Abstract
BEST1 encodes Bestrophin-1 (Best1), a homo-oligomeric, integral membrane protein localized to the basolateral plasma membrane of the retinal pigment epithelium. Mutations in BEST1 cause five distinct retinal degenerative diseases, including adult vitelliform macular dystrophy (AVMD), autosomal recessive bestrophinopathy (ARB), autosomal dominant vitreoretinochoroidopathy (ADVIRC), and retinitis pigmentosa (RP). The mechanisms underlying these diseases and why mutations cause one disease over another are, for the most part, unknown. To gain insights into these four diseases, we expressed 28 Best1 mutants fused to YFP in polarized MDCK monolayers and, via confocal microscopy and immunofluorescence, live-cell FRET, and reciprocal co-immunoprecipitation experiments, screened these mutants for defects in localization and oligomerization. All 28 mutants exhibited comparable FRET efficiencies to and co-immunoprecipitated with WT Best1, indicating unimpaired oligomerization. RP- and ADVIRC-associated mutants were properly localized to the basolateral plasma membrane of cells, while two AVMD and most ARB mutants were mislocalized. When co-expressed, all mislocalized mutants caused mislocalization of WT Best1to intracellular compartments. Our current and past results indicate that mislocalization of Best1 is not an absolute feature of any individual bestrophinopathy, occurring in AVMD, BVMD, and ARB. Furthermore, some ARB mutants that do not also cause dominant disease cause mislocalization of Best1, indicating that mislocalization is not a cause of disease, and that absence of Best1 activity from the plasma membrane is tolerated. Lastly, we find that the ARB truncation mutants L174Qfs*57 and R200X can form oligomers with WT Best1, indicating that the first ~174 amino acids of Best1 are sufficient for oligomerization to occur.
Keywords: Bestrophin, MDCK; retinal pigment epithelium; fluorescence resonance energy transfer; vitelliform dystrophy; retinitis pigmentosa
1. Introduction
Mutations in the gene BEST1, encoding the protein Bestrophin-1 (Best1), are a common cause of inherited retinal degeneration in man. Over 200 distinct mutations (http://www-huge.uniregensburg.de/BEST1_database) have been reported to cause five clinically distinct retinopathies (Boon et al., 2009; Marmorstein et al., 2009). These are Best vitelliform macular dystrophy (BVMD) (Marquardt et al., 1998; Petrukhin et al., 1998), autosomal recessive bestrophinopathy (ARB) (Burgess et al., 2008), adult-onset vitelliform macular dystrophy (AVMD) (Kramer et al., 2000), autosomal dominant vitreoretinochoroidopathy (ADVIRC) (Yardley et al., 2004), and retinitis pigmentosa (RP) (Davidson et al., 2009). These diseases, collectively termed the “bestrophinopathies,” are clinically distinguished from one another by type of inheritance, effects on the electrooculogram and electroretinogram, age of disease onset, and location and extent of retinal lesions (Boon et al., 2009; Marmorstein et al., 2009).
Best1 is a 68 kDa integral membrane protein that forms homo-oligomers (Stanton et al., 2006). Within the eye, the protein is localized to the basolateral plasma membrane of the retinal pigment epithelium (RPE) (Gouras et al., 2009; Marmorstein et al., 2000). Best1 is an anion channel initially reported as a candidate Ca2+ activated Cl− channel (Sun et al., 2002). However, RPE cells from Best1−/− and Best1W93C knock-in mice exhibit normal Cl− conductances (Marmorstein et al., 2006; Zhang et al., 2010). Recently it was shown, however, that Best1 carries a glutamate conductance in hippocampal astrocytes that is not present in Best1−/− mice (Woo et al., 2012), providing clear evidence that Best1 functions as an anion channel. Similarly, Best1 was found to mediate tonic inhibition in cerebellar glial cells through direct permeation of GABA (Lee et al., 2010). Work from the Hartzell laboratory using a heterologous expression system suggests that Best1 may function as a HCO3- channel, with BVMD-causing mutations abolishing HCO3- conductance (Qu and Hartzell, 2008). Although no published study has analyzed the ability of Best1 to mediate HCO3- transport physiologically, we have shown that the closely related ortholog Bestrophin-2 (Best2) mediates a HCO3- conductance in colonic goblet cells that is absent in Best2−/− mice (Yu et al., 2010). Best2 is also indispensable for generating sweat, a process that may be driven by Best2 mediated HCO3- flux (Cui et al., 2012). These data strongly support the ability of Best1 to function as an anion channel, though the anion conductances which are mediated by Best1 in the RPE remain to be determined.
Best1 is also a known modulator of intracellular Ca2+ signaling in RPE cells. Studies in induced pluripotent stem cell (iPSC) derived RPE cells from BVMD patients (Singh et al., 2013), Best1−/− mice (Marmorstein et al., 2006), and Best1 knock-in mice harboring the BVMD causing mutation W93C (Zhang et al., 2010) all demonstrate that Best1 is a potent regulator of intracellular Ca2+ levels in RPE following stimulation with ATP. When co-expressed in heterologous systems, Best1 is capable of regulating the activity of and physically interacting with L-type voltage-dependent Ca2+ channels (Burgess et al., 2008; Milenkovic et al., 2011a; Reichhart et al., 2010; Rosenthal et al., 2006; Yu et al., 2008). Recent work also suggests, however, that a sub-population of Best1 may be expressed in the endoplasmic reticulum adjacent to the basolateral plasma membrane, where its activity may serve to influence Ca2+ stores (Gomez et al., 2013; Neussert et al., 2010).
All disease-associated mutations of Best1 studied to date result in a loss of anion channel activity and/or altered Ca2+ signaling (Hartzell et al., 2008; Marmorstein et al., 2009; Xiao et al., 2010). How these aspects of Best1 dysfunction vary amongst disease phenotypes and what underlies these dysfunctions are, for the most part, unknown. It was previously shown that numerous BVMD mutants are mislocalized in stably transfected Madin-Darby Canine Kidney (MDCK) epithelial cells (Milenkovic et al., 2011b). MDCK cells do not express endogenous Best1, however, and Best1 is known to form homo-oligomers (Stanton et al., 2006; Sun et al., 2002). For this reason, we recently analyzed BVMD mutant localization in MDCK cells as well as fetal human RPE (fhRPE) cells and RPE in the mouse eye, as both fetal human and mouse RPE express endogenous Best1 (Bakall et al., 2003; Johnson et al., 2013). We found that MDCK cells are a valid, predictive model for Best1 localization in the RPE. We also discovered that the three BVMD mutants tested form oligomers with WT Best1 and that WT and mutant Best1 can influence the localization of each other (Johnson et al., 2013). Since defects in localization and oligomerization are known to underlie other channelopathies (Jentsch et al., 2005; Pedemonte and Galietta, 2012), we were curious how mutations causal for the remaining bestrophinopathies impacted these parameters. In this study, we examined how 28 mutations associated with AVMD, ARB, RP, and ADVIRC (http://www-huge.uni-regensburg.de/BEST1_database) impact the localization and oligomerization of Best1. We conclude that none of the bestrophinopathies are associated with oligomeric defects and, while numerous AVMD and ARB mutants are mislocalized, failure to traffic to the plasma membrane alone is insufficient to distinguish between these retinopathies and other pathogenic mechanisms are likely involved. Recessive ARB mutants also caused mislocalization of WT Best1 in intracellular compartments, further indicating that mislocalization is not, on its own, pathogenic and that the functional loss of Best1 from the plasma membrane is well tolerated.
2. Materials and methods
2.1. Cell culture
MDCK II cells (American Type Culture Collection, Manassas, VA, USA) were maintained in a 95% air 5% / CO2 environment at 37°C. Cells were grown in Dulbecco's Modification Eagle's Medium (Cellgro, Manassas, VA, USA) supplemented with 10% fetal bovine serum and penicillin/streptomycin solution (Cellgro, Manassas, VA, USA). For immunofluorescence and co-immunoprecipitation studies, MDCK cells were plated at confluence on 1.0 cm2 Transwell filters (Corning, Corning, NY, USA) and maintained for 5 days prior to infections with replication defective type 5 adenovirus vectors at a multiplicity of infection (MOI) of 30 and used 48 hrs later. For fluorescence resonance energy transfer (FRET) studies, MDCK cells were plated onto coverslips in 35 mm glass bottom culture dishes (MatTek Corporation, Ashland, MA, USA) and subjected to the same protocol. For FRET studies involving the positive and negative control (CFP-YFP and Best1-CFP co-expressed with YFP, respectively), MDCK cells were transfected using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) at ~80% confluence and used 48 hrs later.
