Abstract
The functions of the liver and the pancreas differ; however, chronic inflammation in both organs is associated with fibrosis. Evidence suggests that fibrosis in both organs is partially regulated by organ-specific stellate cells. We explore the proteome of human hepatic stellate cells (hHSC) and human pancreatic stellate cells (hPaSC) using mass spectrometry (MS)-based quantitative proteomics to investigate pathophysiologic mechanisms. Proteins were isolated from whole cell lysates of immortalized hHSC and hPaSC. These proteins were tryptically digested, labeled with tandem mass tags (TMT), fractionated by OFFGEL, and subjected to MS. Proteins significantly different in abundance (P < 0.05) were classified via gene ontology (GO) analysis. We identified 1223 proteins and among them, 1222 proteins were quantifiable. Statistical analysis determined that 177 proteins were of higher abundance in hHSC, while 157 were of higher abundance in hPaSC. GO classification revealed that proteins of relatively higher abundance in hHSC were associated with protein production, while those of relatively higher abundance in hPaSC were involved in cell structure. Future studies using the methodologies established herein, but with further upstream fractionation and/or use of enhanced MS instrumentation will allow greater proteome coverage, achieving a comprehensive proteomic analysis of hHSC and hPaSC.
Keywords: Fibrosis, Proteomics, Pancreas, Tandem mass tag
Introduction
Myofibroblast-like stellate cells are found in both the liver and the pancreas and are implicated in major diseases of both organs [1]. Hepatic stellate cells (HSC) are found in the space of Disse, between Kupffer cells and hepatocytes [2]. Analogously, pancreatic stellate cells (PaSCs) are located in the periacinar space of the exocrine pancreas, intercalating duct and acinar cells [3]. Stellate cells represent 5–8% of the total number of liver cells [4] and 4% of pancreas cells [5,6]. Despite the low abundance of these cells in both organs, the various interactions of stellate cells with their microenvironments implicate these cells as key components of pathologic fibrosis and fibrotic diseases, such as chronic pancreatitis and liver cirrhosis [7].
While numerous cellular characteristics of HSC have been studied, to date, analogous studies of similar depth and scale of human PaSC (hPaSC) have been limited. For instance, in mid-2012, a PubMed search of “hepatic stellate cells” located 3374 articles, while a search of “pancreatic stellate cells” resulted in only 409 articles. Nevertheless, the nearly ten-fold greater number of publications for HSC is an underestimation as abstracts with alternate names for HSC – such as, Ito cells, lipocytes, pericytes, peri- and parasinusoidal cells – were not included in this estimation [8]. As such, PaSCs (often also abbreviated as PSC) are currently an underexplored resource to developing a better understanding of diseases of the exocrine pancreas. Given the similar mechanisms regulating HSC and PaSC, applying techniques that have been successful in liver research may elucidate the pathogenesis and pathophysiology of PaSC mediated-fibrosis, which is associated with chronic pancreatitis. A thorough understanding of the molecular mechanisms governing HSC function in regeneration, and potential analogous processes in PaSC, could revolutionize the treatment of pancreatitis and pancreatic cancer.
HSC and PaSC share similar morphological and functional features, despite originating from different organs. Both cell types exist in a quiescent state, which upon exposure to exogenous signals, e.g., cytokines and growth factors, transdifferentiate into an activated state [7]. Following activation, the stellate cells also produce smooth muscle actin and secrete extracellular matrix (ECM) components, pro-fibrotic cytokines, and pro-mitotic cytokines [1]. As such, studies have implicated HSC and PaSC as integral to the development of fibrosis [9–14], which is central to the pathogenesis and progression of liver cirrhosis and chronic pancreatitis/pancreatic cancer.
Extensive proteomic analyses of HSC have been performed [15–20]. However, few large-scale proteomic studies focusing on PaSC have been published [21–23]. Furthermore, transcriptomic analysis of HSC and PaSC has demonstrated 99% sequence identity [3,24], but no analogous quantitative proteomic comparison has been published is available in the literature. While HSC and PaSC share similar structural and functional characteristics, differences are expected as each cell type resides in its unique microenvironment in organs with vastly different functions. In this study, we aim to compare the proteomes of a human HSC (hHSC) cell line, LX-2, and a human PaSC (hPaSC) cell line, RLT-PSC, using state-of-the-art MS-based quantitative (tandem mass tag isobaric labeling) proteomics. Similar to a previously published transcriptomic analysis comparing hHSC and hPaSC [25], we identified the majority of proteins as common between the cell types, but pronounced differences in certain protein classes were also identified.
Results
MS analysis revealed differences in the abundance of hundreds of proteins between hHSC and hPaSC
We used MS-based quantitative proteomic strategy to compare the proteomes of hHSC and hPaSC (as outlined in Figure 1). Tandem mass tag (TMT) labeling upstream of mass spectrometric analysis allows for multiplexed relative quantification of proteins in multiple samples [26]. In total, we identified 1223 proteins with a TMT labeling efficiency of greater than 99%, i.e., over 99% of the proteins identified were quantified. Only a single protein, branched-chain-amino-acid aminotransferase, was identified exclusively in hHSC (Figure 2), being present in all three hHSC replicates with 2 unique peptides, but not in any hPaSC samples. Branched chain aminotransferase functions as a catalyst that synthesizes the branched chain amino acids isoleucine, leucine, and valine [27]. Although the possible reasons for the identification of this protein in hHSC and not hPaSC are yet to be determined, branched-chain-amino-acid aminotransferase has been studied previously in relation to liver development [28]. As such, branched-chain-amino-acid aminotransferase may merit further investigation.
