Abstract
The poly(ADP-ribose) polymerase (PARP) protein family generates ADP-ribose (ADPr) modifications onto target proteins using NAD+ as substrate. Based on the composition of three NAD+ coordinating amino acids, the H-Y-E motif, each PARP is predicted to generate either poly(ADP-ribose) (PAR) or mono(ADP-ribose) (MAR). However, the reaction product of each PARP has not been clearly defined, and is an important priority since PAR and MAR function via distinct mechanisms. Here we show that the majority of PARPs generate MAR, not PAR, and demonstrate that the H-Y-E motif is not the sole indicator of PARP activity. We identify automodification sites on seven PARPs, and demonstrate that MAR and PAR generating PARPs modify similar amino acids, suggesting that the sequence and structural constraints limiting PARPs to MAR synthesis do not limit their ability to modify canonical amino acid targets. In addition, we identify cysteine as a novel amino acid target for ADP-ribosylation on PARPs.
Introduction
The primary function of the seventeen-member poly(ADP-ribose) polymerase (PARP) protein family is to generate ADP-ribose (ADPr) modifications onto target proteins using NAD+ as substrate1. The best understood PARP functions involve poly(ADP-ribose) (PAR) synthesis and include physiological functions in cell division2-6, transcriptional regulation (reviewed in 7) and regulation of protein degradation8,9. PAR also functions during cell stress responses such as DNA damage10, heat shock11,12, and the cytoplasmic stress response13. Recently it was shown that certain PARPs, such as PARP10 and PARP14, only generate mono(ADP-ribose) (MAR)14. However it is not clear if other PARP family members are also limited to MAR synthesis. Computational analysis of amino acid sequence in combination with this data led to the prediction that each PARP can generate either PAR or MAR, but not both14.
The functional distinction between PAR and MAR synthesis is important since the type of ADPr modification generated has critical impacts on the potential mechanisms of function. MAR modifications add single ADPr units on to proteins whereas PAR polymers can be up to 200 units in length when generated in vitro, and can contain both linear and branched glycosidic linkages15. Although both can regulate the function of target proteins via direct covalent modification, PAR can also recruit binding proteins that contain a 20-amino acid PAR binding motif16 as well as characterized PAR binding domains including Macro17, PBZ18 and WWE19 domains. This allows PAR to function as a reversible high-density protein binding scaffold for the nucleation of multiprotein complexes of great complexity. Therefore, identifying the type of ADPr modifications generated by each PARP is critical to understanding specific mechanisms of PARP function.
Although less is known about MAR, our recent family-wide analysis of PARP function and previous work by other labs has identified important functions for PARPs predicted to generate MAR. These include regulation of transcription (PARP3, 7 and 14)20-22, signal transduction pathways (PARP10 and 14)23-25, the unfolded protein response (PARP16)26, the actin cytoskeleton (PARP14)27, and membrane organelles (PARP8 and 16)27. The diversity of pathways that appear to be regulated by MAR demonstrates the general importance of MAR in cell function and suggests that a deeper understanding of the molecule is in order.
The primary predictors of PARP enzymatic activity are thought to be the amino acids that catalyze the ADP-ribose transfer reaction. The PARP catalytic domain contains a signature H-Y-E motif originally identified in various bacterial mono-ADP-ribosyltransferase (mART) toxins that also mediate ADP-ribose transfer. Histidine and tyrosine residues are required for binding of the substrate NAD+ and the glutamate for catalysis28,29. Since most PARPs contain an isoleucine, leucine or tyrosine in place of the catalytic glutamate they are predicted to generate MAR14 (Supplementary Table 1). Additionally, PARP9 and 13 also contain amino acid substitutions for the NAD+ binding histidine and are predicted to be inactive14.