2.2. Plasmid constructs
Best1 with a 6x-c-myc tag at the C-terminus (Best1-c-myc) in the pRK5 plasmid (kindly provided by Dr. Jeremy Nathans (Sun et al., 2002)) as well as replication defective adenoviral vectors carrying Best1-CFP and Best1-YFP have been previously described (Johnson et al., 2013). Mutations causal for AVMD, ARB, ADVIRC, and RP were generated in Best1-YFP in the viral shuttle vector pacAd5CMVK-NpA (provided by the Gene Transfer Vector Core, University of Iowa, Iowa City, IA, USA) via site-directed mutagenesis using a kit according to the manufacturer's instructions (Agilent Technologies, Santa Clara, CA, USA). For those mutations resulting in early stop codons, the region prior to the stop codon was excised and placed in-frame with YFP in the viral shuttle vector. Best1 tagged with c-myc was inserted into the shuttle vector pacAd5CMVK-NpA and, along with the newly generated mutants, sent to the Gene Transfer Vector Core at the University of Iowa for generation, amplification, purification, and titration of replication defective adenoviral vectors. Replication defective adenoviral vectors carrying the untagged Best1 mutants L140V, Y227C, and D228N were used for initial localization studies and generated according to the method of Hardy et al. (Hardy et al., 1997) as described previously (Marmorstein et al., 2004).
2.3. Immunofluorescence
Immunofluorescence was performed as before (Johnson et al., 2013). In brief, transduced MDCK cells were stained for nuclei and/or the endogenous, apical plasma membrane protein gp135 using 4′,6-diamidino-2-phenylindole and the mouse, monoclonal antibody 3F4 (generous gift of Dr. George Ojakian, SUNY Health Science Center at Brooklyn, NY, USA), respectively. Images were obtained using a 40X oil immersion objective on a Leica SP5 confocal microscope. For co-localization analysis, MDCK cells were stained for c-myc using a rabbit, polyclonal antibody specific to c-myc (Sigma-Aldrich, St. Louis, MO, USA).
2.4. FRET
FRET was performed as described previously (Johnson et al., 2013). Live MDCK cells were imaged using a 40X oil immersion objective and the 514 nm laser on a Leica SP5 confocal microscope was used to bleach the acceptor, Best1-YFP. Excitations were 458 nm and 514 nm and emissions were collected from 465 to 505 nm and 525 to 600 nm for the donor (CFP) and acceptor (YFP), respectively. Fluorescence for the donor, Best1-CFP, was measured before and after bleaching in ImageJ and FRET efficiency (%E) was calculated as follows:
2.5. Immunoprecipitation and western blotting
Reciprocal co-immunoprecipitation experiments were done as before (Johnson et al., 2013) using adenoviral vectors. All experiments were performed on polarized MDCK monolayers grown on Transwell filters (Corning, Corning, NY, USA). Cells were infected at confluence at a MOI of 30 and, 48 hrs later, filters were harvested for immunoprecipitation. Cells were immunoprecipitated using rabbit, polyclonal antibodies specific to either YFP (Clontech, Mountain View, CA, USA) or c-myc (Sigma-Aldrich, St. Louis, MO, USA). Western blotting was then performed using a mouse, monoclonal antibody specific to the opposite tag, either cmyc (9E10; Invitrogen, Grand Island, NY, USA) or YFP (JL-8; Clontech, Mountain View, CA, USA). For the immunoprecipitation and blotting of WT Best1-YFP compared to the four truncation mutants, MDCK cells were immunoprecipitated for YFP using a rabbit, polyclonal antibody (Clontech, Mountain View, CA, USA) and western blotted back using a mouse, monoclonal antibody specific to YFP (JL-8; Clontech, Mountain View, CA, USA).
2.6. Patch clamp electrophysiology
Currents were recorded from HEK293 cells using the whole-cell configuration of the patch-clamp technique as described in (Adomaviciene et al., 2013). Cells were transiently transfected with 2 μg of Best1 (WT or tagged) cDNA using Fugene HD (Roche) according to manufacturer's instructions and used 2 days after transfection. Intracellular recording solution contained (in mM): 20 CsCl, 110 Cs-aspartate, 2 MgCl2, 10 glucose, 10 HEPES, 10 EGTA, 7.2 CaCl2 (giving free Ca2+ concentration of ~0.5 M), pH was 7.2 (adjusted with CsOH). Extracellular recording solution contained (in mM): 140 NaCl, 2 CaCl2, 1 MgCl2, 10 glucose, 30 mannitol, 10 HEPES, pH was 7.4 (adjusted with NaOH). Recording started at least 3 minutes after whole-cell configuration was established in order to allow the intracellular solution to equilibrate with the intracellular milieu (Adomaviciene et al., 2013). Current versus voltage relationships were constructed by measuring the amplitude of the steady-state current in response to 450 ms test pulses from -120 mV to +80 mV in 20 mV increments. The holding potential was -50 mV. Currents were sampled at 10 kHz and filtered at 5 kHz. Membrane current densities were calculated by dividing the current by the cell capacitance. ANOVA with Bonferroni's post-test was used for statistical analysis and p<0.05 was considered significant. Data are given as mean±SEM.
3. Results
3.1. The C-terminal addition of CFP and YFP tags to Best1 does not abolish Best1 anion channel activity
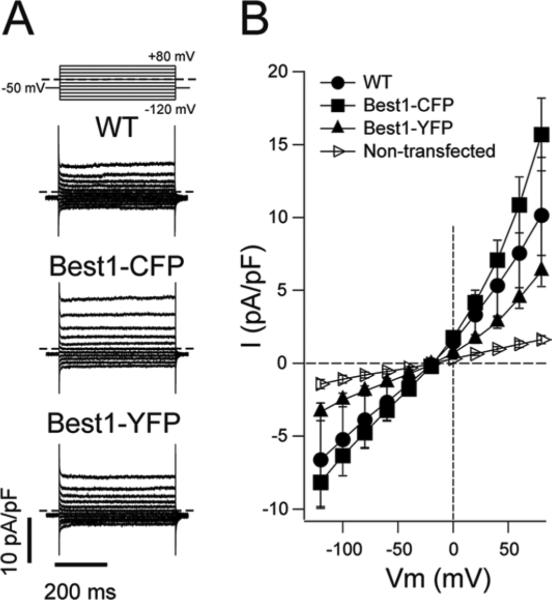
It was previously shown that the addition of a 6X c-myc tag to the C-terminus of Best1 does not impact the ability of Best1 to mediate anion transport in heterologous cells (Sun et al., 2002). To determine if the addition of CFP or YFP to the C-terminus of Best1 impacts its anion channel activity, we performed whole cell patch clamp on human embryonic kidney (HEK293) cells transfected with untagged Best1, Best1-CFP, or Best1-YFP. As shown in Figure 1, cells expressing Best1-CFP or Best1-YFP exhibited a Cl− conductance that is absent in untransfected cells and is generally comparable to cells expressing untagged Best1. This demonstrates that the addition of CFP or YFP to the C-terminus of Best1 does not abolish its anion channel activity. Although cells expressing Best1-CFP or Best1-YFP exhibited robust Cl− conductances compared to untransfected cells, currents in cells expressing Best1-CFP were slightly higher than those in cells expressing untagged Best1. Conversely, Cl− conductances were slightly lower in cells expressing Best1-YFP compared to those expressing untagged Best1. Thus, it is possible that the addition of these tags may exert mild effects on Best1's anion channel activity.
Figure 1. Whole-cell chloride currents mediated by untagged, CFP-tagged and YFP-tagged Best1 channels.
(A) Whole-cell currents recorded from HEK293 cells expressing untagged Best1 (WT) or Best1 tagged with CFP or YFP, as indicated. Dashed lines represent the zero current level. Voltage protocol is shown in the upper panel. (B) Mean whole-cell current density versus voltage relationship. The number of experiments was 5-8 in each case. Tagged and untagged Best1 currents were significantly higher than those observed in untransfected cells, indicating that the C-terminal addition of CFP or YFP does not abolish the associated anion channel activity of Best1.
3.2. ADVIRC and RP mutants are properly localized while numerous AVMD and ARB mutants are mislocalized
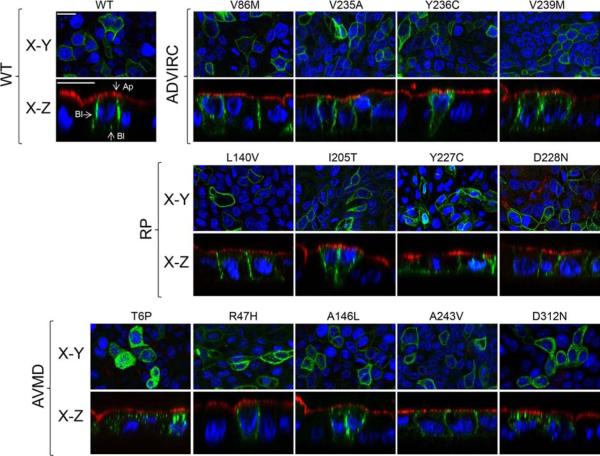
Using adenovirus-mediated gene transfer, WT and mutant Best1 were expressed in polarized monolayers of MDCK cells grown on Transwell filters. Cells were stained for the apical protein marker gp135 as well as nuclei for positional referencing and confocal X-Y and X-Z scans were used to examine the localization of Best1. To help ensure that the localization phenotypes observed were due to mutant defects and not to mis-sorting caused by excess protein, we used a MOI for each adenoviral vector intended to limit potential artifacts due to overexpression. As a result, expression levels varied with some cells in the confocal scans not expressing WT or mutant protein. Consistent with its localization in RPE (Marmorstein et al., 2000), Best1-YFP localized to the basolateral plasma membrane, with very little of the protein accumulating inside cells. No co-localization with the apical plasma membrane was observed (Fig. 2). The ADVIRC mutants V86M, V235A, Y236C, and V239M, the RP mutants L140V, I205T, Y227C, and D228N, and the AVMD mutants R47H, A146L, and A243V all exhibited a similar localization to WT Best1 (Fig. 2). The AVMD mutants T6P and D312N, however, were found predominantly in intracellular compartments (Fig. 2).