Figure 1.
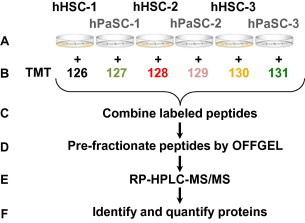
Experimental workflow A. Cell cultures of hHSC and hPaSC were grown in triplicate. B. Proteins were extracted, digested with trypsin and labeled with a specific TMT reagent. C. Resulting peptides were pooled at equal concentrations. D. Peptides were fractionated via OFFGEL separation. E. Reversed-phase HPLC was performed prior to tandem MS analysis. F. Bioinformatics and statistical analyses were performed to identify and quantify the proteins.
Figure 2.
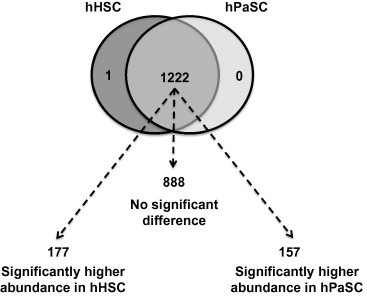
Few proteins were exclusive to one cell type Venn diagram shows that only a single protein was exclusively identified in hHSC sample, while the remaining 1222 proteins were identified in both cell types. Of the 1222 quantified proteins, our statistical analysis showed 888 proteins as not significantly different in abundance between the two cell types, 177 with significantly higher abundance in the hHSC sample and 157 with significantly higher abundance in the hPaSC sample.
t-tests were performed on the remaining 1222 proteins to determine significant differences in abundance between the two cell types (P < 0.05). The initial analysis identified 642 proteins present in significantly different quantities. To correct for multiple testing, we applied the Benjamini–Hochberg method [29], which reduced the number of differentially abundant proteins to 534. Of these, we identified 334 differentially abundant proteins with a ±1.5-fold change in either cell type compared to the other. In total, 177 proteins were enriched in hHSC, 157 were enriched in hPaSC, while the abundance of the remaining 888 proteins, which account for nearly 73% of the total proteins identified in both cell types, were not significantly different between these cell types (Figure 2).
To graphically represent these t-test data, volcano plot – log10(P value) vs. log2(fold change of hHSC/hPaSC) – was constructed to graphically display the quantitative data (Figure 3A). Points above the non-axial horizontal line represent proteins with significantly different abundances (P < 0.05). Points to the left of the left-most non-axial vertical line denote protein fold changes of hHSC/hPaSC less than −1.5, while points to the right of the right-most non-axial vertical line denote protein fold changes of hHSC/hPaSC greater than 1.5. The number of these differentially abundant proteins (P < 0.05 and fold change >|1.5|) was binned according to the specific fold-change of abundance between hHSC and hPaSC (Figure 3B). The hHSC/hPaSC ratio allowed direct comparison of protein abundance in hPaSC to that in hHSC. We note that highest numbers of proteins with significant differences showed 1.5–2-fold change in abundance, while the 2–3-fold bin contained the second highest number of proteins. Only 46 proteins demonstrated a 3 or greater fold change in either of the two cell types.
Figure 3.
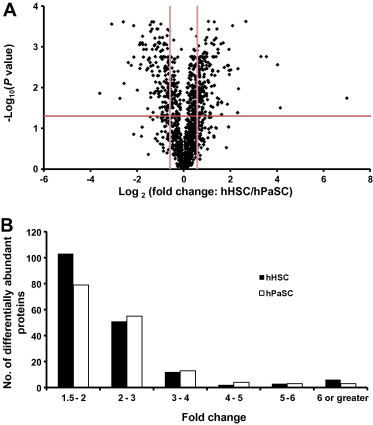
Volcano plot and histogram illustrate differentially abundant proteins A. Volcano plot illustrates significantly differentially abundant proteins. The −log10 (Benjamini–Hochberg corrected P value) is plotted against the log2 (fold change: hHSC/hPaSC). The non-axial vertical lines denote ±1.5-fold change while the non-axial horizontal line denotes P = 0.05, which is our significance threshold (prior to logarithmic transformation). B. Histogram displaying the tally of differentially abundant proteins within a specific range of fold changes. The fold change for hHPC is determined by hHPC/hPaSC, while that for hPaSC is 1/(hHSC/hPaSC).
GO analysis revealed differences in the classification of proteins enriched in hHSC and hPaSC
Differentially abundant proteins were subjected to GO classification via the Panther Classification System database [30] to investigate biological processes, molecular function and cellular compartment (Figure 4). To limit the number of classifications, the analysis returned only those classifications with at least 5 proteins and a difference of at least 1% between the cell types.
Figure 4.
