Key Points
A high frequency of RAS/RAF mutations and recurrent mutations in PDGFRA and JAK3 were found in relapsed multiple myeloma patients.
Patients with NRAS, but not KRAS, mutation exhibited significantly reduced sensitivity to bortezomib but not high-dose dexamethasone.
Abstract
Various translocations and mutations have been identified in myeloma, and certain aberrations, such as t(4;14) and del17, are linked with disease prognosis. To investigate mutational prevalence in myeloma and associations between mutations and patient outcomes, we tested a panel of 41 known oncogenes and tumor suppressor genes in tumor samples from 133 relapsed myeloma patients participating in phase 2 or 3 clinical trials of bortezomib. DNA mutations were identified in 14 genes. BRAF as well as RAS genes were mutated in a large proportion of cases (45.9%) and these mutations were mutually exclusive. New recurrent mutations were also identified, including in the PDGFRA and JAK3 genes. NRAS mutations were associated with a significantly lower response rate to single-agent bortezomib (7% vs 53% in patients with mutant vs wild-type NRAS, P = .00116, Bonferroni-corrected P = .016), as well as shorter time to progression in bortezomib-treated patients (P = .0058, Bonferroni-corrected P = .012). However, NRAS mutation did not impact outcome in patients treated with high-dose dexamethasone. KRAS mutation did not reduce sensitivity to bortezomib or dexamethasone. These findings identify a significant clinical impact of NRAS mutation in myeloma and demonstrate a clear example of functional differences between the KRAS and NRAS oncogenes.
Introduction
Myeloma patients exhibit significant variation in clinical course and survival. Although myeloma is generally considered incurable, clinical disease staging systems and gene-expression classifiers identify patients with notably better or worse prognoses.1-4 The introduction of new therapies including bortezomib, thalidomide, and lenalidomide have improved patient survival,5 but outcomes still vary significantly. Tumor heterogeneity is a key factor influencing this variation2,6 and genomic analyses have highlighted 6 to 10 distinct biological subtypes of myeloma.4,6 One study reporting whole-genome mutational analyses in 38 myeloma patients identified somatic mutations in well-characterized oncogenes (KRAS, NRAS, BRAF), as well as mutations not previously reported in myeloma or other tumors (ie, XBP1, FAM46C, DIS3).7
In most tumor types exhibiting mutation of a RAS gene family member (HRAS, KRAS, or NRAS), the mutational activation of 1 member predominates.8 In solid tumors, including colorectal,9 lung,8 and pancreatic cancer,8 KRAS is mutated much more frequently than NRAS; the reverse is true in some hematologic cancers such as acute lymphoblastic and chronic myelomonocytic leukemias, and Hodgkin lymphoma.8,10 Myeloma does not exhibit such predominance; instead, there appears to be high and approximately equal rates (∼20%) of KRAS and NRAS mutation.11,12 Although much is known about RAS signaling biology, specific functions distinguishing the 3 RAS family members are not fully understood. The genes exhibit differences in structure and expression pattern, while the proteins can have distinct subcellular localizations that are influenced by a short, highly-divergent C-terminal sequence.8,13
The normal functions of RAS family members in the mouse are not redundant; KRAS is required to complete midgestation development,14 while mice lacking both NRAS and HRAS reach adulthood.15 These and other mouse models also highlight the similar functions of RAS family members; HRAS can replace KRAS function in mouse development if expressed from the KRAS locus,16 and NRAS mutants die in utero if 1 wild-type KRAS allele is deleted indicating that KRAS compensates for loss of NRAS function in normal mouse development.14 Mutational analyses also suggest that oncogenic forms of RAS family members activate similar cellular pathways. BRAF mutations are mutually exclusive to KRAS and/or NRAS mutation in various tumors including melanoma and colorectal cancer,7,17,18 suggesting each RAS family member provides a similar oncogenic signal to the RAF/MAPK pathway. This is further supported by analysis of the subset of cancers that mutate multiple RAS family members, including acute lymphoblastic leukemia,19 myeloma,7,12 and thyroid cancer20; in each of these, mutations in 1 family member (NRAS, HRAS, or KRAS) are mutually exclusive to mutations in the other family members.8 Myeloma may be a particularly appropriate setting in which to study the clinical11,12 and cellular21 functions of the different RAS family genes because NRAS and KRAS are both known to be mutated at a high frequency.
We assayed a panel of known cancer genes, including NRAS, KRAS, and HRAS, in a cohort of relapsed myeloma patients who were treated in phase 2 or 3 studies of the proteasome inhibitor bortezomib. We report both the mutation spectrum in 133 myeloma cases and associations between mutations and clinical outcome.
Patients and methods
Clinical studies, and sample collection and enrichment
DNA from bone marrow aspirates was available for 133 myeloma patients who participated in bortezomib clinical trials,22-24 including the phase 3 Assessment of Proteasome Inhibition for EXtending Remissions (APEX) trial in which relapsed myeloma patients were randomized to treatment with single-agent bortezomib or high-dose dexamethasone.22 Germline DNA was not collected from these patients. Review boards at all participating institutions approved the studies, and all patients provided written informed consent for the trial and for pharmacogenomics analysis. Studies were conducted in accordance with the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice guidelines. Details of this research cohort subset, clinical data, and tumor sample enrichment have been summarized previously.25
Mutational analysis
Tumor DNA was amplified and screened for mutations in a panel of cancer genes using MassARRAY/Sequenom mass spectrometry–based methodology.26 Building upon OncoCarta technology, a custom panel was developed with Sequenom that detected 514 reported mutations across 41 distinct oncogenes and tumor suppressor genes (supplemental Table 1, available on the Blood Web site) using matrix-assisted laser desorption/ionization–time of flight mass spectrometry.27 A minimum observed mutant allele frequency of 8% was required for a positive mutant call; automated quality control was performed and all mutation calls were verified by manual inspection of spectra peaks28 (supplemental Methods). Data were further annotated to identify and remove 3 reported variants in 2 genes no longer regarded to be somatic mutations. A breakdown of observed mutations by coding change is given in supplemental Table 2.