Figure 2. Localization of Best1 and ADVIRC-, RP-, as well as AVMD-associated mutants in MDCK cells as determined by confocal microscopy.
Representative X-Y and X-Z scans of WT or mutant Best1-YFP (green) are shown. WT or mutant Best1 were expressed in confluent, polarized monolayers of MDCK cells via adenovirus-mediated gene transfer and stained for the endogenous apical protein gp135 (red) and nuclei (blue) for positional referencing. All ADVIRC (V86M, V235A, Y236C, and V239M) and RP (L140V, I205T, Y227C, and D228N) mutants localized to the basolateral plasma membrane. For AVMD-associated mutants, T6P and D312N were intracellular while R47H, A146L, and A243V were properly localized. Ap and Bl stand for apical and basolateral, respectively. Scale bars: 20 μm.
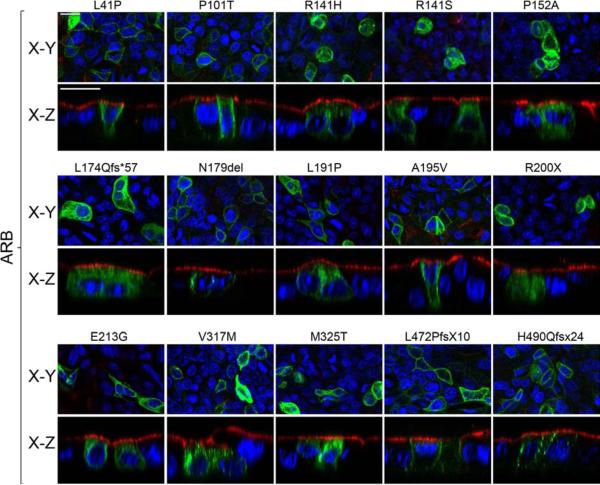
We next examined the localization of mutants associated with ARB (Fig. 3). We found that the mutants L41P, P101T, N179del, A195V, L472PfsX10, and H490QfsX24 were localized to the basolateral plasma membrane (Fig. 3). In contrast, the mutants R141H, R141S, P152A, L174Qfs*57, L191P, R200X, E213G, V317M, and M325T accumulated in intracellular compartments (Fig. 3). These data are in agreement with findings by Davidson et al., which showed that, when transiently transfected into MDCK cells, L41P and A195V were properly localized while P152A, V317M, and M325T were not (Davidson et al., 2011). Although differences were observed, most mislocalized AVMD and ARB mutants were found in the cytoplasm between the apical plasma membrane and the nuclei (Figs. 2 and 3), suggesting that many may be mislocalized to the same intracellular compartments.
Figure 3. Localization of ARB-associated mutants in MDCK cells as determined by confocal microscopy.
Representative X-Y and X-Z scans of mutant Best1-YFP (green) are shown. Mutant Best1 was expressed in confluent, polarized monolayers of MDCK cells via adenovirus-mediated gene transfer and stained for the endogenous apical protein gp135 (red) and nuclei (blue) for positional referencing. L41P, P101T, N179del, A195V, L472PfsX10, and H490QfsX24 were localized to the basolateral plasma membrane. In contrast, R141H, R141S, P152A, L174Qfs*57, L191P, R200X, E213G, V317M, and M325T were found in intracellular compartments. Scale bars: 20 μm.
3.3. Mislocalized ARB and AVMD mutants co-localize with WT Best1
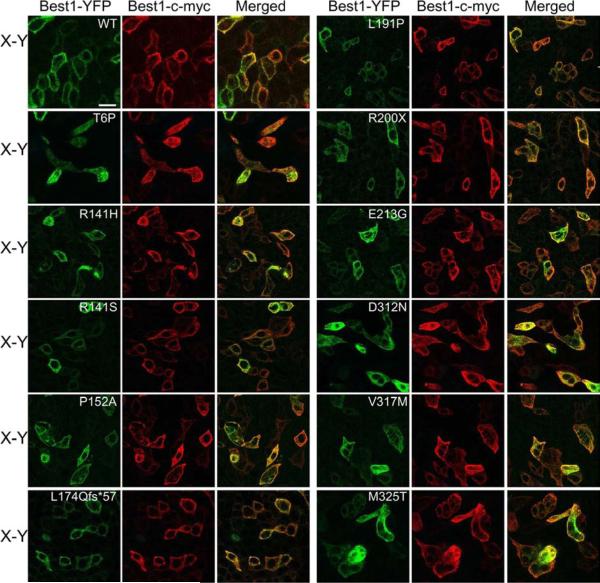
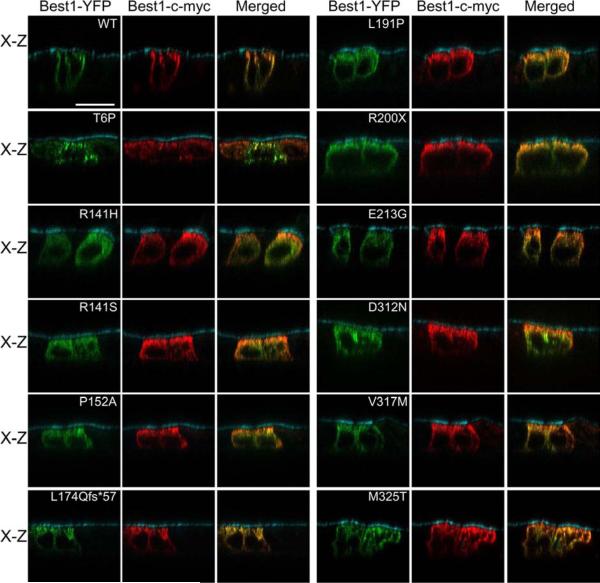
We have previously shown that, in polarized MDCK cells, the BVMD mutant W93C is mislocalized on its own and is rescued to the plasma membrane by the presence of WT Best1. Conversely, the BVMD mutant V9M mutant is mislocalized on its own and, when co-expressed together, dominantly mislocalizes WT Best1 (Johnson et al., 2013). Since we know that WT and mutant Best1 can influence the localization of each other , we co-expressed mislocalized, mutant Best1 fused to YFP with WT Best1-c-myc in MDCK cells via adenovirus-mediated gene transfer (Figs. 4 and 5). Cells were stained for c-myc as well as the apical protein marker gp135 for positional referencing. Co-localization between Best1-c-myc and mutant Best1-YFP was analyzed using confocal X-Y (Fig. 4) and X-Z scans (Fig. 5). WT Best1-YFP and Best1-c-myc co-localized in the basolateral plasma membrane (Figs. 4 and 5). Interestingly, all mislocalized mutants (T6P, R141H, R141S, P152A, L174Qfs*57, L191P, R200X, E213G, D312N, V317M, and M325T) co-localized with WT Best1-c-myc predominantly in intracellular compartments in both the X-Y (Fig. 4) and X-Z (Fig. 5) scans. For some mutants (e.g., R141H, R141S, L174Qfs*57, and R200X), cells were occasionally observed that showed some co-localization at the cell periphery in the X-Y scans (Fig. 4), suggesting that WT Best1 may exert at least a partial rescue effect.
Figure 4. Co-localization X-Y scans between mislocalized AVMD- and ARB-associated mutants and WT Best1 as determined by confocal microscopy.
Representative merged X-Y scans of WT and mutant Best1-YFP co-expressed with Best1-c-myc are shown. Confluent, polarized monolayers of MDCK cells were made to express both Best1-c-myc and WT or mutant Best1-YFP (green) via adenovirus-mediated gene transfer. Cells were stained for c-myc (red) and, for positional referencing, the endogenous apical protein gp135 (cyan). Best1-YFP co-localized with Best1-c-myc in the basolateral plasma membrane, while all 11 mislocalized mutants (T6P, R141H, R141S, P152A, L174Qfs*57, L191P, R200X, E213G, D312N, V317M, and M325T Best1) co-localized with Best1-c-myc predominantly in intracellular compartments. Scale bar: 20 μm.
Figure 5. Co-localization X-Z scans between mislocalized AVMD- and ARB-associated mutants and WT Best1 as determined by confocal microscopy.