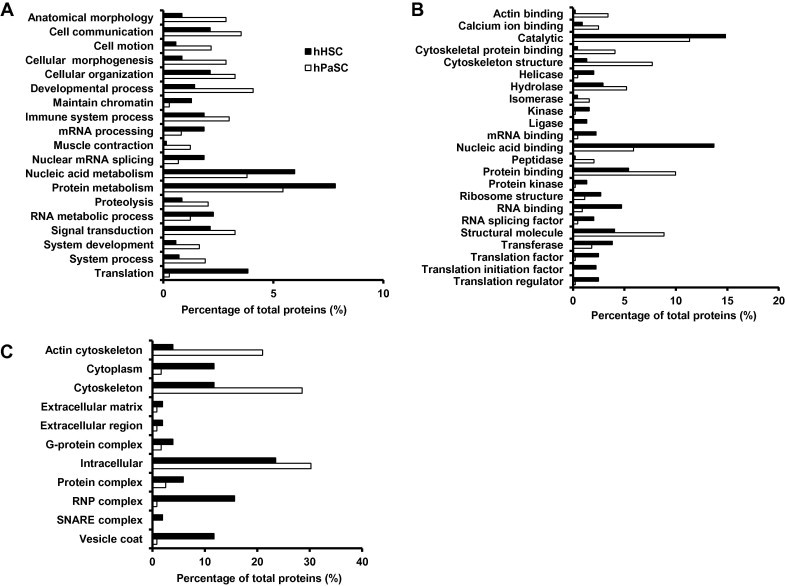
GO analysis illustrates classes of proteins differing between cell types Proteins with significant differences between the two cell types were subjected to GO classification in terms of biological process (A), molecular function (B) and cellular compartment (C). We set a threshold of 5 proteins per classification and a difference of at least 1% between the two cell types examined.
Proteins significantly enriched in hHSC were primarily related to protein production and regulation. For example, biological process classification revealed that proteins involved in nucleic acid metabolism, protein metabolism, and translation were more abundant in hHSC than in hPaSC (Figure 4A). In agreement, the predominant molecular functions of hHSC-enriched proteins included mRNA binding, nucleic acid binding, and RNA splicing factors (Figure 4B). Moreover, in terms of cellular compartment, the hHSC-enriched proteins were comprised of a substantially higher percentage of ribonucleoprotein (RNP) complexes than hPaSC-enriched proteins (Figure 4C). The specific proteins enriched in the hHSC samples include a large number of ribosomal subunits, eukaryotic initiation and translation factors, and histones (Table S1), all of which are linked to cellular transcription and translation processes.
The majority of the proteins that were significantly enriched in hPaSC were related to cellular structure. In terms of biological processes, these proteins were classified mainly in anatomical morphology, cell communication, cellular morphogenesis, cellular organization, and developmental processes (Figure 4A). As expected, the biological processes correlated well with their molecular function classification (Figure 4B): cytoskeletal protein binding, cytoskeletal structure, protein binding and structural molecules. These molecular functions indicate a direct involvement in cell structure. Moreover, the cellular compartment classification of these proteins revealed that the most populous categories were actin cytoskeleton, cytoskeleton and intracellular, consistent with the aforementioned biological processes and molecular functions (Figure 4C). The specific proteins enriched in the hPaSC samples include annexins, actinins, actin-related proteins, filamins, myosins and tropomyosins (Table S2), all of which are actin binding proteins involved in cytoskeletal structure and function.
Comparison of proteomic data with previously published transcriptomic data
Using a 23,000 feature oligonucleotide microarray, a previous transcriptomic analysis discovered 29 genes of significant differential expression (fold difference > 2), between hHSC and hPaSC [16]. Although it is difficult to directly compare our differential proteomic analysis to the aforementioned transcriptomic analysis [25], due to confounders related to cell lines, sample preparation techniques, and statistical methods used, these studies revealed similar observations. In general, both analyses indicated that the majority of the identified proteins/transcripts were present in both cell types and did not demonstrate significant differences in abundance (P > 0.05).
While only 29 transcripts (9 in hHSC and 20 in hPaSC, approximately 0.1% of the dataset) were determined to be enriched in the transcriptomic analysis [31], we identified a total of 334 proteins (177 in hHSC and 157 in hPaSC, approximately 5% of our dataset) that were enriched in either sample. Our protein counts, however, were determined using a 1.5-fold difference threshold. Using a 2-fold threshold, as was done in [25], the number of enriched proteins we identified would be reduced to 64 in hHSC and 76 in hPaSC (Figure 3B). Some of the differences between the transcriptomics and proteomics data may be attributed to regulation at the level of protein translation [32].
Interestingly, for 5 of the 29 genes determined to be differentially expressed in the transcriptomic analysis, their corresponding encoded proteins were also identified as differentially expressed in our proteomics analysis. These proteins included actin, gamma-enteric smooth muscle (ACTG2), destrin, four and a half LIM domains protein 1 (FHL1), L-lactate dehydrogenase (LDH) and ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCH-L1) (Table 1). Except LDH, the remaining 4 proteins were all lower in abundance in hHSC compared to hPaSC, and as such may be the first proteins upon which to follow-up. Proteins corresponding to the remaining 24 genes were not identified in our proteomic analysis, and may be below the detection limit of our analytical strategy.
Table 1.
Differentially-expressed transcripts with proteins identified in current proteomic analysis
| Uniprot | Entry name | Protein name | Transcriptomics | Proteomics |
|---|---|---|---|---|
| hHSC/hPaSC | hHSC/hPaSC | |||
| P63267 | ACTH_HUMAN | Actin, gamma-enteric smooth muscle | 0.43 | 0.97 |
| P60981 | DEST_HUMAN | Destrin | 0.47 | 0.56 |
| Q13642 | FHL1_HUMAN | Four and a half LIM domains protein 1 | 0.24 | 0.63 |
| P00338 | LDHA_HUMAN | L-lactate dehydrogenase A chain | 0.43 | 2.48 |
| P09936 | UCHL1_HUMAN | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 0.46 | 0.29 |
Note: hHSC, human hepatic stellate cells; hPaSC, human pancreatic stellate cells.