Consistent with reports of high specificity and sensitivity with Sequenom,28 our results from polymerase chain reaction (PCR) testing of these samples also showed high concordance in mutation calls. We validated a subset of 50 samples using PCR amplification of hotspot regions in the KRAS and NRAS loci. Of 100 PCR validation runs attempted, 96 passed quality control. Using the PCR results as a reference, a highly significant rate of concordance in the frequency of KRAS and NRAS mutations was detected with the 2 technologies, with 100% and 96% sensitivity for KRAS and NRAS assays, respectively, and 92% specificity for both gene assays (supplemental Table 3).
Details of the mutations observed in the 133 tumor samples analyzed, as well as associated patient demographics, number of prior lines of therapy received, drug treatment on study, response to treatment, time to progression (TTP), and overall survival (OS) are provided in supplemental Table 4.
Statistical analyses
Associations between genetic mutations and patients’ response to bortezomib or dexamethasone, classified using European Group for Blood and Marrow Transplantation criteria,29 were tested using a 2-sided Fisher exact test. For each gene, tests were conducted for overrepresentation and/or underrepresentation of response groups within subsets of patients who were positive for mutations within a given gene. Mutations with significant enrichment in responding or nonresponding patient groups were further analyzed to determine whether presence of the mutant gene was associated with TTP or OS. TTP and OS were estimated using Kaplan-Meier analysis. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated using the Cox proportional hazard test and P values were determined using the pairwise log-rank test. Reported P values are unadjusted for multiple testing; however, any result described as statistically significant had a Bonferroni-corrected P value of < .05 after adjusting for the number of genes tested for each association (14 mutations in the response analyses and 2 mutations for TTP and OS).
Results
Mutation detection
Tumor samples were collected from relapsed myeloma patients prior to clinical trial enrollment and subject to focused DNA mutation analysis using the MassARRAY platform.28,30,31 This subset of patients has been described previously25 and shown to exhibit demographic and prognostic characteristics similar to the overall trial populations.22-25
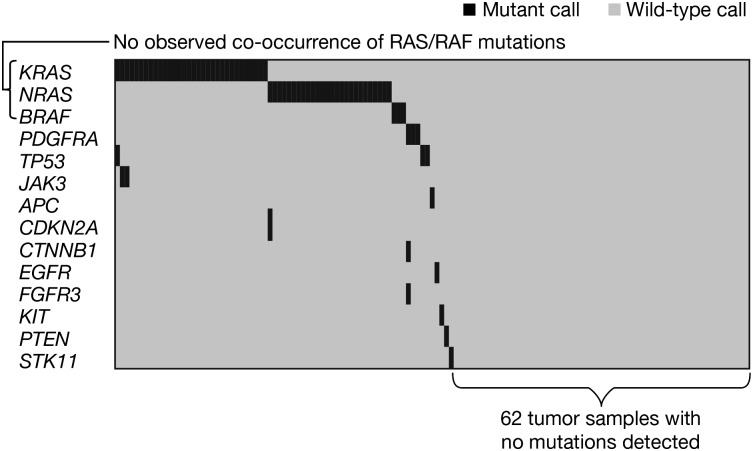
At least 1 mutation was identified in 71 of 133 (53%) tumor samples across 14 genes (Figure 1). Recurrent mutations were detected in KRAS, NRAS, TP53, BRAF, PDGFRA, and JAK3. KRAS and NRAS mutation detection was also independently verified with PCR-based methodology (see “Patients and methods”). Table 1 summarizes all mutations observed alongside those of the whole-genome sequencing analysis of 38 myeloma tumor samples by Chapman et al.7 Mutations were generally >15% of the DNA detected (supplemental Table 5), indicating these are likely a major clone of the enriched tumor, although the possibility of mixed clonality32 cannot be excluded. These data support the high prevalence of oncogenic RAS mutations in myeloma, as reported previously.7,11 There were 32 patients with KRAS mutation (24.1%) and 26 additional patients with NRAS mutation (19.5%). BRAF and TP53 were mutated at low frequency (3 mutations for each) in myeloma, which is largely consistent with results from Chapman et al.7 Nine mutations detected in the present study were not reported by Chapman et al,7 including recurrent mutations in PDGFRA and JAK3, as well as 1 in FGFR3, a gene that undergoes recurrent translocation but is rarely mutated in myeloma.33 Importantly, this assay platform interrogates only tumor DNA, relying on prior knowledge that these “hotspot mutations” are relevant in human cancer,28,30,31 and does not compare tumor DNA to germline DNA.
Figure 1.
Coexpression matrix of all screened samples (N = 133).
Table 1.
Comparison of nonsynonymous mutations
| Gene | Mutants observed | |||
|---|---|---|---|---|
| Present analysis, N = 133 myeloma tumor samples | Previous study, N = 38 myeloma tumor samples | |||
| n | % (95% CI) | n | % (95% CI) | |
| RAS/RAF* | 61 | 45.9 (37-54.3) | 20 | 52.6 (37-68.5) |
| RAS* | 58 | 43.6 (35-52.0) | 19 | 50.0 (34-65.9) |
| KRAS | 32 | 24.1 (17-31.3) | 10 | 26.3 (12-40.3) |
| NRAS | 26 | 19.5 (13-26.3) | 9 | 23.7 (10-37.2) |
| TP53 | 3 | 2.3 (0-4.8) | 3 | 7.9 (0-16.5) |
| BRAF | 3 | 2.3 (0-4.8) | 1 | 2.6 (0-7.7) |
| PDGFRA† | 3 | 2.3 (0-4.8) | 0 | 0 |
| JAK3† | 2 | 1.5 (0-3.6) | 0 | 0 |
| EGFR | 1 | 0.8 (0-2.2) | 1 | 2.6 (0-7.7) |
| APC† | 1 | 0.8 (0-2.2) | 0 | 0 |
| CDKN2A† | 1 | 0.8 (0-2.2) | 0 | 0 |
| CTNNB1† | 1 | 0.8 (0-2.2) | 0 | 0 |
| FGFR3† | 1 | 0.8 (0-2.2) | 0 | 0 |
| KIT† | 1 | 0.8 (0-2.2) | 0 | 0 |
| PTEN† | 1 | 0.8 (0-2.2) | 0 | 0 |
| STK11† | 1 | 0.8 (0-2.2) | 0 | 0 |
| ERBB3 | 0 | 0 | 1 | 2.6 (0-7.7) |
| FGFR2 | 0 | 0 | 1 | 2.6 (0-7.7) |
| JAK2 | 0 | 0 | 1 | 2.6 (0-7.7) |
| PTPN11 | 0 | 0 | 1 | 2.6 (0-7.7) |
| RB1 | 0 | 0 | 1 | 2.6 (0-7.7) |
Comparison of nonsynonymous mutations detected by Sequenom screening of 133 myeloma tumor samples in the present analysis, and in a previous study7 of 38 myeloma tumor samples.