Secondary structural features of the PARP catalytic domain are also predicted to influence catalytic activity. In addition to the NAD+ binding residues of the H-Y-E motif, the Donor loop (D-loop) shapes the substrate binding pocket and interacts with NAD+30. This loop varies in size and rigidity within the PARP family and analysis of the binding of small molecule PARP inhibitors to PARP catalytic domains identified the D-loop as a structural element that contributes to differential inhibitor binding30. Therefore, the shape of the substrate binding pocket, partly lined by the D-loop, could contribute to differences in NAD+ binding among the PARPs and impact catalytic activity or enzyme kinetics. Another structural component of the PARP catalytic domain is the acceptor pocket, partly lined by the loop between β sheets 4 and 5 and referred to as the acceptor loop. This loop is implicated in the binding of either protein substrate or ADPr acceptor for bacterial mARTs or eukaryotic PARPs respectively and varies in length among PARP proteins14,31-33. Therefore, the ability to bind to an incoming ADPr unit on a PAR polymer could vary based on the structure of the acceptor loop, impacting the ability to elongate a PAR chain or create a branched modification.
Whether or not these amino acid or structural constraints thought to limit PARP activity to MAR synthesis affect selectivity of amino acid targets is unknown. Most current data pertaining to amino acid selectivity is based on the PAR generating PARP1, however even here amino acid selectivity is unclear. Although most studies identify glutamate and aspartate residues as the primary targets of modification34-37, lysines have also been identified38,39.
Here we take a systematic approach to examine PARP enzymatic activity, assaying automodification of each member of the PARP family of proteins with high resolution. Our results suggest that the primary enzymatic activity for the PARPs is MAR synthesis and that the amino acid identity of the H-Y-E motif is not the sole indicator of PAR or MAR synthesis. They further demonstrate that MAR and PAR generating PARPs modify both acidic amino acids and lysines and identify ADPr modifications on cysteine residues of PARPs. In conjunction with our recent work identifying new PARP functions from a PARP family wide analysis, these results demonstrate the importance of MAR as a product of PARP activity as the majority of PARP phenotypes result from knockdown of MAR generating PARPs27.
Results
PARP Enzymatic Activity Assays
Recent work has provided experimental evidence demonstrating PAR or MAR activity for several of the PARPs (Supplementary Table 1). The most commonly used approach has been resolution of automodified PARPs on SDS-PAGE gels13,14,22,26,40-47. PARPs that appear as distinct bands are identified as MARylating, and those that resolve as smears due to heterogeneity of attached polymers are identified as PARylating (Supplementary Table 1). One problem with this approach is that it cannot distinguish between the addition of single ADPr units or addition of short oligomers of ADPr since each ADPr unit adds only 0.6 kDa to a protein and protein resolution on SDS-PAGE is both mass and charge dependent making resolution of small modifications unpredictable.
To provide a more accurate analysis of PARP reaction products we expressed and purified each full-length PARP as N-terminal GFP fusions in human 293F cells, then performed standard automodification assays48 previously shown to effectively measure enzymatic activity since the majority of PARP activity is self-directed49. Three approaches were used to analyze the reaction products: standard PAGE based automodification assays (Fig. 1), hydrolysis with enzymes specific for PAR or MAR and chemical treatments to release ADPr modifications at the site of protein linkage (Fig. 2, 3). The released products generated by approach 2 and 3 were then analyzed using high resolution TBE-acrylamide sequencing gels or thin layer chromatography (TLC) (Fig. 2, 3).