Discussion
We have shown that the proteomes of hHSC and hPaSC are similar at our depth of analysis. Using whole cell lysates, OFFGEL fractionation, and TMT MS-based quantitative proteomics we quantified over 1200 proteins. Of these proteins, we determined several hundred to be significantly enriched in one of the two cell types, although over 70% of the proteins did not differ significantly between the two cell types. GO classification revealed that the majority of proteins of higher abundance in hHSC are related to protein production. Such a result may imply that cellular proliferation is at a higher rate in hHSC than hPaSC. Conversely the majority of proteins of higher abundance in hPaSC play a role in cellular structure, which may indicate that a different set of proteins are used in hPaSC to maintain or regulate changes in cellular morphology. The mechanisms underlying such distinct differences are unclear, and further studies are necessary to determine its causes and implications.
A comparison of previously published transcriptomic data with our proteomic data yielded similar general trends. The majority of the proteins were of comparable abundance in each cell type, and all but one protein was identified in both cell types. However, the transcriptomic data showed no differences in over 99% of the genes investigated, whereas over 70% of the total proteins in our proteomic data did not demonstrate significant differences. Discordance between mRNA and protein analyses is common. Studies have shown similar low correlation between transcriptomic and proteomic data, particularly when the depth of proteomic data is limited [25,33,34]. Although central dogma of molecular biology illustrates the flow of information from mRNA to proteins, other factors – for example, translational efficiency, alternative splicing, complex formation and degradation rates and localization – can affect the level of proteins independent of the transcript. This may have been the case with L-lactate dehydrogenase, whose transcript was half as abundant in hHSC compared to hPaSC, but at the protein level was 2.5 times as abundant in the hHSC compared to hPaSC. Limited by the data collected, we cannot determine the true source of this disparity. Greater proteome coverage could provide further evidence supporting the similarities identified in our proteomics study between hHSC and hPaSC, and may uncover additional proteins which are significantly different in the two cell types, for example those discovered by transcriptomic analysis.
The five proteins listed in Table 1 may serve as promising candidates for follow-up experiments, as many have been previously associated with hepatic and/or pancreatic disease. Smooth muscle actin has been implicated in fibrosis in both hepatic [35–37] and pancreatic [38] disease, as this protein is a marker of activation of stellate cells from quiescence [39,40]. Likewise, the actin binding protein destrin is associated with perineural invasion of pancreatic cancer [41], and has been associated with effects on hepatic tumor cells in the presence of an anti-tumor drug [42]. Four and a half LIM domains protein 1 is a low abundant protein in the liver and pancreas and is commonly linked to myopathies [43], but its role in pathophysiological aspects of hepatic and pancreatic disease is currently unknown. L-lactate dehydrogenase A (LDHA) is involved in tissue breakdown and turnover, which has been shown to be elevated in cancer cells, including those of pancreatic cancer [44]. In addition, ubiquitin carboxy-terminal hydrolase L1 is a deubiquitinating enzyme highly specific to neurons [45] and has a role in tumor development [46], however, its role in pancreatic and hepatic disease remains undetermined. These proteins may be investigated further to study their involvement in the pathophysiology of pancreatic and/or hepatic disease.
Although immortalized cell lines are accepted for initial investigation, particularly for the ease of propagation and homogeneity, freshly isolated cells from their in vivo environment may better reflect the true cellular characteristics. In fact, disparities with the transcriptomics dataset [31] and our proteomics data may also be a result of our proteomics data being based on immortalized cell lines, while the transcriptomics data were from freshly isolated cells. As such, while the work herein focuses on immortalized hHSC and hPaSC cell lines, performing these experiments with freshly isolated cells would avoid potential confounding by artifacts introduced by the immortalization process and thus reveal more similarities with the previously collected transcriptomics data. Such studies would be valuable as these freshly isolated cells are genotypically unaltered from their in vivo states and are a more accurate representation of in vivo stellate cells.
The differences identified between the two cell types may be partially attributed to the organ of origin for these cells, as well as the evolutionary divergence of these two cell types. Questions surround the potential of a common origin of HSC and PaSC, evidence suggests that PaSC and HSC have common precursor cells in the neural crest [4]. These precursor cells give rise to astrocytes, smooth muscle cells and neurons. In intermediate stages, the precursor cells express nestin, glial fibrillary acidic protein (GFAP) and smooth muscle actin, all of which are protein markers of stellate cells [47]. However, other data have shown that pluripotent stem cells from the adult pancreas can differentiate into cell types characteristic of pancreatic endocrine, exocrine and stellate cells [48], and HSC may be differentiated from bone marrow [49], which would indicate that the microenvironment of the cells, rather than a common precursor, may be responsible for the phenotypic commonalities between the two cell types. Further research exploring the origin of HSC and PaSC would provide important data about hepatic and pancreatic development, fibrosis and carcinogenesis.
In summary, PaSC hold great promise in the study of chronic pancreatitis and pancreatic cancer. These cells represent a valuable resource in improving our understanding of pancreatic disease pathogenesis, pathophysiology and therapy. Tapping into the decades of prior HSC studies may rapidly expedite PaSC research. We have used quantitative MS to identify nearly 900 proteins that are of similar abundance in hHSC and hPaSC, in addition to over 300 which were enriched in one of the cell lines. Similarities were expected as both cells share structural characteristics, transdifferentiate into ECM-secreting activated cells upon exogenous insult and play key roles in fibrosis of their respective organs. Transcriptomic studies have also provided evidence of transcript similarity at the mRNA level [25]. However, fundamental differences remain as these cells are part of larger organs with dissimilar functions and are situated in very different microenvironments. Without further evidence, it is premature to correlate these proteomic differences between liver and pancreas with the regenerative capacity of the liver and the absence of such in the pancreas. However, application and methodological enhancements of the quantitative proteomics techniques described herein may facilitate future studies of pancreatic regenerative potential via in-depth characterization of hHSC and hPaSC proteomes. Further research is necessary to fully characterize the differences and similarities between hHSC and hPaSC proteomes and to elucidate further the roles of the differentially-expressed proteins in healthy and diseased livers and pancreata.