RAS/RAF and RAS are aggregate categories including KRAS, NRAS, and BRAF mutants, and KRAS and NRAS mutants, respectively.
Mutants detected in the present study that were not reported in the previous study by Chapman et al7 and were absent and/or rarely observed in previous reports of myeloma tumor genetic profiling.
Characterization of 133 distinct patients enabled analysis of co-occurrence or mutual exclusivity of mutations, and inference of which genes may function in the same pathway. Figure 1 shows that mutations in KRAS, NRAS, and BRAF were mutually exclusive in this myeloma sample set (mutual exclusivity, P* = .00005; calculated using a permutation test [see supplemental Methods]), indicating these well-characterized oncogenes likely act as a single integrated signaling pathway critical to growth and/or survival of approximately half of myeloma cases. Interestingly, the 3 mutations identified in PDGFRA were also mutually exclusive to RAS/RAF mutations in this data set. It appears that tumors with RAS/RAF pathway mutation generally had few additional mutations, while the remainder of mutant cases often exhibited multiple oncogene mutations in the same sample (Figure 1). Larger data sets are required to definitively address this possibility.
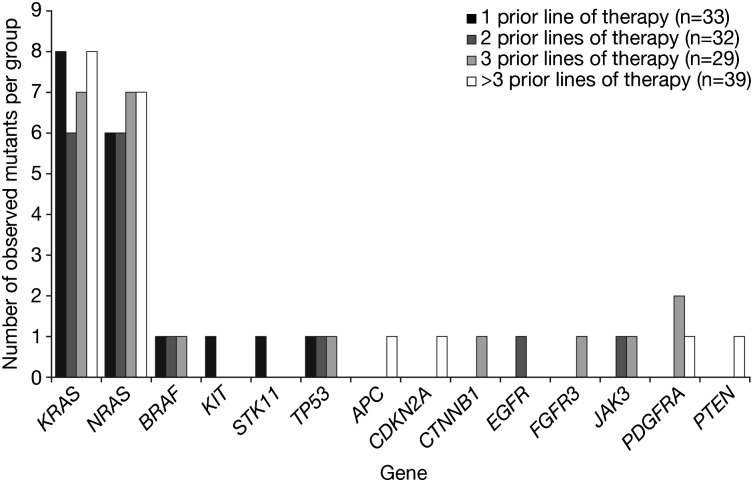
Patients in the present analysis had received 1 to 14 prior lines of therapy. To examine whether the number of prior therapies received was associated with an increased number of mutations, we assessed the frequency of mutations per gene in patients grouped by number of prior lines of therapy (1 to >3 prior therapies). KRAS and NRAS mutations occurred at the same frequency in each group (Figure 2). Similarly, the other genetic mutations detected did not show increased frequency of mutation with increasing number of prior lines of therapy. A focused assessment of known mutagenic therapies yielded similar results. When analysis was limited to samples from patients who had received 0 (N = 20) or >3 (N = 35) prior mutagenic chemotherapies (alkylators, anthracyclines, and/or high-dose chemotherapy preceding transplant), we found no significant difference in the rates of KRAS (P = .51) or NRAS (P = .76) mutations between the groups (supplemental Table 6). Linear regression modeling also failed to detect a significant association between the number of prior mutagenic therapies received and the observed frequency of KRAS and NRAS mutations (P = .28). These data, together with the comparable frequencies of KRAS and NRAS mutation reported in other studies,7,11 suggest that prior drug exposure was not a key contributor to the observed mutational spectrum at particular sites within the specific genes analyzed in this study.
Figure 2.
Frequency of mutations detected per gene by number of prior lines of therapy received before sample collection (1, 2, 3, or >3 prior lines). Depending upon institutional practice, a line of therapy may have included 1 agent, a combination, or multiagent induction followed by bone marrow ablation and stem cell transplantation.
Mutation status impacts subsequent drug response
Response data for evaluable patients are summarized in supplemental Table 7. Each gene was analyzed for an association with response (any response, compared with stable disease or progressive disease) (Table 2). In 74 patients treated with bortezomib in the APEX trial, 1 of 15 patients with NRAS mutant tumors responded (7%, 95% CI: 0%-19%) compared with 31 of 59 (53%, 95% CI: 40%-65%) with wild-type NRAS. In contrast, 9 of 17 KRAS mutant patients (53%, 95% CI: 29%-77%) responded to bortezomib, which was similar to the response rate observed in KRAS wild-type patients (23 of 57, 40%, 95% CI: 28%-53%). In 31 dexamethasone-treated patients, 3 of 6 with NRAS mutant tumors responded (50%, 95% CI: 10%-90%), compared with 8 of 25 (32%, 95% CI: 14%-50%) responding NRAS wild-type patients. KRAS mutation status did not significantly impact response to dexamethasone with 3 of 9 (33%, 95% CI: 3%-64%) and 8 of 22 (36%, 95% CI: 16%-56%) responses in KRAS mutant and KRAS wild-type patients, respectively. The only statistically significant association was between presence of NRAS mutation and lack of response to bortezomib (P = .00116, Bonferroni-corrected P = .016). Supplemental Figure 1 shows the overall mutation profile across bortezomib-treated and dexamethasone-treated patients separately, plus best response.
Table 2.