Representative merged X-Z scans of WT and mutant Best1-YFP co-expressed with Best1-c-myc are shown. Confluent, polarized monolayers of MDCK cells were made to express both Best1-c-myc and WT or mutant Best1-YFP (green) via adenovirus-mediated gene transfer. Cells were stained for c-myc (red) and, for positional referencing, the endogenous apical protein gp135 (cyan). Best1-YFP co-localized with Best1-c-myc in the basolateral plasma membrane, while all 11 mislocalized mutants (T6P, R141H, R141S, P152A, L174Qfs*57, L191P, R200X, E213G, D312N, V317M, and M325T Best1) co-localized with Best1-c-myc predominantly in intracellular compartments. Scale bar: 20 μm.
3.4. ADVIRC, RP, AVMD, and ARB mutants form oligomers with WT Best1
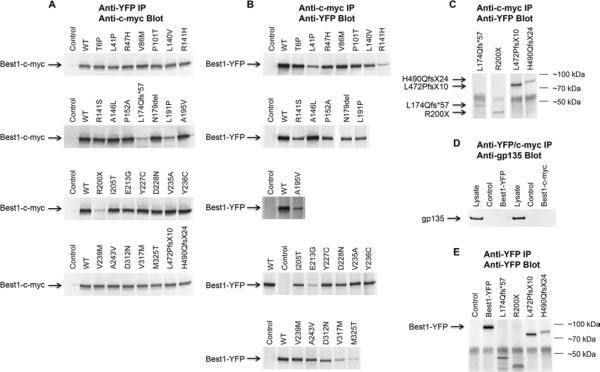
That mislocalized AVMD and ARB mutants robustly co-localize with WT Best1 (Figs. 4 and 5) suggests that these diseases are not associated with oligomeric defects. To test this further, we performed reciprocal, co-immunoprecipitation experiments between Best1-c-myc and all 28 mutants fused to YFP using antibodies specific to each of these tags. Confluent, polarized MDCK cells were made to express both forms of Best1 using replication defective adenoviruses. As we reported previously, Best1 with a 6x-c-myc tag (~75 kDa) and Best1 with a YFP tag (~95 kDa) were capable of reciprocal co-immunoprecipitation with each other (Fig. 6A-C). In contrast, Best1-c-myc and Best1-YFP were unable to co-immunoprecipitate with gp135, a 135 kDa apical plasma membrane protein endogenous to MDCK cells (Fig. 6D). Best1-c-myc and all 28 mutants fused to YFP immunoprecipitated the other tagged form of Best1 (Fig. 6A-C), indicative of physical interaction and hetero-oligomer formation between WT and mutant Best1. We also immunoprecipitated and western blotted for Best1-YFP and the four truncation mutants (L174Qfs*57, R200X, L472PfsX10, and H490QfsX24) fused to YFP. We find that all four mutants were expressed and migrated at their predicted Mr (Fig. 6C, E).
Figure 6. Reciprocal co-immunoprecipitation of Best1-c-myc and WT or mutant Best1-YFP in MDCK cells.
(A) MDCK cells were transduced to express both Best1-c-myc (~75 kDa) and WT or mutant Best1-YFP (~95 kDa) using adenovirus-mediated gene transfer. Co-expressing cells were lysed and Best1 immunoprecipitated using an anti-YFP antibody and western blotted using an antibody specific to c-myc. Control lanes were loaded with immunoprecipitates prepared from uninfected MDCK cells. All other lanes were loaded with immunoprecipitated Best1. (B) Excluding the four ARB truncation mutants, MDCK cells were transduced to express both Best1-c-myc (~75 kDa) and WT or mutant Best1-YFP (~95 kDa) using adenovirus-mediated gene transfer. Co-expressing cells were lysed and Best1 immunoprecipitated using an anti-c-myc antibody and western blotted using an antibody specific to YFP. (C) Lanes were loaded with MDCK cells co-expressing WT Best1-c-myc and YFP-tagged ARB truncation mutants (L174Qfs*57, R200X, L472PfsX10, and H490QfsX24 Best1-YFP ). Cells were immunoprecipitated using an antibody specific to c-myc and blotted back using an antibody specific to YFP. (D) MDCK cells were infected with either Best1-c-myc or Best1-YFP via adenovirus-mediated gene transfer. Best1 was immunoprecipitated using anti-cmyc or anti-YFP antibodies and blotted back using an antibody specific to gp135 (135 kDa). Lysate lanes were loaded using lysates from cells infected with either Best1-c-myc or Best1-YFP. (E) MDCK cells were infected with Best1-YFP or with one of the truncation mutants (L174Qfs*57, R200X, L472PfsX10, and H490QfsX24 Best1-YFP). Best1 was immunoprecipitated using an anti-YFP antibody and blotted back using a separate anti-YFP antibody.
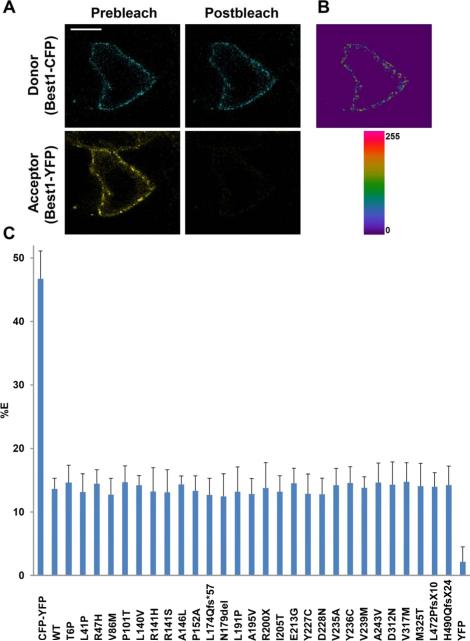
To validate that Best1 mutants form oligomers with WT Best1 rather than aggregates due to protein overexpression, we performed live-cell, confocal, FRET acceptor photobleaching in polarized MDCK cells. This experimental approach has been successfully used to assess and quantify the interaction of channels at the plasma membrane (Johnson et al., 2013; Sheridan et al., 2011). Confluent cells were made to co-express Best1-CFP and either WT or mutant Best1-YFP via adenovirus-mediated gene transfer. FRET was quantified by the increase in donor (CFP) fluorescence following photobleaching of the acceptor (YFP), as indicated in the methods. In all cases the FRET efficiencies significantly differed (p < 0.001) from the positive and negative controls, which were a CFP-YFP fusion protein and Best1-CFP co-expressed with YFP, respectively (Fig. 7). FRET efficiency for the positive control, CFP-YFP, was near-maximal at 46.74% ± 4.37 (mean ± s.d., n = 23). Pre- and post-bleach photos highlighting this near-maximal FRET efficiency for CFP-YFP are shown in Supplementary Figure 1. FRET efficiency for the negative control, Best1-CFP co-expressed with YFP, was at background at 2.14% ± 2.36 (n = 24). As we observed previously (Johnson et al., 2013), both Best1-CFP and Best1-YFP exhibited FRET in the plasma membrane (Fig. 7A, B). When co-expressed with Best1-CFP, all 28 mutants associated with ADVIRC, AVMD, ARB, and RP exhibited FRET efficiencies that did not differ significantly from the FRET efficiency of Best1-CFP and WT Best1-YFP (Fig. 7C). The p-values for FRET between WT and mutant Best1 ranged from 0.1249 (n= 21, P101T) to 0.8811 (n= 24, R200X). FRET between Best1-CFP and Best1-YFP was 13.64% ± 1.67 (n = 21). The mutant FRET efficiencies ranged from 12.45% ± 3.56 (n = 20, N179del) to 14.77% ± 2.33 (n = 20, A146L) and, for all experiments, the value of n was greater than or equal to 19 (Fig. 7C). Along with our co-localization and reciprocal co-immunoprecipitation experiments, these data strongly suggest that none of the 28 mutants associated with these four bestrophinopathies exhibit defects in oligomerization. The results of our localization and oligomerization screen are summarized in Table 1.
Figure 7. Live-cell, confocal FRET acceptor photobleaching of Best1-CFP and WT or mutant Best1-YFP in MDCK cells.
(A) Representative X-Y scan of Best1-CFP (blue, donor) and Best1-YFP (yellow, acceptor) co-expressed in polarized MDCK cells using adenovirus-mediated gene transfer. Using a 514 nm laser on a confocal microscope, live-cell acceptor photobleaching was performed, generating the resultant image in (B), which highlights regions in the plasma membrane where donor intensity was increased post-bleaching. (C) For all experiments, n ≥ 19. FRET efficiencies (% E) were determined for Best1-CFP paired with WT or mutant Best1-YFP via adenovirus-mediated gene transfer. As a positive control, MDCK cells were transfected with a CFP-YFP fusion protein. As a negative control, MDCK cells were cotransfected with Best1-CFP and YFP. WT and mutant Best1 had % E's that did not significantly differ from each other, but did significantly differ from the negative and positive controls (p < 0.001). Scale bar: 10 μm. Error bars indicate ± S.D.
Table 1.