Materials and methods
Materials
Dulbecco’s modified Eagle’s-F12 medium (DMEM/F12; 11330) was purchased from Gibco (Carlsbad, CA). Fetal bovine serum (FBS; F0392) was purchased from Sigma (St. Louis, MO). CellStripper (25-056-CL) was purchased from Mediatech (Manassas, VA). TMTsixplex Isobaric Mass Tagging Kit was purchased from Thermo Scientific (Rockford, IL). Sequencing-grade modified trypsin (V5111) was obtained from Promega (Madison, WI). SeeBluePlus2 Pre-Stained standard (LC5925), lithium dodecyl sulfate (LDS) sample buffer (NP0008), NuPAGE 4–12% Bis-Tris polyacrylamide gels (NP0335), SimplyBlueCoomassie stain (LC0665) and 2-(N-morpholino) ethanesulfonic acid-sodium dodecyl sulfate (MES-SDS) electrophoresis buffer (NP002) were from Invitrogen (Carlsbad, CA). Other reagents and solvents were from Sigma–Aldrich (St. Louis, MO) and Burdick & Jackson (Morristown, NJ), respectively.
Cell lines and maintenance
The hHSC cell line LX-2 [50] was a kind gift from Dr. Scott Friedman (Mount Sinai School of Medicine) and the hPaSC cell line RLT-PSC [51] was a gift from Dr. Ralf Jesnowski (German Cancer Research Center). Cell growth and propagation was carried out as previously described [21,22].
Cell lysis and protein extraction
One milliliter of TBSp (50 mM Tris, 150 mM NaCl, pH 7.4 supplemented with 1× Roche Complete protease inhibitors) containing 1% Triton X-100 and 0.5% SDS was added to each cell pellet. Cells were homogenized by 12 passes through a 27 gauge (1.25 inches long) needle and incubated on ice with gentle agitation for 1 h. The homogenate was then sedimented by ultracentrifugation at 100,000 × g for 60 min at 4 °C, and the supernatant was collected. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (23225, ThermoFisher Scientific).
Tryptic digestion and TMT labeling
Each cell lysate was diluted to a concentration of 150 μg of total protein per 100 μl of 100 mM triethyl ammonium bicarbonate (TEAB) buffer. Protein digests were performed as specified in the manufacturer’s instructions for the Thermo Scientific’s TMTsixplex Isobaric Mass Tagging Kit (catalog #90064B). Similarly, TMT labeling was performed according to the manufacturer’s instructions. Each biological replicate was labeled with a unique tag. hHSC samples received tags 126, 128, and 130, while hPaSC samples received tags 127, 129, and 131 (as illustrated in Figure 1). Prior to fractionation, samples were desalted using OASIS HLB reversed-phase cartridges (Waters, 186000383) as outlined in the manufacturer’s instructions. The methanol-eluted peptides were vacuum centrifuged to dryness.
OFFGEL peptide fractionation
Peptides were reconstituted in a 0.2% ampholyte solution in water (GE Healthcare, 17-6000-87). Peptides were separated on a 24-cm IEF strip (GE Healthcare 17-6002-44) using standard program OG24PE00 on an Agilent OFFGEL 3100 system, collected from OFFGEL wells and deposited in separate tubes. 150 μL of 0.1% TFA was added to each well and then incubated for 15 min. This extraction was added to the tube containing the first aspirate. Each sample was desalted in TopTip SCX columns (Glygen, TT2SSC) using a binding solution containing 0.1% formic acid and 20% acetonitrile, and releasing solution containing 5% ammonium hydroxide and 30% methanol releasing solution. The eluent was vacuum centrifuged to dryness.
LC–MS/MS analysis
Peptides were reconstituted in 5% formic acid and 5% acetonitrile in water prior to fractionation by nanoflow reversed-phase ultra-high pressure liquid chromatography (nanoLC, Eksigent) in-line with a linear trap quadrupole mass spectrometer (LTQ, Thermo Scientific). The reversed-phase liquid chromatography columns (15 cm × 100 μm ID) were packed in-house (Magic C18, 5 μm, 100 Å, Michrom BioResources). Samples were analyzed with a 60-min linear gradient (5–35% acetonitrile with 0.2% formic acid) and data were acquired in a data-dependent manner, with 6 MS/MS scans for every full scan spectrum. Precursor ion activation and dissociation was accomplished via pulsed Q dissociation (PQD) to allow the observation of low m/z fragments that are usually excluded from standard collision-induced dissociation (CID). The collision energy was set at 32, the isolation wavelength at 2 Th, the activation Q at 0.55 and the acquisition time at 0.4 ms.