Response to bortezomib and dexamethasone according to NRAS and KRAS mutation status
| Patients treated with bortezomib | Patients treated with dexamethasone | |||||
|---|---|---|---|---|---|---|
| R | NR | Total | R | NR | Total | |
| NRAS | ||||||
| Mutant | 1 | 14 | 15 | 3 | 3 | 6 |
| Wild-type | 31 | 28 | 59 | 8 | 17 | 25 |
| Total | 32 | 42 | P = .00116* | 11 | 20 | P = .64 |
| KRAS | ||||||
| Mutant | 9 | 8 | 17 | 3 | 6 | 9 |
| Wild-type | 23 | 34 | 57 | 8 | 14 | 22 |
| Total | 32 | 42 | P = .41 | 11 | 20 | P = 1.0 |
CR, complete response; MR, minimal response; NR, nonresponse (stable disease + PD); PD, progressive disease; PR, partial response.
R category equals complete, partial, or minimal response22; NR category equals stable or progressive disease. Similar results if R equals complete and partial response (see supplemental Table 13).
P = .016 after Bonferroni correction; P values calculated from 2-sided Fisher exact test.
In light of the highly conserved structure of the RAS genes, it is unclear why response to bortezomib was significantly different for NRAS, but not KRAS, mutation. One possibility is that specific codons are mutated in the different genes. Oncogenic mutations in RAS genes cluster either at codon 12, 13, or 61 and recent reports suggest specific cellular signaling may result from activation at different hotspots.34 Therefore, the apparent NRAS specificity could be confounded by hotspot bias if 1 mutant codon in NRAS conferred resistance and mutation within that codon was absent from KRAS mutants in this data set. The observed effects of NRAS mutation in the present study might have been explained by the fact that codon 61 is mutated more frequently in NRAS than in KRAS10; however, as summarized in supplemental Table 8, this possibility is not supported by the current data set. The NRAS mutant cohort included 12 cases within codon 61 (1 response) and 3 within codon 12 (no responses). Among the response-evaluable KRAS mutant patients, there were 10 cases of mutation within codon 61 and 8 cases of mutation within codons 12 or 13; both groups exhibited a 50% response rate to bortezomib (supplemental Table 8). For both genes, these codon-specific mutation rates are consistent with prior reports in myeloma.12,21 Thus, the lower rate of response to single-agent bortezomib was linked to mutations in the NRAS gene specifically, rather than the location of the mutation in either RAS gene.
Mutation status impacts TTP
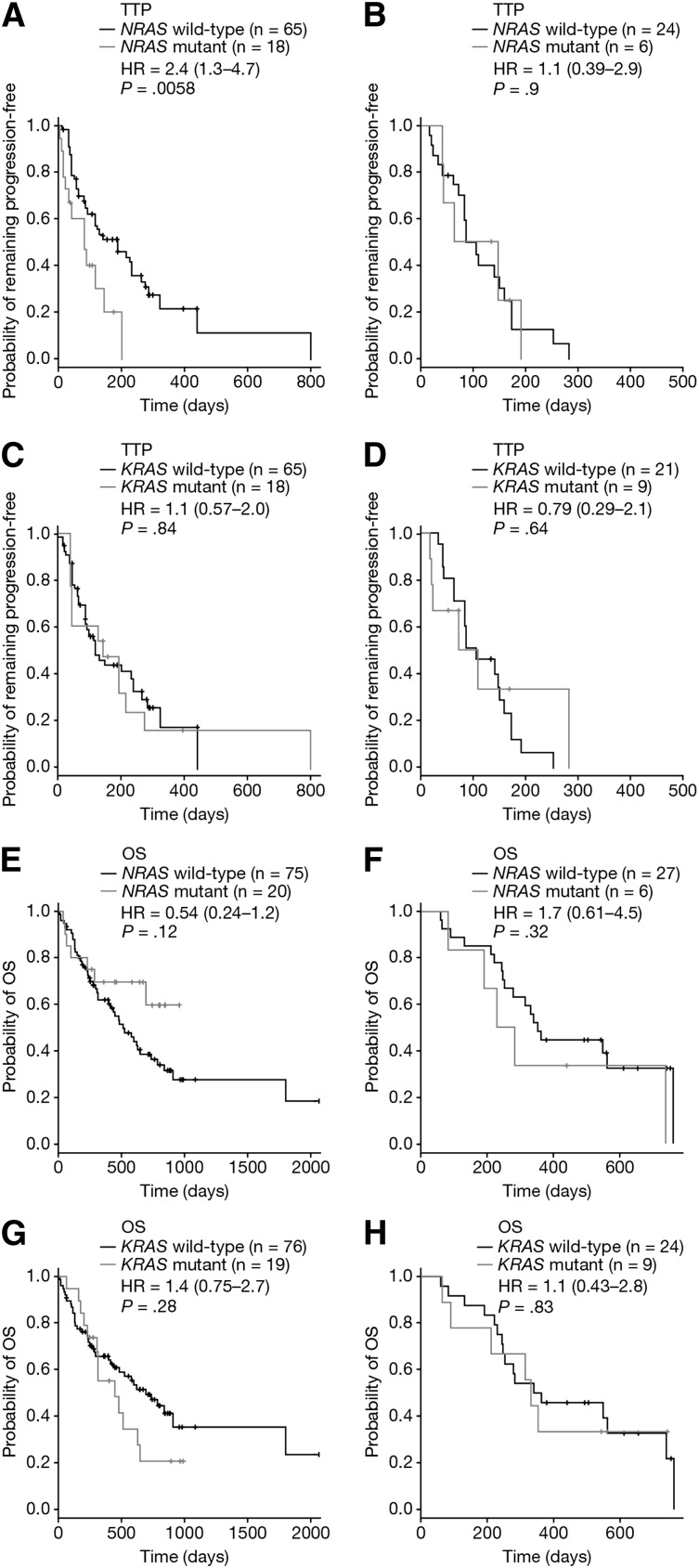
The effect of KRAS or NRAS mutation on TTP was tested in 113 evaluable patients with available mutation data; of these 113 patients, 83 had received bortezomib and 30 had received dexamethasone (supplemental Table 7). This analysis revealed a significantly shorter TTP in bortezomib-treated patients with NRAS mutation compared with NRAS wild-type (HR = 2.4 [95% CI: 1.3-4.7]; P = .0058; Bonferroni-corrected P = .012; Figure 3A). Analysis of 30 dexamethasone-treated patients with both TTP and mutation data showed no significant difference in TTP based upon NRAS status (HR = 1.1 [95% CI: 0.39-2.9]; P = .9; Figure 3B). There was also no difference in TTP when patients were stratified by KRAS mutation, whether treated with bortezomib (HR = 1.1 [95% CI: 0.57-2.0]; P = .84; Figure 3C) or dexamethasone (HR = 0.79 [95% CI: 0.29-2.1]; P = .64; Figure 3D). Thus, the reduced sensitivity of NRAS mutant patients was observed for both response and TTP, and in both cases the effect was detected for single-agent bortezomib and not high-dose dexamethasone.