Summary of localization, co-localization, and oligomerization findings for mutant and WT Best1.
| Mutant | Associated Diseases | Properly Localized | Co-localizes with WT Best1 at the Plasma Membrane | Forms Oligomers with WT Best1 | Diminished Chloride Conductance |
|---|---|---|---|---|---|
| WT | N/A | + | + | + | N/A |
| T6P | AVMD, BVMD | - | - | + | +,Σ |
| L41P | ARB, BVMD | + | N/A | + | +*,¥ |
| R47H | AVMD | + | N/A | + | Untested |
| V86M | ADVIRC | + | N/A | + | Untested |
| P101T | ARB, BVMD | + | N/A | + | Untested |
| L140V | RP, ARB | + | N/A | + | +*,‡ |
| R141H | ARB, BVMD | - | - | + | +**,† |
| R141S | ARB | - | - | + | Untested |
| A146L | AVMD | + | N/A | + | Untested |
| P152A | ARB | - | - | + | +**,† |
| L174Qfs*57 | ARB | - | - | + | Untested |
| N179del | ARB | + | N/A | + | Untested |
| L191P | ARB | - | - | + | Untested |
| A195V | ARB, BVMD | + | N/A | + | +**,¥ |
| R200X | ARB | - | - | + | Untested |
| I205T | RP | + | N/A | + | +*,‡ |
| E213G | ARB | - | - | + | Untested |
| Y227C | RP, BVMD | + | N/A | + | Untested |
| D228N | RP | + | N/A | + | Untested |
| V235A | ADVIRC | + | N/A | + | Untested |
| Y236C | ADVIRC | + | N/A | + | Untested |
| V239M | ADVIRC | + | N/A | + | Untested |
| A243V | AVMD | + | N/A | + | +**,Ω |
| D312N | AVMD, ARB | - | - | + | +*,¥ |
| V317M | ARB | - | - | + | +**,¥ |
| M325T | ARB | - | - | + | +*,¥ |
| L472PfsX10 | ARB | + | N/A | + | Untested |
| H490QfsX24 | ARB, BVMD | + | N/A | + | Untested |
* = Mutant significantly suppresses the chloride conductance of WT Best1.
** = Mutant does not significantly alter the chloride conductance of WT Best1.
+ = Properly localized/Co-localizes at the plasma membrane/Forms oligomers with WT Best1/Exhibits a diminished chloride conductance
- = Mislocalized/Co-localizes in intracellular compartments
† Previously reported by (Burgess et al., 2008)
‡ Previously reported by (Davidson et al., 2009)
¥ Previously reported by (Davidson et al., 2011)
Σ Previously reported by (Milenkovic et al., 2011b)
Ω Previously reported by (Yu et al., 2006)
4. Discussion
Here, we present the results of a localization and oligomerization screen for 28 Best1 mutants associated with ADVIRC, RP, AVMD, and ARB. We find that the fusion of CFP- or YFP- tags to the C-terminus does not abrogate the associated anion conductance of Best1 in transfected HEK293 cells (Fig. 1). This is analogous to previous findings, which show that the addition of a 6x-c-myc tag to the C-terminus of Best1 does not impact its anion channel activity in heterologous cells (Sun et al., 2002). Our findings are summarized along with previous data for the 28 Best1 mutants tested here regarding anion channel activity (Burgess et al., 2008; Davidson et al., 2011; Davidson et al., 2009; Milenkovic et al., 2011b; Yu et al., 2006) and associated disease phenotypes (http://www-huge.uni-regensburg.de/BEST1_database) in Table 1. We find that missense mutants associated with RP and ADVIRC are all properly localized (Fig. 2), while mutants associated with AVMD and ARB can, like BVMD mutants (Johnson et al., 2013; Milenkovic et al., 2011b), be properly localized or mislocalized (Figs. 2 and 3). Interestingly, the ARB mutants L472PfsX10 and H490QfsX24 localized to the basolateral plasma membrane, indicating that amino acids 481-585 are not necessary for plasma membrane localization (Fig. 3). Our laboratories (Figs. 2 and 3) (Johnson et al., 2013) as well as other laboratories (Davidson et al., 2011; Davidson et al., 2009; Milenkovic et al., 2011b) have investigated a number of Best1 mutants in polarized MDCK cells. When properly localized and mislocalized mutants are highlighted in the Best1 topology models proposed by Tsunenari et al. (Tsunenari et al., 2003) (Fig. S2) and Milenkovic et al. (Milenkovic et al., 2007) (Fig. S3), there is no distinct correlation between the localization of the protein and the location of the mutation within the protein. There are, however, high concentrations of mislocalization-inducing mutants in the intracellular N-terminus (Figs. S2 and S3) leading up to the first putative transmembrane domain, and (Figs. S2 and S3) immediately following the emergence of the final transmembrane domain into the cytosol. These data suggest that these regions may contain signals relevant to Best1 trafficking.
Consistent with our data, Milenkovic et al. also looked at the localization of mutant T6P and reported it to be mislocalized in stably transfected, polarized MDCK cells (Milenkovic et al., 2011b). They also examined mutants at amino acids positions 227 and 243, finding that Y227N was mislocalized but that A243T was localized to the plasma membrane (Milenkovic et al., 2011b). Mullins et al. found that Y227N in an eye form a donor with BVMD was mislocalized (Mullins et al., 2005). According to our data, both Y227C and A243V are properly localized (Fig. 2). It is interesting that the Y227N, but not the Y227C, mutation results in mislocalization of the protein. It was recently reported, however, that different mutations at this position can have disparate effects on the localization of Best1, with Y227N and Y227F being mislocalized and properly localized, respectively (Doumanov et al., 2013). Although Y227N and Y227C are mutations thought to be causal for disease, Y227F has, to date, not been reported in association with disease. Singh et al. also examined the localization of the BVMD mutant A146K in iPS cell-derived RPE, finding it be properly localized (Singh et al., 2013), similar to our results for the AVMD mutant A146L (Fig. 2). In transiently transfected MDCK cells, Davidson et al. reported that the RP mutant I205T was properly localized but that the RP mutants L140V and D228N were mislocalized (Davidson et al., 2009).
The latter two results conflict with our findings, which suggest that all of the RP-associated mutants localize to the basolateral plasma membrane (Fig. 2). We have observed that, unless Best1 is expressed in a highly confluent, polarized monolayer of MDCK cells, it will not reliably localize to the plasma membrane. This has been shown for other epithelial proteins, which require the formation of confluent, polarized monolayers for proper sorting to occur (Low et al., 2000; Rodriguez-Boulan and Salas, 1989; Wang et al., 1990a, b). It has been reported, for example, that the vasopressin type-2 receptor does not localize to the plasma membrane in transiently transfected cells, but does so in stably transfected cells (van Beest et al., 2006). Thus, the different results for these 2 mutants may be due to the use of transient transfection, which is typically performed in cells not fully polarized, versus the use of adenovirus-mediated gene transfer in highly polarized monolayers of MDCK cells.
All mutants tested could reciprocally co-immunoprecipitate and exhibit FRET with WT Best1, including the ARB truncation mutations L174Qfs*57 and R200X (Figs. 6 and 7). This is unlikely to be due to overexpression, as significant plasma membrane FRET was observed between WT Best1 and properly localized mutants (Fig. 7). Furthermore, even cells expressing small quantities of WT and mutant Best1 robustly co-localized with each other (Figs. 4 and 5) and exhibited significant levels of FRET. These data suggest that the first ~174 amino acids are sufficient for the formation of oligomers with WT Best1 and that, like previously tested BVMD mutants (Johnson et al., 2013), oligomeric defects are not associated with ADVIRC, RP, AVMD, and ARB. According to both extant Best1 topology models (Figs. S2 and S3) (Milenkovic et al., 2007; Tsunenari et al., 2003), the first ~174 amino acids are comprised of the intracellular N-terminus, the first 2 transmembrane domains, and all or a significant portion of an intracellular loop following transmembrane domain 2. Studies on the closely related Bestrophin-2 indicate that transmembrane 2 likely forms the channel pore of the bestrophins (Qu et al., 2006; Qu and Hartzell, 2004). Given that the channel pore is frequently the epicenter for channel formation (Clarke and Gulbis, 2012), it is quite feasible that, in heterozygous patients, the mutants L174Qfs*57 and R200X multimerize with WT Best1. Moreover, we have previously shown that mouse and human Best1 can exhibit FRET and reciprocally co-immunoprecipitate each other, despite having significant disparity in their primary amino acid sequence outside of their respective bestrophin domains (Johnson et al., 2013). Cumulatively, these data indicate that Best1 homo-oligomer formation requires at most the first 174 amino acids of the bestrophin domain.