Bioinformatics and data analysis
All data generated were searched against the Human UniProt database (downloaded November 11, 2011) using the Mascot search engine (v.2.3; Matrix Science, Boston MA). Table S3 includes a complete list of proteins identified. One miscleavage per peptide was allowed and mass tolerances of ±1 Da (monoisotopic) for precursors and of ±0.8 Da for fragment ions were used, as is default for LTQ data analysis. Amino acid modifications: fixed: carbamidomethyl (Cys); variable: TMT6plex (N-term) and TMT6plex (K). Mascot search results are combined using in-house-developed software. In compliance with recommendations [52–54] proposed by the major proteomic journals, we utilized protein identification validation methods that minimize false positives and report only high confidence identifications. Our false discovery rate (FDR) was 1% at the peptide level, as determined by searching the same dataset against the target database and a decoy database; the latter featuring the reversed amino acid sequences of all the entries in the Human UniProt database [55,56]. Median intensities of reporter ions for each protein were determined normalized across all samples.
Statistical analysis
t-tests, Benjamini–Hochberg corrections [29] and volcano plots [57] were produced in Excel 2010 (Microsoft; Redmond, WA). Three biological replicates were performed for each cell type and proteomic differences were evaluated for statistical significance (P < 0.05) by student t-tests, and corrected for multiple testing using the Benjamini–Hochberg correction. Means were calculated for the three biological replicates and fold-changes were determined by dividing the mean intensity value of the liver samples by that of the pancreas samples for each protein. The fold change was transformed using the log2 function, so that the data is centered around zero, while the Benjamini–Hochberg corrected P value was −log10 transformed for volcano plot scaling.
Authors’ contributions
JP, PB, HS and DC conceived the study and participated in its design and coordination. JP carried out the experiments. JP and VK drafted the original manuscript. All authors revised the manuscript and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Acknowledgements
Funds were provided by the following NIH grants: 1 F32 DK085835-01A1 (JP), 1 R21 DK081703-01A2 (DC), 5 P30 DK034854-24 (Harvard Digestive Diseases Center; DC), as well as a grant from the American College of Gastroenterology: ACG – 042103580 (JP). We would like to thank the Burrill family for their generous support through the Burrill Research Grant. We would also like to thank members of the Steen Laboratory at Boston Children’s Hospital, in particular John FK Sauld and Ali Ghoulidi for their technical assistance and critical reading of the manuscript. In addition, we thank members of the Center for Pancreatic Disease at Brigham and Women’s Hospital, particularly Shadeah Suleiman for her technical assistance.
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Contributor Information
Joao A. Paulo, Email: joao_paulo@post.harvard.edu.
Hanno Steen, Email: hanno.steen@tch.harvard.edu.
Appendix Supplementary. material
Table S1. Proteins of higher abundance in hHSC as determined via TMT analysis for individual samples.
Table S2. Proteins of higher abundance in hPaSC as determined via TMT analysis for individual samples.
Table S3. All proteins identified in the TMT analysis.
References
- 1.Erkan M., Adler G., Apte M.V., Bachem M.G., Buchholz M., Detlefsen S. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut. 2012;61:172–178. doi: 10.1136/gutjnl-2011-301220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omary M.B., Lugea A., Lowe A.W., Pandol S.J. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117:50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geerts A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis. 2001;21:311–335. doi: 10.1055/s-2001-17550. [DOI] [PubMed] [Google Scholar]
- 5.Bachem M.G., Schneider E., Gross H., Weidenbach H., Schmid R.M., Menke A. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115:421–432. doi: 10.1016/s0016-5085(98)70209-4. [DOI] [PubMed] [Google Scholar]
- 6.Apte M.V., Haber P.S., Applegate T.L., Norton I.D., McCaughan G.W., Korsten M.A. Periacinar stellate shaped cells in rat pancreas: identification, isolation, and culture. Gut. 1998;43:128–133. doi: 10.1136/gut.43.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kordes C., Sawitza I., Häussinger D. Hepatic and pancreatic stellate cells in focus. Biol Chem. 2009;390:1003–1012. doi: 10.1515/BC.2009.121. [DOI] [PubMed] [Google Scholar]
- 8.Suematsu M., Aiso S. Professor Toshio Ito: a clairvoyant in pericyte biology. Keio J Med. 2001;50:66–71. doi: 10.2302/kjm.50.66. [DOI] [PubMed] [Google Scholar]
- 9.Moreira R. Hepatic stellate cells and liver fibrosis. Arch Pathol Lab Med. 2007;131:1728–1734. doi: 10.5858/2007-131-1728-HSCALF. [DOI] [PubMed] [Google Scholar]
- 10.Gressner A.M., Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-beta as major players and therapeutic targets. J Cell Mol Med. 2006;10:76–99. doi: 10.1111/j.1582-4934.2006.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safadi R., Friedman S.L. Hepatic fibrosis – role of hepatic stellate cell activation. MedGenMed. 2002;4:27. [PubMed] [Google Scholar]