Figure 3.
TTP and OS stratified by RAS mutation status. TTP stratified by NRAS mutation status in (A) bortezomib-treated patients and (B) dexamethasone-treated patients. TTP stratified by KRAS mutation status in (C) bortezomib-treated patients and (D) dexamethasone-treated patients. OS stratified by NRAS mutation status in (E) bortezomib-treated patients and (F) dexamethasone-treated patients. OS stratified by KRAS mutation status in (G) bortezomib-treated patients and (H) dexamethasone-treated patients. Each panel shows Cox proportional HR estimates (plus 95% CI) and P values based on a pairwise log-rank test.
OS and prognosis
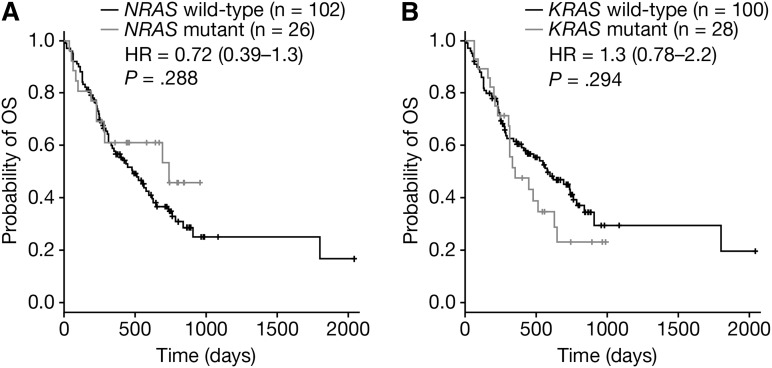
In this study, 128 patients were OS-evaluable; 95 bortezomib-treated and 33 dexamethasone-treated (supplemental Table 7). No significant difference in OS was observed for NRAS mutant patients treated with either bortezomib (HR = 0.54 [95% CI: 0.24-1.2]; P = .12; Figure 3E) or dexamethasone (HR = 1.7 [95% CI: 0.61-4.5]; P = .32; Figure 3F). The OS of KRAS mutant patients was also not significantly worse than for KRAS wild-type patients, whether patients were treated with bortezomib (HR = 1.4 [95% CI: 0.75-2.7]; P = .28; Figure 3G) or dexamethasone (HR = 1.1 [95% CI: 0.43-2.8]; P = .83; Figure 3H). Pooling bortezomib- and dexamethasone-treated patients and evaluating OS according to NRAS (Figure 4A) or KRAS (Figure 4B) mutation status showed neither mutation significantly impacted on OS (HRNRAS = 0.72 [95% CI: 0.39-1.3]; P = .288 and HRKRAS = 1.3 [95% CI: 0.78-2.2]; P = .294). Consistent with Liu et al,12 KRAS and NRAS mutation status was not linked to disease stage (supplemental Table 9). Although these OS data indicate that RAS mutation is not associated with disease prognosis in this data set, prior studies showing RAS to be prognostic have used larger sample sizes.
Figure 4.
OS stratified by RAS mutation status in a pooled data set. OS stratified by (A) NRAS and (B) KRAS mutant status in a pooled data set including both bortezomib-treated and dexamethasone-treated patients. Each panel shows Cox proportional HR estimates (plus 95% CI) and P values based on a pairwise log-rank test.
NRAS mutation and myeloma subtype
The only association found between the frequency of KRAS and NRAS mutations in different TC myeloma subtypes was an enrichment of NRAS mutations within the D1/hyperdiploid subtype (supplemental Table 10); however, this enrichment is not definitive (P = .014; Bonferroni-corrected P = .098) and warrants further investigation in other data sets. A logistic regression analysis was done, including both NRAS mutant and D1/hyperdiploid TC subtype status as covariates, to assess whether D1/hyperdiploid subtype might confound the inferred relationship between NRAS mutation and bortezomib response. Only the association between NRAS mutation and lack of response to bortezomib retained statistical significance in the adjusted model (P = .024) (supplemental Methods, supplemental Table 11). We also determined that NRAS mutant cases showed a reduced response rate relative to NRAS wild-type in both D1/hyperdiploid and non-D1/hyperdiploid TC subtype patients (supplemental Table 12).
Discussion
This report describes a mutational analysis of 133 purified myeloma samples, with interrogation of specific mutations across a panel of 41 genes previously implicated in oncogenesis. Limitations include the small sample size of certain mutant subgroups and lack of germline DNA samples to demonstrate that the observed mutations are somatic. Despite these caveats, these data inform both the prevalence of these oncogenic mutations and indicate an impact on patient outcomes after therapy. The similarities in the prevalence of mutations, particularly in genes of the RAS/RAF pathway, to those reported by Chapman et al7 are notable. Chapman et al sequenced tumor genomes in 38 myeloma patients, with an equal mix of newly diagnosed and relapsed myeloma samples included in their data set. Samples in the present study were from relapsed patients with highly variable exposure to prior chemotherapy; however, the frequency of observed mutations did not correlate with the number of prior therapies received suggesting that prior exposure to chemotherapy drugs was not a key contributor to the observed mutational spectrum at specific sites within the genes studied here. Importantly, 62 of 133 tumor samples analyzed in the present study did not show mutations in any of the cancer genes in the panel, highlighting the limitations of studies that preselect genes of interest. These findings, along with data from Chapman et al7 and Walker et al,35 suggest that additional genes (such as FAM46C, XBP1, EIF3B, RPL10, and DIS3) are likely mutated in the tumors of this data set. Additional genomic analyses in myeloma are needed to confirm our observations; in particular, the previously unreported mutations should be further investigated to better define their prevalence, and the observed clinical associations require confirmation in another cohort of patients.