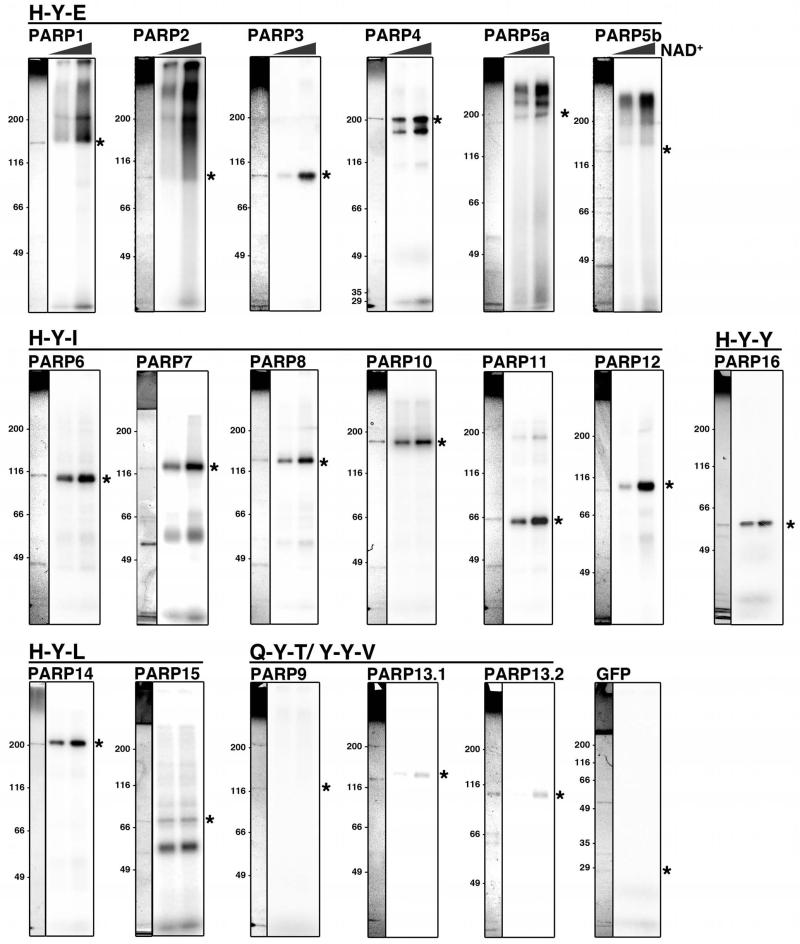
Figure 1. H-Y-E motif is not the sole indicator for PAR synthesis activity.
GFP-PARPs were immunoprecipitated from 293F cells and subjected to NAD+ incorporation reactions in vitro with 5 or 10μM cold NAD+ supplemented with a constant ratio of 32P-NAD+. Automodifed PARPs were resolved on SDS-PAGE gels and subjected to autoradiography. Representative coomassie stained gels for each PARP purification are shown to the left of the autoradiogram and the expected molecular weight of the PARP indicated by an asterisk. Assays were repeated at least twice. Of the H-Y-E motif containing PARPs, PARP1, 2, 5a and 5b generated polymer, as evidenced by the smear of signal starting from the molecular weight of the PARP. The remaining PARPs resolve as a discreet band, indicating that they do not generate poly(ADP-ribose). See also Supplementary Fig. 1-3.
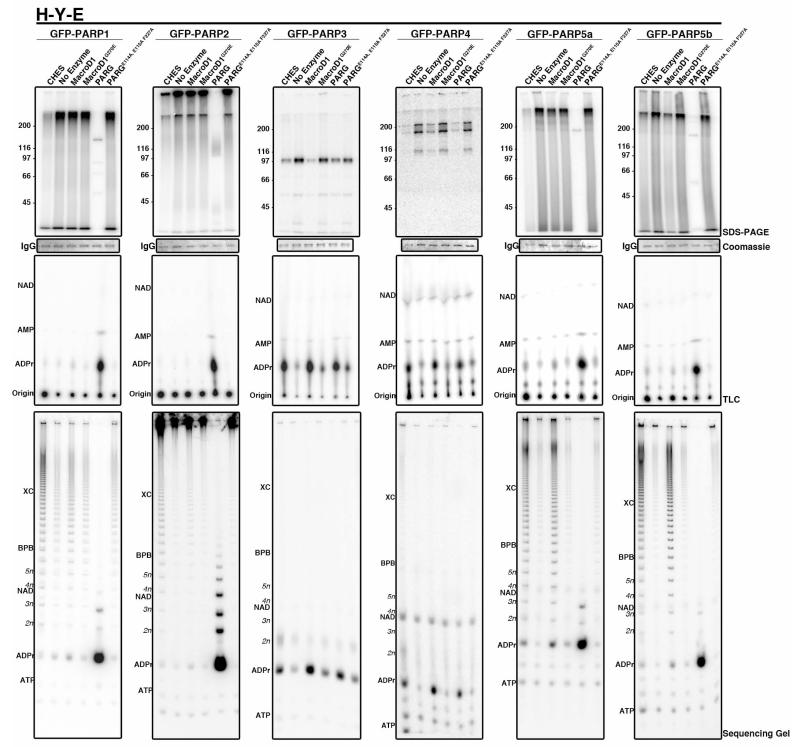
Figure 2. Enzymatic and chemical release of H-Y-E PARPs indicates that additional features of PARP catalytic domain impact to enzymatic activity.