- 12.Bataller R., Brenner D.A. Hepatic stellate cells as a target for the treatment of liver fibrosis. Semin Liver Dis. 2001;21:437–451. doi: 10.1055/s-2001-17558. [DOI] [PubMed] [Google Scholar]
- 13.Haber P.S., Keogh G.W., Apte M.V., Moran C.S., Stewart N.L., Crawford D.H. Activation of pancreatic stellate cells in human and experimental pancreatic fibrosis. Am J Pathol. 1999;155:1087–1095. doi: 10.1016/S0002-9440(10)65211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman S.L. Stellate cell activation in alcoholic fibrosis – an overview. Alcohol Clin Exp Res. 1999;23:904–910. [PubMed] [Google Scholar]
- 15.Deng X., Liang J., Lin Z.X., Wu F.S., Zhang Y.P., Zhang Z.W. Natural taurine promotes apoptosis of human hepatic stellate cells in proteomics analysis. World J Gastroenterol. 2010;16:1916–1923. doi: 10.3748/wjg.v16.i15.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosselut N., Housset C., Marcelo P., Rey C., Burmester T., Vinh J. Distinct proteomic features of two fibrogenic liver cell populations: hepatic stellate cells and portal myofibroblasts. Proteomics. 2010;10:1017–1028. doi: 10.1002/pmic.200900257. [DOI] [PubMed] [Google Scholar]
- 17.Molleken C., Sitek B., Henkel C., Poschmann G., Sipos B., Wiese S. Detection of novel biomarkers of liver cirrhosis by proteomic analysis. Hepatology. 2009;49:1257–1266. doi: 10.1002/hep.22764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim P.K., Kim M.R., Kim H.J., Yoo H.S., Kim J.S., Cho E.H. Proteome analysis of the rat hepatic stellate cells under high concentrations of glucose. Proteomics. 2007;7:2184–2188. doi: 10.1002/pmic.200700051. [DOI] [PubMed] [Google Scholar]
- 19.Gressner O.A., Weiskirchen R., Gressner A.M. Biomarkers of liver fibrosis: clinical translation of molecular pathogenesis or based on liver-dependent malfunction tests. Clin Chim Acta. 2007;381:107–113. doi: 10.1016/j.cca.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 20.Kawada N. Analysis of proteins dominantly expressed in hepatic stellate cells of activated phenotype. Methods Mol Med. 2005;117:371–379. doi: 10.1385/1-59259-940-0:371. [DOI] [PubMed] [Google Scholar]
- 21.Paulo J.A., Urrutia R., Banks P.A., Conwell D.L., Steen H. Proteomic analysis of a rat pancreatic stellate cell line using liquid chromatography tandem mass spectrometry (LC-MS/MS) J Proteomics. 2011;75:708–717. doi: 10.1016/j.jprot.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulo J.A., Urrutia R., Banks P.A., Conwell D.L., Steen H. Proteomic analysis of an immortalized mouse pancreatic stellate cell line identifies differentially-expressed proteins in activated vs nonproliferating cell states. J Proteome Res. 2011;10:4835–4844. doi: 10.1021/pr2006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wehr A.Y., Furth E.E., Sangar V., Blair I.A., Yu K.H. Analysis of the human pancreatic stellate cell secreted proteome. Pancreas. 2011;40:557–566. doi: 10.1097/MPA.0b013e318214efaf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathison A., Liebl A., Bharucha J., Mukhopadhyay D., Lomberk G., Shah V. Pancreatic stellate cell models for transcriptional studies of desmoplasia-associated genes. Pancreatology. 2010;10:505–516. doi: 10.1159/000320540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchholz M., Kestler H.A., Holzmann K., Ellenrieder V., Schneiderhan W., Siech M. Transcriptome analysis of human hepatic and pancreatic stellate cells: organ-specific variations of a common transcriptional phenotype. J Mol Med (Berl) 2005;83:795–805. doi: 10.1007/s00109-005-0680-2. [DOI] [PubMed] [Google Scholar]
- 26.Thompson A., Schafer J., Kuhn K., Kienle S., Schwarz J., Schmidt G. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 27.Hutson S. Structure and function of branched chain aminotransferases. Prog Nucleic Acid Res Mol Biol. 2001;70:175–206. doi: 10.1016/s0079-6603(01)70017-7. [DOI] [PubMed] [Google Scholar]
- 28.Torres N., Vargas C., Hernandez-Pando R., Orozco H., Hutson S.M., Tovar A.R. Ontogeny and subcellular localization of rat liver mitochondrial branched chain amino-acid aminotransferase. Eur J Biochem. 2001;268:6132–6139. doi: 10.1046/j.0014-2956.2001.02563.x. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y., Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 30.Thomas P.D., Kejariwal A., Campbell M.J., Mi H., Diemer K., Guo N. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffin T.J., Gygi S.P., Ideker T., Rist B., Eng J., Hood L. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Foss E.J., Radulovic D., Shaffer S.A., Ruderfer D.M., Bedalov A., Goodlett D.R. Genetic basis of proteome variation in yeast. Nat Genet. 2007;39:1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- 33.Fu J., Keurentjes J.J., Bouwmeester H., America T., Verstappen F.W., Ward J.L. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet. 2009;41:166–167. doi: 10.1038/ng.308. [DOI] [PubMed] [Google Scholar]
- 34.Ghazalpour A., Bennett B., Petyuk V.A., Orozco L., Hagopian R., Mungrue I.N. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mak K.M., Chu E., Lau K.H., Kwong A.J. Liver fibrosis in elderly cadavers: localization of collagen types I, III, and IV, alpha-smooth muscle actin, and elastic fibers. Anat Rec (Hoboken) 2012;295:1159–1167. doi: 10.1002/ar.22504. [DOI] [PubMed] [Google Scholar]