This study identified an association between NRAS mutation and reduced sensitivity to bortezomib. In the context of single-agent bortezomib therapy, our findings indicate that mutations in NRAS but not KRAS have a negative impact on both response rate (P = .00116, Bonferroni-corrected P = .016) and TTP (HR = 2.4; P = .0058, Bonferroni-corrected P = .012). Interestingly, this impact was not observed in NRAS mutant patients who were randomized to high-dose dexamethasone in APEX. Although the number of dexamethasone-treated patients is relatively small and additional work is warranted, these data indicate that the impact of NRAS may be drug-specific. Several potential confounders were ruled out as explanations for the NRAS-specific or bortezomib-specific effects, including codon bias in RAS genes, International staging system (ISS) disease stage, and number of prior therapies; the NRAS mutation effect was independent of each of these. Additional experiments should be conducted to rule out potential unrecognized confounders and directly test the effect of NRAS status on sensitivity to bortezomib. This research could be pursued in 2 ways. First, independent clinical data sets should be examined in a manner which permits direct testing of NRAS mutational status and sensitivity to bortezomib. This might be done, for example, with small institutional data sets, alternative clinical trial data sets, or the CoMMpass registry (https://research.themmrf.org/) of 1000 newly diagnosed myeloma cases. Second, cell biology experiments designed to manipulate the NRAS (and KRAS) gene should be conducted to investigate gene activation and relative sensitivity to bortezomib. These experiments have been initiated and will be the subject of our subsequent reports. In light of the complexity of testing antimyeloma therapeutic agents, particularly proteasome inhibitors, in tissue-culture systems, the latter approach will involve some significant caveats.
NRAS mutation did not significantly impact OS in patients treated with bortezomib or dexamethasone despite the shortened TTP in these patients following bortezomib. A partial explanation for this finding could be the apparent enrichment for D1/hyperdiploid cases in the NRAS mutant group, as hyperdiploid patients generally exhibit good prognosis.36 As this enrichment may not be statistically robust (P = .014; Bonferroni-corrected P = .098), further investigation is needed to discern whether there is a genuine association between NRAS mutation and hyperdiploidy in myeloma. It is also possible that postbortezomib therapies may have been particularly effective in NRAS mutant patients, such that their OS did not lag relative to OS in NRAS wild-type patients. In the phase 2 SUMMIT and CREST trials, 41% of patients who achieved stable disease or progressive disease on bortezomib alone subsequently received bortezomib plus dexamethasone.37 It will be important to determine whether there is a specific drug or drug combination that improves outcome in NRAS mutant patients, or if there is an alternative explanation for the lack of impact on OS in NRAS mutant patients. Interestingly, the recent report of clinical response to a BRAF inhibitor in a BRAF mutant myeloma case38 suggests this pathway may be targeted in myeloma. In this context, a better understanding of how KRAS and NRAS signaling impact BRAF and the clinical outcome of each patient group is critical.
Although several studies have reported on the frequency and potential prognostic impact of RAS mutations in myeloma, results are somewhat variable. For example, Bezieau et al found NRAS or KRAS mutation in 54.5% of myeloma cases at diagnosis and 81% at relapse, but at lower frequencies in monoclonal gammopathy of undetermined significance (MGUS) and indolent myeloma.11 Furthermore, KRAS mutations were more frequent than NRAS mutations.11 Another study reported mutation rates of 17% for NRAS and 6% for KRAS39; in that study and in the study of Liu et al,12 KRAS mutation was associated with poor survival prognosis in myeloma. Other studies reported RAS mutation frequencies of 21% to 25%.39-41 Variation in the reported frequency and clinical consequence of these mutations may relate to differences in assay sensitivity, tumor cell purity, or the patient cohorts studied. In the current study of relapsed myeloma, RAS mutations did not confer poor prognosis for survival either individually or collectively. It is possible this data set is of insufficient size to detect a prognostic effect on OS; however, an alternative explanation may be that differences in the salvage therapies received by patients included in studies prior to this study might have impacted OS.
Although some data suggest potentially distinct biological consequences for mutation of the related RAS family members, studies demonstrating a clear clinical distinction between NRAS and KRAS are lacking. In general, the mutual exclusivity of mutations of NRAS and KRAS in varied tumor types suggests that they provide similar or identical oncogenic signals. The significant mutual exclusivity value reported here (P = .00005) further supports this notion. However, this data set reveals very distinct clinical consequences for the NRAS mutant subset, as these patients but not the KRAS mutant cohort, were found to be insensitive to single-agent bortezomib. Thus, while NRAS and KRAS may be capable of equal signaling through the RAF/MAPK pathway, our results suggest that NRAS mutation also provides a distinct, prosurvival signal that mutational activation of KRAS does not.
As reviewed recently,17 potential differences in downstream signaling from KRAS, NRAS, and HRAS are only just emerging.17,42 However, specific data support this suggestion of an NRAS survival signaling pathway. In the interleukin 6–dependent ANBL6 myeloma cell line, while both NRAS and KRAS mutations resulted in continued DNA synthesis in the absence of interleukin 6, only NRAS mutations resulted in both proliferation and protection from apoptosis.21 In a colorectal model system,42 NRAS was less proliferative and more antiapoptotic than KRAS, and recent data suggest this apoptotic suppression by NRAS involves the STAT3 pathway.43 Other data that support potential distinctions between these genes include the observation that NRAS-activating mutations can induce B-cell neoplasia and may be a required event in myeloma pathogenesis or evolution from MGUS.44,45
In summary, the reduced efficacy of bortezomib in NRAS mutant myeloma patients provides important clinical support for the existence of an additional function of NRAS and is consistent with a model in which NRAS provides survival signals. These data suggest that an important component of bortezomib antitumor activity acts at a level upstream of NRAS survival signaling and thus cannot effectively kill myeloma cells with this mutation. Characterization of the poor response and TTP in NRAS mutant myeloma patients treated with single-agent bortezomib also sets the stage for potentially improving clinical outcomes by assessing new drugs and drug combinations in specific patient subsets.