For ADVIRC, all 4 associated mutants localized to the basolateral plasma membrane (Fig. 2), indicating that mislocalization is not a component of disease pathogenesis. Since none of the ADVIRC mutants have been tested for anion conductance (Table 1), it is difficult to speculate how these missense mutations might impact Best1 function. It has been previously reported, however, that ADVIRC mutations lie adjacent to intron-exon boundaries. Assays of in vitro splicing indicate that these mutations disrupt mRNA splicing and generate both missense and exon-deletion mutants (Burgess et al., 2009; Yardley et al., 2004). Although the existence of mis-spliced Best1 mRNA has yet to be demonstrated in tissue, this may explain why these mutations uniquely cause ADVIRC. Similar to ADVIRC, all 4 RP-associated mutants were properly localized (Fig. 2). In transfected HEK293 cells, tested RP mutants (Table 1) exhibit diminished Cl− conductances compared to WT Best1 and can attenuate the Cl− current of WT protein (Davidson et al., 2009). However, numerous BVMD, AVMD, and ARB mutants are also marked by diminished anion channel activity and, when co-expressed with WT Best1, impair the Cl− conductance of WT Best1 as well (Table 1) (Davidson et al., 2011; Hartzell et al., 2008; Xiao et al., 2010). While lack of anion transport may contribute to disease pathogenesis, there are likely other mechanisms involved which result in one bestrophinopathy over another. It is interesting that both RP and ADVIRC missense mutants properly localize to the basolateral plasma membrane (Fig. 2) as both RP and ADVIRC affect the peripheral retina and are marked by retinal pigmentary abnormalities (Boon et al., 2009; Davidson et al., 2009). Thus, the pathogenic mechanisms underlying RP and ADVIRC may be similar and RP and ADVIRC due to BEST1 mutation may be a manifestation of the same disease. Further studies are warranted to test this hypothesis.
2 of the 5 AVMD mutants were mislocalized and the remaining 3 trafficked to the basolateral plasma membrane (Fig. 2). AVMD mutants exhibit a diminished Cl− conductance in transfected HEK293 cells and, when co-expressed, can dominantly suppress the channel activity of WT Best1 or exert no effect on WT conductance (Table 1) (Davidson et al., 2011; Milenkovic et al., 2011b; Yu et al., 2006). Interestingly, T6P and A243V have been independently reported to cause BVMD (Kramer et al., 2000; Petrukhin et al., 1998) and mutations at positions T6, A146, A243, and D312 have all been reported to cause both BVMD and AVMD (Kramer et al., 2000; Petrukhin et al., 1998; Schatz et al., 2010; Singh et al., 2013) . We find that, just like previously tested BVMD mutants (Johnson et al., 2013), AVMD mutants can be either properly localized or mislocalized (Fig. 2). It has been postulated that AVMD may represent a more mild form of BVMD, since its distinguishing characteristic is that it is typically a later-onset disease and patients typically have a normal or near-normal electrooculogram (Boon et al., 2009; Marmorstein et al., 2009). As such, it is not surprising that, in terms of localization and Cl− conductances (Table 1), AVMD and BVMD are mechanistically similar.
Our results for ARB are perhaps the most curious, as they were largely unexpected. It was previously hypothesized that, due to the recessive nature of ARB, this disease may be caused by defective oligomerization (Xiao et al., 2010). Namely, the heterozygous state would be recessive due to an inability of mutant Best1 to interact with and impair WT Best1 (Xiao et al., 2010). This is the case for recessive myotonia caused by mutations in CLC-1, in which some truncation mutants are unable to dimerize with WT subunits (Jentsch et al., 2005). Although we found this theory to be plausible, particularly for the 4 ARB-associated truncation mutants, our results indicate that all 16 ARB-associated mutants tested could form oligomers with WT Best1 (Figs. 6 and 7). Furthermore, several ARB-associated variants exhibit a diminished Cl− conductance in transfected HEK293 cells (Davidson et al., 2011). Some, like L41P, significantly impair the conductance of WT Best1 as well, while others, like V317M and R141H, fail to impact the Cl− currents of WT protein (Davidson et al., 2011). This finding is puzzling, as our data show that WT and V317M or R141H Best1 co-localize together predominantly in intracellular compartments (Figs. 4 and 5). As of yet, there is no distinct correlation between localization or diminished Cl− conductance and disease phenotype (Table 1).
It is also curious that all mislocalized mutants, even those associated with the recessive disease ARB, largely mislocalized WT Best1 when co-expressed together (Figs. 4 and 5). This would lead us to believe that the disease pathogenesis of the bestrophinopathies is wholly independent of Best1's localization to the plasma membrane. For example, 1 copy of an ARB allele, such as M325T, is insufficient to cause disease (Burgess et al., 2008). Our data show, however, that M325T causes the mislocalization of both WT and mutant protein when co-expressed. That mislocalized ARB mutants mislocalize WT Best1 but do not induce a disease phenotype in heterozygous patients, indicates that an inability to traffic to the membrane cannot, on its own, cause disease. This is further indicated by our past (Johnson et al., 2013) and current data, which show that defects in localization are unable to differentiate between the five retinal degenerative diseases associated with Best1. Moreover, patients homozygous for properly localized ARB mutants – such as L472PfsX10 (Fig. 3) – still exhibit an ARB phenotype (Bitner et al., 2011), demonstrating that mislocalization is not a necessary component of ARB pathogenesis. Given the sum of findings for all Best1 mutants we propose that failure of Best1 to traffic to the plasma membrane is not sufficient to cause disease, and that the absence of Best1 activity at the plasma membrane is well tolerated.
This conclusion is contingent upon the assumption that ARB mutants are truly recessive. Although a minority, some ARB mutants have been independently reported as dominant BVMD or AVMD mutants (http://www-huge.uni-regensburg.de/BEST1_database). For example, D312N is associated with both ARB and AVMD and L41P is associated with both ARB and BVMD (Table 1). The majority of ARB mutants remain exclusively associated with ARB, however, and our data show that these (i.e., M325T) induce the mislocalization of WT Best1 (Figs. 4 and 5). Furthermore, we have previously shown that the BVMD mutant W93C is mislocalized on its own, but is rescued to the plasma membrane in the presence of WT Best1 (Johnson et al., 2013). Despite the differences in trafficking, patients heterozygous or homozygous for the mutation W93C exhibit a comparable disease phenotype (Bakall et al., 2007). We would thus still argue that Best1 mislocalization is not by itself pathogenic. It remains possible, however, that these mutants may cause dominant disease is some cases, and that other factors may cause them to present as recessive disease in others. Further efforts are warranted to understand why some mutations are associated with multiple disease phenotypes.
Lastly, we did not look at specific patterns of localization – we only looked to see whether or not a mutant was localized to the basolateral plasma membrane or was mislocalized to intracellular compartments. It was previously reported by Davidson et al. that some ARB mutants are targeted to the proteasome while others are not (Davidson et al., 2011). Similarly, we observed that, although most mislocalized mutants exhibited similar patterns of localization, others were distinct. For example, in the X-Z scans, the mutant R200X was found diffuse throughout the cytoplasm while the mutant T6P was more punctate and concentrated between the nuclei and the apical plasma membrane. It is possible that these mutants traffic to different compartments and also differ in their handling (e.g., degraded via lysosomes, proteasomes, or autophagosomes). Differences such as these could further distinguish individual mutations, and potentially differentiate mislocalization in different diseases. Moreover, we did not assess the functional consequences (e.g., cell loss) of mislocalized mutants in RPE cells. Although compartmental trafficking of mutants and the functional consequences of mislocalization are beyond the scope of this study, they should be the subject of future investigations.
In summary, we find that AVMD, ARB, ADVIRC, and RP are associated with disparate effects on Best1 localization, but not oligomerization. The etiology of these diseases remains poorly understood and further efforts to understand the unique pathogenesis of each one are warranted. We postulate that unique changes in Ca2+ homeostasis, phagocytosis, binding partners (e.g., voltage-dependent Ca2+ channels (Milenkovic et al., 2011a; Reichhart et al., 2010; Yu et al., 2008), regulatory kinases (Marmorstein et al., 2002; Xiao et al., 2009), etc.), and/or other factors may underlie the differential pathogenesis of one disease over another.
Supplementary Material
Highlights.
Mislocalization of Best1 is not pathogenic.
The absence of Best1 at the plasma membrane is well tolerated.
Oligomeric defects are not associated with the bestrophinopathies.
The first 174 amino acids of Best1 are sufficient for oligomerization to occur.
Amino acids 472-585 are not necessary for proper localization of Best1.
Acknowledgements
We thank Dr. Robert Tarran (University of North Carolina, Chapel Hill, NC, USA) for generously providing us with a positive FRET control, CFP fused to YFP, Dr. Jeremy Nathans (John Hopkins University, Baltimore, MD, USA) for the gift of Best1-6x-c-myc, and Dr. George Ojakian (SUNY Health Science Center at Brooklyn, Brooklyn, NY, USA) for supplying generous amounts of monoclonal antibody 3F4 which recognizes gp135.
Funding
This work was supported by grants from the NIH (EY13160 to ADM, EY014465 to LYM, and NIGMS 5T32 GM08400 to Graduate Training in Systems and Integrative Physiology at the University of Arizona, Tucson, AZ), the Macula Vision Research Foundation, and unrestricted grants from Research to Prevent Blindness to the Departments of Ophthalmology at the University of Arizona, Tucson, AZ, and the Mayo Clinic, Rochester, MN.
