H, Cook AD, Hertzog PJ, Hamilton JA. GM-CSF- and M-CSF-dependent macrophage phenotypes display differential dependence on type I interferon signaling. J Leukoc Biol. 2009;86:411–421. doi: 10.1189/jlb.1108702. [DOI] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Gratchev A, Kzhyshkowska J, Kannookadan S, Ochsenreiter M, Popova A, Yu X, Mamidi S, Stonehouse-Usselmann E, Muller-Molinet I, Gooi L, et al. Activation of a TGF-beta-specific multistep gene expression program in mature macrophages requires glucocorticoid-mediated surface expression of TGF-beta receptor II. J Immunol. 2008;180:6553–6565. doi: 10.4049/jimmunol.180.10.6553. [DOI] [PubMed] [Google Scholar]
- Gundra UM, Girgis NM, Ruckerl D, Jenkins S, Ward LN, Kurtz ZD, Wiens KE, Tang MS, Basu-Roy U, Mansukhani A, et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood. 2014 doi: 10.1182/blood-2013-08-520619. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013;34:216–223. doi: 10.1016/j.it.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Singh AR, Zulcic M, Bao L, Messer K, Ideker T, Dutkowski J, Durden DL. Rac2 controls tumor growth, metastasis and m1–m2 macrophage differentiation in vivo. PLoS One. 2014;9:e95893. doi: 10.1371/journal.pone.0095893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, Udalova IA. IRF5 promotes inflammatory macrophage polarization and TH1–TH17 responses. Nat Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, Thomas B, Fabbri M, Crawshaw A, Ho LP, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121:e57–e69. doi: 10.1182/blood-2012-06-436212. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- Mills CD. M1 and M2 Macrophages: Oracles of Health and Disease. Critical Reviews in Immunology. 2012;32:463–488. doi: 10.1615/critrevimmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.1701141. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Obstacles and opportunities for understanding macrophage polarization. J Leukoc Biol. 2011a;89:557–563. doi: 10.1189/jlb.0710409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011b;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, Di Liberto D, Dieli F, Ghisletti S, Natoli G, De Baetselier P, et al. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci U S A. 2009;106:14978–14983. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, Murray PJ. Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J Immunol. 2001;166:2173–2177. doi: 10.4049/jimmunol.166.4.2173. [DOI] [PubMed] [Google Scholar]
- Sans-Fons MG, Yeramian A, Pereira-Lopes S, Santamaria-Babi LF, Modolell M, Lloberas J, Celada A. Arginine transport is impaired in C57Bl/6 mouse macrophages as a result of a deletion in the promoter of Slc7a2 (CAT2), and susceptibility to Leishmania infection is reduced. J Infect Dis. 2013;207:1684–1693. doi: 10.1093/infdis/jit084. [DOI] [PubMed] [Google Scholar]
- Schroder K, Irvine KM, Taylor MS, Bokil NJ, Le Cao KA, Masterman KA, Labzin LI, Semple CA, Kapetanovic R, Fairbairn L, et al. Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proc Natl Acad Sci U S A. 2012;109:E944–E953. doi: 10.1073/pnas.1110156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey KA, Cole LE, Keegan AD, Vogel SN. Francisella tularensis live vaccine strain induces macrophage alternative activation as a survival mechanism. J Immunol. 2008;181:4159–4167. doi: 10.4049/jimmunol.181.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey KA, Lai W, Pletneva LM, Karp CL, Divanovic S, Blanco JC, Vogel SN. Role of the lipoxygenase pathway in RSV-induced alternatively activated macrophages leading to resolution of lung pathology. Mucosal Immunol. 2014;7:549–557. doi: 10.1038/mi.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey KA, Pletneva LM, Puche AC, Keegan AD, Prince GA, Blanco JC, Vogel SN. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha-, TLR4-, and IFN-beta-dependent. Mucosal Immunol. 2010;3:291–300. doi: 10.1038/mi.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieweke MH, Allen JE. Beyond stem cells: self-renewal of differentiated macrophages. Science. 2013;342:1242974. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–349. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–513. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub DD, Cox GW. Murine Th1 and Th2 cell clones differentially regulate macrophage nitric oxide production. J Leukoc Biol. 1995;58:80–89. doi: 10.1002/jlb.58.1.80. [DOI] [PubMed] [Google Scholar]
- Warren MK, Vogel SN. Bone marrow-derived macrophages: development and regulation of differentiation markers by colony-stimulating factor and interferons. J Immunol. 1985;134:982–989. [PubMed] [Google Scholar]
- Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Horl WH, Hengstschlager M, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S, Rao J, Zhu J, Busuttil RW, Kupiec-Weglinski JW, Lu L, Wang X, Zhai Y. Myeloid PTEN Deficiency Protects Livers from Ischemia Reperfusion Injury by Facilitating M2 Macrophage Differentiation. J Immunol. 2014;192:5343–5353. doi: 10.4049/jimmunol.1400280. [DOI] [PMC free article] [PubMed] [Google Scholar]