Acknowledgments
The authors acknowledge all of the patients who participated in bortezomib trials, as well as the efforts of all of the investigators and research staff who have contributed to these trials and to sample collection and enrichment, particularly Drs D. H. Irwin, A. E. Traynor, B. Mavromatis, and S. Hegewisch-Becker. G.M. acknowledges the assistance and foresight of S. Kim, A. Bolt, J. Brown, and M. Kaufmann, as well as the contributions of B. Bryant and A. Boral. M. Quinlan provided valuable comments on the manuscript. The authors also acknowledge Steve Hill and Emma Landers, medical writers with FireKite, for writing support during the development of this manuscript, which was funded by Millennium: The Takeda Oncology Company.
This work was supported by Takeda Pharmaceuticals International Co.
Footnotes
Presented in abstract form at the 53rd annual meeting of the American Society of Hematology, San Diego, CA, December 10-13, 2011.46
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.M., D.I.L., A.B., E.K., W.T., N.C., A.J.D., P.G.R., and D.-L.E. were involved in the conception and design of the study; G.M., E.K., H.B., S.J., P.G.R., E.A.S., P.S., J.F.S.M., and M.S. collected and assembled the data; G.M., A.D.B., S.J.B., E.K., W.T., B.L., R.N., J.B.B., H.v.d.V., D.R., S.J., J.R.B., P.G.R., E.A.S., R.Z.O., S.L., K.C.A., P.S., J.F.S.M., D.-L.E., and M.S. analyzed and interpreted the data; J.R.B. and K.C.A. provided study materials; and all authors contributed to the content of the manuscript and approved the final version to be published.
Conflict-of-interest disclosure: G.M., D.I.L., A.D.B., S.J.B., A.B., E.K., H.B., W.T., B.L., R.N., N.C., J.B.B., A.J.D., D.-L.E. and M.S. disclose employment with Takeda Pharmaceuticals International Co. D.I.L. discloses stock in Johnson & Johnson and Sequenom. A.J.D. discloses stock in Takeda. H.v.d.V. discloses employment with Janssen R&D and holds stock with Johnson & Johnson. D.R. discloses employment with and stock in Janssen. S.J. consults/advises for, and receives honoraria from, Millennium: The Takeda Oncology Company, Merck, and Celgene. P.G.R. consults/advises for Millennium: The Takeda Oncology Company. R.Z.O. consults/advises for Abbott Laboratories, Bristol-Myers Squibb, Celgene, Centocor Ortho-Biotech, Cephalon, Millennium: The Takeda Oncology Company, Novartis, and Onyx. S.L. consults/advises for Millennium: The Takeda Oncology Company, Celgene, Bristol-Myers Squibb, Novartis, Onyx, and Sanofi. K.C.A. consults/advises for Takeda, Celgene, Onyx, Gilead, and Sanofi-Aventis, and receives remuneration from Acetylon and OncoPep. P.S. consults/advises for and receives research funding from Millennium: The Takeda Oncology Company, Janssen, Celgene, and Onyx. J.F.S.M. consults/advises for and receives honoraria from Millennium: The Takeda Oncology Company, Janssen, Celgene, Onyx, and Novartis. D.-L.E. holds stock in Johnson & Johnson. J.R.B. consults/advises for and receives honoraria and research funding from Millennium: The Takeda Oncology Company. The remaining authors declare no competing financial interests.
Correspondence: George Mulligan, Translational Medicine, Takeda Pharmaceuticals International Co, 35 Landsdowne St, Cambridge, MA 02139; e-mail: george.mulligan@takeda.com.
References
- 1.Munshi NC, Anderson KC, Bergsagel PL, et al. International Myeloma Workshop Consensus Panel 2. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011;117(18):4696–4700. doi: 10.1182/blood-2010-10-300970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corre J, Avet-Loiseau H. The impact of genomics on the management of myeloma. J Natl Compr Canc Netw. 2011;9(10):1200–1206. doi: 10.6004/jnccn.2011.0097. [DOI] [PubMed] [Google Scholar]
- 3.Decaux O, Lodé L, Magrangeas F, et al. Intergroupe Francophone du Myélome. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myélome. J Clin Oncol. 2008;26(29):4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 4.Shaughnessy JD, Jr, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 5.Kumar SK, Rajkumar SV, Dispenzieri A, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111(5):2516–2520. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 7.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2(3):344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irahara N, Baba Y, Nosho K, et al. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19(3):157–163. doi: 10.1097/PDM.0b013e3181c93fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bezieau S, Devilder MC, Avet-Loiseau H, et al. High incidence of N and K-Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Hum Mutat. 2001;18(3):212–224. doi: 10.1002/humu.1177. [DOI] [PubMed] [Google Scholar]
- 12.Liu P, Leong T, Quam L, et al. Activating mutations of N- and K-ras in multiple myeloma show different clinical associations: analysis of the Eastern Cooperative Oncology Group Phase III Trial. Blood. 1996;88(7):2699–2706. [PubMed] [Google Scholar]
- 13.Prior IA, Hancock JF. Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol. 2012;23(2):145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson L, Greenbaum D, Cichowski K, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11(19):2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteban LM, Vicario-Abejón C, Fernández-Salguero P, et al. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001;21(5):1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potenza N, Vecchione C, Notte A, et al. Replacement of K-Ras with H-Ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep. 2005;6(5):432–437. doi: 10.1038/sj.embor.7400397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellano E, Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2(3):216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sclafani F, Gullo G, Sheahan K, Crown J. BRAF mutations in melanoma and colorectal cancer: a single oncogenic mutation with different tumour phenotypes and clinical implications. Crit Rev Oncol Hematol. 2013;87(1):55–68. doi: 10.1016/j.critrevonc.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Paulsson K, Horvat A, Strömbeck B, et al. Mutations of FLT3, NRAS, KRAS, and PTPN11 are frequent and possibly mutually exclusive in high hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2008;47(1):26–33. doi: 10.1002/gcc.20502. [DOI] [PubMed] [Google Scholar]