Abbreviations used
- Best1
Bestrophin-1
- BVMD
Best vitelliform macular dystrophy
- AVMD
Adult-onset vitelliform macular dystrophy
- ARB
Autosomal recessive bestrophinopathy
- ADVIRC
Autosomal dominant vitreoretinochoroidopathy
- RP
Retinitis pigmentosa
- RPE
Retinal pigment epithelium
- MOI
Multiplicity of infection
- HEK293
Human embryonic kidney
- Best2
Bestrophin-2
- MDCK
Madin-Darby canine kidney
- iPSC
Induced pluripotent stem cell
- YFP
Yellow fluorescent protein
- CFP
Cyan fluorescent protein
- FRET
Fluorescence resonance energy transfer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that they have no conflict of interest.
References
- Adomaviciene A, Smith KJ, Garnett H, Tammaro P. Putative pore-loops of TMEM16/anoctamin channels affect channel density in cell membranes. J Physiol. 2013;591:3487–3505. doi: 10.1113/jphysiol.2013.251660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakall B, Marmorstein LY, Hoppe G, Peachey NS, Wadelius C, Marmorstein AD. Expression and localization of bestrophin during normal mouse development. Invest Ophthalmol Vis Sci. 2003;44:3622–3628. doi: 10.1167/iovs.03-0030. [DOI] [PubMed] [Google Scholar]
- Bakall B, Radu RA, Stanton JB, Burke JM, McKay BS, Wadelius C, Mullins RF, Stone EM, Travis GH, Marmorstein AD. Enhanced accumulation of A2E in individuals homozygous or heterozygous for mutations in BEST1 (VMD2). Exp Eye Res. 2007;85:34–43. doi: 10.1016/j.exer.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Bitner H, Mizrahi-Meissonnier L, Griefner G, Erdinest I, Sharon D, Banin E. A homozygous frameshift mutation in BEST1 causes the classical form of Best disease in an autosomal recessive mode. Invest Ophthalmol Vis Sci. 2011;52:5332–5338. doi: 10.1167/iovs.11-7174. [DOI] [PubMed] [Google Scholar]
- Boon CJ, Klevering BJ, Leroy BP, Hoyng CB, Keunen JE, den Hollander AI. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog Retin Eye Res. 2009;28:187–205. doi: 10.1016/j.preteyeres.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Burgess R, MacLaren RE, Davidson AE, Urquhart JE, Holder GE, Robson AG, Moore AT, Keefe RO, Black GC, Manson FD. ADVIRC is caused by distinct mutations in BEST1 that alter pre-mRNA splicing. J Med Genet. 2009;46:620–625. doi: 10.1136/jmg.2008.059881. [DOI] [PubMed] [Google Scholar]
- Burgess R, Millar ID, Leroy BP, Urquhart JE, Fearon IM, De Baere E, Brown PD, Robson AG, Wright GA, Kestelyn P, Holder GE, Webster AR, Manson FD, Black GC. Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am J Hum Genet. 2008;82:19–31. doi: 10.1016/j.ajhg.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke OB, Gulbis JM. Oligomerization at the membrane: potassium channel structure and function. Adv Exp Med Biol. 2012;747:122–136. doi: 10.1007/978-1-4614-3229-6_8. [DOI] [PubMed] [Google Scholar]
- Cui CY, Childress V, Piao Y, Michel M, Johnson AA, Kunisada M, Ko MS, Kaestner KH, Marmorstein AD, Schlessinger D. Forkhead transcription factor FoxA1 regulates sweat secretion through Bestrophin 2 anion channel and Na-K-Cl cotransporter 1. Proc Natl Acad Sci U S A. 2012;109:1199–1203. doi: 10.1073/pnas.1117213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AE, Millar ID, Burgess-Mullan R, Maher GJ, Urquhart JE, Brown PD, Black GC, Manson FD. Functional characterization of bestrophin-1 missense mutations associated with autosomal recessive bestrophinopathy. Invest Ophthalmol Vis Sci. 2011;52:3730–3736. doi: 10.1167/iovs.10-6707. [DOI] [PubMed] [Google Scholar]
- Davidson AE, Millar ID, Urquhart JE, Burgess-Mullan R, Shweikh Y, Parry N, O'Sullivan J, Maher GJ, McKibbin M, Downes SM, Lotery AJ, Jacobson SG, Brown PD, Black GC, Manson FD. Missense mutations in a retinal pigment epithelium protein, bestrophin-1, cause retinitis pigmentosa. Am J Hum Genet. 2009;85:581–592. doi: 10.1016/j.ajhg.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumanov JA, Zeitz C, Dominguez Gimenez P, Audo I, Krishna A, Alfano G, Diaz ML, Moskova-Doumanova V, Lancelot ME, Sahel JA, Nandrot EF, Bhattacharya SS. Disease-causing mutations in BEST1 gene are associated with altered sorting of bestrophin-1 protein. Int J Mol Sci. 2013;14:15121–15140. doi: 10.3390/ijms140715121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez NM, Tamm ER, Straubeta O. Role of bestrophin-1 in store-operated calcium entry in retinal pigment epithelium. Pflugers Arch. 2013;465:481–495. doi: 10.1007/s00424-012-1181-0. [DOI] [PubMed] [Google Scholar]
- Gouras P, Braun K, Ivert L, Neuringer M, Mattison JA. Bestrophin detected in the basal membrane of the retinal epithelium and drusen of monkeys with drusenoid maculopathy. Graefes Arch Clin Exp Ophthalmol. 2009;247:1051–1056. doi: 10.1007/s00417-009-1091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps ML. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell HC, Qu Z, Yu K, Xiao Q, Chien LT. Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies. Physiol Rev. 2008;88:639–672. doi: 10.1152/physrev.00022.2007. [DOI] [PubMed] [Google Scholar]
- Jentsch TJ, Poet M, Fuhrmann JC, Zdebik AA. Physiological functions of CLC Cl- channels gleaned from human genetic disease and mouse models. Annu Rev Physiol. 2005;67:779–807. doi: 10.1146/annurev.physiol.67.032003.153245. [DOI] [PubMed] [Google Scholar]
- Johnson AA, Lee YS, Stanton JB, Yu K, Hartzell CH, Marmorstein LY, Marmorstein AD. Differential effects of Best disease causing missense mutations on bestrophin-1 trafficking. Hum Mol Genet. 2013;22:4688–4697. doi: 10.1093/hmg/ddt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer F, White K, Pauleikhoff D, Gehrig A, Passmore L, Rivera A, Rudolph G, Kellner U, Andrassi M, Lorenz B, Rohrschneider K, Blankenagel A, Jurklies B, Schilling H, Schutt F, Holz FG, Weber BH. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur J Hum Genet. 2000;8:286–292. doi: 10.1038/sj.ejhg.5200447. [DOI] [PubMed] [Google Scholar]
- Lee S, Yoon BE, Berglund K, Oh SJ, Park H, Shin HS, Augustine GJ, Lee CJ. Channel-mediated tonic GABA release from glia. Science. 2010;330:790–796. doi: 10.1126/science.1184334. [DOI] [PubMed] [Google Scholar]
- Low SH, Miura M, Roche PA, Valdez AC, Mostov KE, Weimbs T. Intracellular redirection of plasma membrane trafficking after loss of epithelial cell polarity. Mol Biol Cell. 2000;11:3045–3060. doi: 10.1091/mbc.11.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein AD, Cross HE, Peachey NS. Functional roles of bestrophins in ocular epithelia. Prog Retin Eye Res. 2009;28:206–226. doi: 10.1016/j.preteyeres.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein AD, Marmorstein LY, Rayborn M, Wang X, Hollyfield JG, Petrukhin K. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2000;97:12758–12763. doi: 10.1073/pnas.220402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein AD, Stanton JB, Yocom J, Bakall B, Schiavone MT, Wadelius C, Marmorstein LY, Peachey NS. A model of best vitelliform macular dystrophy in rats. Invest Ophthalmol Vis Sci. 2004;45:3733–3739. doi: 10.1167/iovs.04-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein LY, McLaughlin PJ, Stanton JB, Yan L, Crabb JW, Marmorstein AD. Bestrophin interacts physically and functionally with protein phosphatase 2A. J Biol Chem. 2002;277:30591–30597. doi: 10.1074/jbc.M204269200. [DOI] [PubMed] [Google Scholar]
- Marmorstein LY, Wu J, McLaughlin P, Yocom J, Karl MO, Neussert R, Wimmers S, Stanton JB, Gregg RG, Strauss O, Peachey NS, Marmorstein AD. The light peak of the electroretinogram is dependent on voltage-gated calcium channels and antagonized by bestrophin (best-1). J Gen Physiol. 2006;127:577–589. doi: 10.1085/jgp.200509473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt A, Stohr H, Passmore LA, Kramer F, Rivera A, Weber BH. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best's disease). Hum Mol Genet. 1998;7:1517–1525. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- Milenkovic VM, Krejcova S, Reichhart N, Wagner A, Strauss O. Interaction of bestrophin-1 and Ca2+ channel beta-subunits: identification of new binding domains on the bestrophin-1 C-terminus. PLoS One. 2011a;6:e19364. doi: 10.1371/journal.pone.0019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic VM, Rivera A, Horling F, Weber BH. Insertion and topology of normal and mutant bestrophin-1 in the endoplasmic reticulum membrane. J Biol Chem. 2007;282:1313–1321. doi: 10.1074/jbc.M607383200. [DOI] [PubMed] [Google Scholar]