- 36.Zakaria S., Youssef M., Moussa M., Akl M., El-Ahwany E., El-Raziky M. Value of alpha-smooth muscle actin and glial fibrillary acidic protein in predicting early hepatic fibrosis in chronic hepatitis C virus infection. Arch Med Sci. 2010;6:356–365. doi: 10.5114/aoms.2010.14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nouchi T., Tanaka Y., Tsukada T., Sato C., Marumo F. Appearance of alpha-smooth-muscle-actin-positive cells in hepatic fibrosis. Liver. 1991;11:100–105. doi: 10.1111/j.1600-0676.1991.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 38.Matsubara K., Suda K., Suzuki F., Kumasaka T., Shiotsu H., Miyano T. Alpha-Smooth muscle actin immunoreactivity may change in nature in interlobular fibrosis of the pancreas in patients with congenital biliary dilatation. Pathol Int. 2004;54:498–502. doi: 10.1111/j.1440-1827.2004.01656.x. [DOI] [PubMed] [Google Scholar]
- 39.Apte M., Pirola R., Wilson J. The fibrosis of chronic pancreatitis: new insights into the role of pancreatic stellate cells. Antioxid Redox Signal. 2011;15:2711–2722. doi: 10.1089/ars.2011.4079. [DOI] [PubMed] [Google Scholar]
- 40.Xu Z., Vonlaufen A., Phillips P.A., Fiala-Beer E., Zhang X., Yang L. Role of pancreatic stellate cells in pancreatic cancer metastasis. Am J Pathol. 2010;177:2585–2596. doi: 10.2353/ajpath.2010.090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klose T., Abiatari I., Samkharadze T., De Oliveira T., Jäger C., Kiladze M. The actin binding protein destrin is associated with growth and perineural invasion of pancreatic cancer. Pancreatology. 2012;12:350–357. doi: 10.1016/j.pan.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Z., Wang K., Wei J., Lu X., Liu B. Proteomic analysis of anti-tumor effects by tetrandrine treatment in HepG2 cells. Phytomedicine. 2010;17:1000–1005. doi: 10.1016/j.phymed.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 43.Ng E.K., Lee S.M., Li H.Y., Ngai S.M., Tsui S.K., Waye M.M. Characterization of tissue-specific LIM domain protein (FHL1C) which is an alternatively spliced isoform of a human LIM-only protein (FHL1) J Cell Biochem. 2001;82:1–10. doi: 10.1002/jcb.1110. [DOI] [PubMed] [Google Scholar]
- 44.Rong Y, Wu W, Ni X, Kuang T, Jin D, Wang D et al. Lactate dehydrogenase A is overexpressed in pancreatic cancer and promotes the growth of pancreatic cancer cells. Tumour Biol 2013. doi: http://dx.doi.org/10.1007/s13277-013-0679-1. [DOI] [PubMed]
- 45.Gong B., Cao Z., Zheng P., Vitolo O.V., Liu S., Staniszewski A. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 46.Lien H.C., Wang C.C., Huang C.S., Yang Y.W., Kuo W.H., Yao Y.T. Ubiquitin carboxy-terminal hydrolase L1 may be involved in the development of mammary phyllodes tumors. Virchows Arch. 2013;462:155–161. doi: 10.1007/s00428-012-1366-0. [DOI] [PubMed] [Google Scholar]
- 47.Tsai R.Y., McKay R.D. Cell contact regulates fate choice by cortical stem cells. J Neurosci. 2000;20:3725–3735. doi: 10.1523/JNEUROSCI.20-10-03725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seaberg R.M., Smukler S.R., Kieffer T.J., Enikolopov G., Asghar Z., Wheeler M.B. Clonal identification of multipotent precursors from adult mouse pancreas that generate neural and pancreatic lineages. Nat Biotech. 2004;22:1115–1124. doi: 10.1038/nbt1004. [DOI] [PubMed] [Google Scholar]
- 49.Baba S., Fujii H., Hirose T., Yasuchika K., Azuma H., Hoppo T. Commitment of bone marrow cells to hepatic stellate cells in mouse. J Hepatol. 2004;40:255–260. doi: 10.1016/j.jhep.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 50.Xu L., Hui A., Albanis E., Arthur M., O’Byrne S., Blaner W. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jesnowski R., Furst D., Ringel J., Chen Y., Schrodel A., Kleeff J. Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by matrigel and N-acetylcysteine. Lab Invest. 2005;85:1276–1291. doi: 10.1038/labinvest.3700329. [DOI] [PubMed] [Google Scholar]
- 52.Carr S., Aebersold R., Baldwin M., Burlingame A., Clauser K., Nesvizhskii A. The need for guidelines in publication of peptide and protein identification data: working group on publication guidelines for peptide and protein identification data. Mol Cell Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Taylor G.K., Goodlett D.R. Rules governing protein identification by mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:3420. doi: 10.1002/rcm.2225. [DOI] [PubMed] [Google Scholar]
- 54.Wilkins M.R., Appel R.D., Van Eyk J.E., Chung M.C., Gorg A., Hecker M. Guidelines for the next 10 years of proteomics. Proteomics. 2006;6:4–8. doi: 10.1002/pmic.200500856. [DOI] [PubMed] [Google Scholar]
- 55.Elias J.E., Gibbons F.D., King O.D., Roth F.P., Gygi S.P. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat Biotechnol. 2004;22:214–219. doi: 10.1038/nbt930. [DOI] [PubMed] [Google Scholar]
- 56.Moore R.E., Young M.K., Lee T.D. Method for screening peptide fragment ion mass spectra prior to database searching. J Am Soc Mass Spectrom. 2000;11:422–426. doi: 10.1016/S1044-0305(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 57.Cui X., Churchill G.A. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 2003;4:210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Proteins of higher abundance in hHSC as determined via TMT analysis for individual samples.
Table S2. Proteins of higher abundance in hPaSC as determined via TMT analysis for individual samples.
Table S3. All proteins identified in the TMT analysis.