- 20.Agrawal N, Jiao Y, Sausen M, et al. Exomic sequencing of medullary thyroid cancer reveals dominant and mutually exclusive oncogenic mutations in RET and RAS. J Clin Endocrinol Metab. 2013;98(2):E364–E369. doi: 10.1210/jc.2012-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Billadeau D, Liu P, Jelinek D, Shah N, LeBien TW, Van Ness B. Activating mutations in the N- and K-ras oncogenes differentially affect the growth properties of the IL-6-dependent myeloma cell line ANBL6. Cancer Res. 1997;57(11):2268–2275. [PubMed] [Google Scholar]
- 22.Richardson PG, Sonneveld P, Schuster MW, et al. Assessment of Proteasome Inhibition for Extending Remissions (APEX) Investigators. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352(24):2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 23.Jagannath S, Barlogie B, Berenson J, et al. A phase 2 study of two doses of bortezomib in relapsed or refractory myeloma. Br J Haematol. 2004;127(2):165–172. doi: 10.1111/j.1365-2141.2004.05188.x. [DOI] [PubMed] [Google Scholar]
- 24.Richardson PG, Barlogie B, Berenson J, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348(26):2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 25.Mulligan G, Mitsiades C, Bryant B, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109(8):3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 26.Cullinan A, Cantor C. Sequenom, Inc. Pharmacogenomics. 2008;9(9):1211–1215. doi: 10.2217/14622416.9.9.1211. [DOI] [PubMed] [Google Scholar]
- 27.Ragoussis J, Elvidge GP, Kaur K, Colella S. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry in genomics research. PLoS Genet. 2006;2(7):e100. doi: 10.1371/journal.pgen.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS ONE. 2009;4(11):e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bladé J, Samson D, Reece D, et al. Myeloma Subcommittee of the EBMT. European Group for Blood and Marrow Transplant. Criteria for evaluating disease response and progression in patients with multiple myeloma treated by high-dose therapy and haemopoietic stem cell transplantation. Br J Haematol. 1998;102(5):1115–1123. doi: 10.1046/j.1365-2141.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- 30.Kiessling MK, Oberholzer PA, Mondal C, et al. High-throughput mutation profiling of CTCL samples reveals KRAS and NRAS mutations sensitizing tumors toward inhibition of the RAS/RAF/MEK signaling cascade. Blood. 2011;117(8):2433–2440. doi: 10.1182/blood-2010-09-305128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeng CH, Lee J, van Hummelen P, et al. High-throughput genotyping in metastatic esophageal squamous cell carcinoma identifies phosphoinositide-3-kinase and BRAF mutations. PLoS ONE. 2012;7(8):e41655. doi: 10.1371/journal.pone.0041655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egan JB, Shi CX, Tembe W, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120(5):1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soverini S, Terragna C, Testoni N, et al. Novel mutation and RNA splice variant of fibroblast growth factor receptor 3 in multiple myeloma patients at diagnosis. Haematologica. 2002;87(10):1036–1040. [PubMed] [Google Scholar]
- 34.Metro G, Chiari R, Duranti S, et al. Impact of specific mutant KRAS on clinical outcome of EGFR-TKI-treated advanced non-small cell lung cancer patients with an EGFR wild type genotype. Lung Cancer. 2012;78(1):81–86. doi: 10.1016/j.lungcan.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Walker BA, Wardell CP, Melchor L, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120(5):1077–1086. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 36.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23(26):6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 37.Jagannath S, Richardson PG, Barlogie B, et al. SUMMIT/CREST Investigators. Bortezomib in combination with dexamethasone for the treatment of patients with relapsed and/or refractory multiple myeloma with less than optimal response to bortezomib alone. Haematologica. 2006;91(7):929–934. [PubMed] [Google Scholar]
- 38.Andrulis M, Lehners N, Capper D, et al. Targeting the BRAF V600E mutation in multiple myeloma. Cancer Discov. 2013;3(8):862–869. doi: 10.1158/2159-8290.CD-13-0014. [DOI] [PubMed] [Google Scholar]
- 39.Chng WJ, Gonzalez-Paz N, Price-Troska T, et al. Clinical and biological significance of RAS mutations in multiple myeloma. Leukemia. 2008;22(12):2280–2284. doi: 10.1038/leu.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matozaki S, Nakagawa T, Nakao Y, Fujita T. RAS gene mutations in multiple myeloma and related monoclonal gammopathies. Kobe J Med Sci. 1991;37(1):35–45. [PubMed] [Google Scholar]
- 41.Ortega MM, Faria RM, Shitara ES, et al. N-RAS and K-RAS gene mutations in Brazilian patients with multiple myeloma. Leuk Lymphoma. 2006;47(2):285–289. doi: 10.1080/10428190500300969. [DOI] [PubMed] [Google Scholar]
- 42.Haigis KM, Kendall KR, Wang Y, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Velho S, Vakiani E, et al. Mutant N-RAS protects colorectal cancer cells from stress-induced apoptosis and contributes to cancer development and progression. Cancer Discov. 2013;3(3):294–307. doi: 10.1158/2159-8290.CD-12-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linden MA, Kirchhof N, Carlson CS, Van Ness BG. Targeted overexpression of an activated N-ras gene results in B-cell and plasma cell lymphoproliferation and cooperates with c-myc to induce fatal B-cell neoplasia. Exp Hematol. 2012;40(3):216–227. doi: 10.1016/j.exphem.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen T, Kuehl M, Lodahl M, Johnsen HE, Dahl IM. Possible roles for activating RAS mutations in the MGUS to MM transition and in the intramedullary to extramedullary transition in some plasma cell tumors. Blood. 2005;105(1):317–323. doi: 10.1182/blood-2004-03-0833. [DOI] [PubMed] [Google Scholar]
- 46.Mulligan G, Lichter DI, Di Bacco A, et al. Mutational analysis of tumor samples from patients with relapsed or refractory multiple myeloma (MM) highlights the prevalence of RAS/RAF pathway activation and demonstrates previously unreported mutations in known cancer genes. Blood (ASH Annual Meeting Abstracts) 2011;118(21) Abstract 1377. [Google Scholar]