- Milenkovic VM, Rohrl E, Weber BH, Strauss O. Disease-associated missense mutations in bestrophin-1 affect cellular trafficking and anion conductance. J Cell Sci. 2011b;124:2988–2996. doi: 10.1242/jcs.085878. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Oh KT, Heffron E, Hageman GS, Stone EM. Late development of vitelliform lesions and flecks in a patient with best disease: clinicopathologic correlation. Arch Ophthalmol. 2005;123:1588–1594. doi: 10.1001/archopht.123.11.1588. [DOI] [PubMed] [Google Scholar]
- Neussert R, Muller C, Milenkovic VM, Strauss O. The presence of bestrophin-1 modulates the Ca2+ recruitment from Ca2+ stores in the ER. Pflugers Arch. 2010;460:163–175. doi: 10.1007/s00424-010-0840-2. [DOI] [PubMed] [Google Scholar]
- Pedemonte N, Galietta LJ. Pharmacological Correctors of Mutant CFTR Mistrafficking. Front Pharmacol. 2012;3:175. doi: 10.3389/fphar.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrukhin K, Koisti MJ, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, Vujic M, Bergen AA, McGarty-Dugan V, Figueroa D, Austin CP, Metzker ML, Caskey CT, Wadelius C. Identification of the gene responsible for Best macular dystrophy. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- Qu Z, Chien LT, Cui Y, Hartzell HC. The anion-selective pore of the bestrophins, a family of chloride channels associated with retinal degeneration. J Neurosci. 2006;26:5411–5419. doi: 10.1523/JNEUROSCI.5500-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Hartzell C. Determinants of anion permeation in the second transmembrane domain of the mouse bestrophin-2 chloride channel. J Gen Physiol. 2004;124:371–382. doi: 10.1085/jgp.200409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Hartzell HC. Bestrophin Cl- channels are highly permeable to HCO3. Am J Physiol Cell Physiol. 2008;294:C1371–1377. doi: 10.1152/ajpcell.00398.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhart N, Milenkovic VM, Halsband CA, Cordeiro S, Strauss O. Effect of bestrophin-1 on L-type Ca2+ channel activity depends on the Ca2+ channel beta-subunit. Exp Eye Res. 2010;91:630–639. doi: 10.1016/j.exer.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Salas PJ. External and internal signals for epithelial cell surface polarization. Annu Rev Physiol. 1989;51:741–754. doi: 10.1146/annurev.ph.51.030189.003521. [DOI] [PubMed] [Google Scholar]
- Rosenthal R, Bakall B, Kinnick T, Peachey N, Wimmers S, Wadelius C, Marmorstein A, Strauss O. Expression of bestrophin-1, the product of the VMD2 gene, modulates voltage-dependent Ca2+ channels in retinal pigment epithelial cells. Faseb J. 2006;20:178–180. doi: 10.1096/fj.05-4495fje. [DOI] [PubMed] [Google Scholar]
- Schatz P, Bitner H, Sander B, Holfort S, Andreasson S, Larsen M, Sharon D. Evaluation of macular structure and function by OCT and electrophysiology in patients with vitelliform macular dystrophy due to mutations in BEST1. Invest Ophthalmol Vis Sci. 2010;51:4754–4765. doi: 10.1167/iovs.10-5152. [DOI] [PubMed] [Google Scholar]
- Sheridan JT, Worthington EN, Yu K, Gabriel SE, Hartzell HC, Tarran R. Characterization of the oligomeric structure of the Ca(2+)-activated Cl- channel Ano1/TMEM16A. J Biol Chem. 2011;286:1381–1388. doi: 10.1074/jbc.M110.174847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Shen W, Kuai D, Martin JM, Guo X, Smith MA, Perez ET, Phillips MJ, Simonett JM, Wallace KA, Verhoeven AD, Capowski EE, Zhang X, Yin Y, Halbach PJ, Fishman GA, Wright LS, Pattnaik BR, Gamm DM. iPS cell modeling of Best disease: insights into the pathophysiology of an inherited macular degeneration. Hum Mol Genet. 2013;22:593–607. doi: 10.1093/hmg/dds469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton JB, Goldberg AF, Hoppe G, Marmorstein LY, Marmorstein AD. Hydrodynamic properties of porcine bestrophin-1 in Triton X-100. Biochim Biophys Acta. 2006;1758:241–247. doi: 10.1016/j.bbamem.2006.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tsunenari T, Yau KW, Nathans J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc Natl Acad Sci U S A. 2002;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunenari T, Sun H, Williams J, Cahill H, Smallwood P, Yau KW, Nathans J. Structure-function analysis of the bestrophin family of anion channels. J Biol Chem. 2003;278:41114–41125. doi: 10.1074/jbc.M306150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beest M, Robben JH, Savelkoul PJ, Hendriks G, Devonald MA, Konings IB, Lagendijk AK, Karet F, Deen PM. Polarisation, key to good localisation. Biochim Biophys Acta. 2006;1758:1126–1133. doi: 10.1016/j.bbamem.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Wang AZ, Ojakian GK, Nelson WJ. Steps in the morphogenesis of a polarized epithelium. I. Uncoupling the roles of cell-cell and cell-substratum contact in establishing plasma membrane polarity in multicellular epithelial (MDCK) cysts. J Cell Sci. 1990a;95(Pt 1):137–151. doi: 10.1242/jcs.95.1.137. [DOI] [PubMed] [Google Scholar]
- Wang AZ, Ojakian GK, Nelson WJ. Steps in the morphogenesis of a polarized epithelium. II. Disassembly and assembly of plasma membrane domains during reversal of epithelial cell polarity in multicellular epithelial (MDCK) cysts. J Cell Sci. 1990b;95(Pt 1):153–165. doi: 10.1242/jcs.95.1.153. [DOI] [PubMed] [Google Scholar]
- Woo DH, Han KS, Shim JW, Yoon BE, Kim E, Bae JY, Oh SJ, Hwang EM, Marmorstein AD, Bae YC, Park JY, Lee CJ. TREK-1 and Best1 channels mediate fast and slow glutamate release in astrocytes upon GPCR activation. Cell. 2012;151:25–40. doi: 10.1016/j.cell.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Xiao Q, Hartzell HC, Yu K. Bestrophins and retinopathies. Pflugers Arch. 2010;460:559–569. doi: 10.1007/s00424-010-0821-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Yu K, Cui YY, Hartzell HC. Dysregulation of human bestrophin-1 by ceramide-induced dephosphorylation. J Physiol. 2009;587:4379–4391. doi: 10.1113/jphysiol.2009.176800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardley J, Leroy BP, Hart-Holden N, Lafaut BA, Loeys B, Messiaen LM, Perveen R, Reddy MA, Bhattacharya SS, Traboulsi E, Baralle D, De Laey JJ, Puech B, Kestelyn P, Moore AT, Manson FD, Black GC. Mutations of VMD2 splicing regulators cause nanophthalmos and autosomal dominant vitreoretinochoroidopathy (ADVIRC). Invest Ophthalmol Vis Sci. 2004;45:3683–3689. doi: 10.1167/iovs.04-0550. [DOI] [PubMed] [Google Scholar]
- Yu K, Cui Y, Hartzell HC. The bestrophin mutation A243V, linked to adult-onset vitelliform macular dystrophy, impairs its chloride channel function. Invest Ophthalmol Vis Sci. 2006;47:4956–4961. doi: 10.1167/iovs.06-0524. [DOI] [PubMed] [Google Scholar]
- Yu K, Lujan R, Marmorstein A, Gabriel S, Hartzell HC. Bestrophin-2 mediates bicarbonate transport by goblet cells in mouse colon. J Clin Invest. 2010;120:1722–1735. doi: 10.1172/JCI41129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Xiao Q, Cui G, Lee A, Hartzell HC. The best disease-linked Cl- channel hBest1 regulates Ca V 1 (L-type) Ca2+ channels via src-homology-binding domains. J Neurosci. 2008;28:5660–5670. doi: 10.1523/JNEUROSCI.0065-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Stanton JB, Wu J, Yu K, Hartzell HC, Peachey NS, Marmorstein LY, Marmorstein AD. Suppression of Ca2+ signaling in a mouse model of Best disease. Hum Mol Genet. 2010;19:1108–1118. doi: 10.1093/hmg/ddp583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.