Automodified PARPs were treated with CHES or the indicated wild type and catalytically inactive ADPr hydrolytic enzyme. Signal remaining attached to protein was analyzed by SDS-PAGE (top row), coomassies of either IgG (PARPs 1, 2, 5a and 5b) or PARP (PARP3 and 4) are shown below. Released product was analyzed by TLC (middle row) and sequencing gel (bottom row). Assays were repeated at least twice. PARPs 1, 2, 5a and 5b release PAR ladders upon CHES treatment and are sensitive to T.c. PARG hydrolysis. In contrast, PARP3 and 4 release ADPr upon CHES, MacroD1 and T.c. PARG treatment, indicative of MAR synthesis activity. See also Supplementary Fig. 4-7.
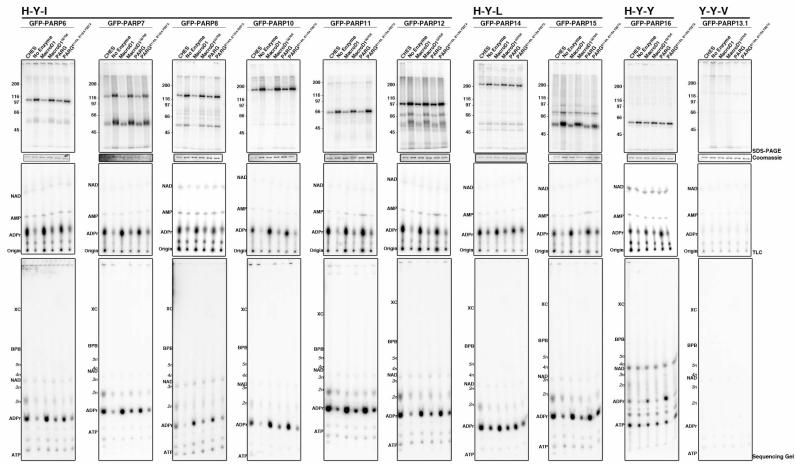
Figure 3. Enzymatic and chemical release shows that MAR synthesis is the primary activity of non-H-Y-E PARPs.
Automodified PARPs were treated with CHES or the indicated wild type and catalytically inactive ADPr hydrolytic enzyme. Signal remaining attached to protein was analyzed by SDS-PAGE (top row), coomassies for each PARP are shown below. Released product was analyzed by TLC (middle row) and sequencing gel (bottom row). Assays were repeated at least twice. CHES, MacroD1 and T.c. PARG treatment of non-H-Y-E PARPS results in ADPr release, indicative of MAR synthesis activity. See also Supplementary Fig. 4, 5 and 8.
Most PARPs do not synthesize PAR upon automodification
Incorporation reactions were performed on protein A magnetic beads using anti-GFP precipitation of the GFP-PARP fusions. Bead-bound GFP-PARPs were incubated with 5 or 10 μM NAD+ supplemented with a constant ratio of 32P-NAD+ (Fig. 1). Total NAD+ concentrations were kept low to increase the ratio of hot:cold NAD+ due to weak signal incorporation by many PARPs relative to PARP1 as identified during initial analysis. PARP1 exhibited robust polymer synthesis even at these low NAD+ concentrations, indicating that NAD+ is not limiting in our reactions (Fig. 1).
Several controls were performed to confirm that assay conditions did not affect PARP enzymatic activity. First we examined the effect of the N-terminal location of the GFP tag on PARP enzymatic activity by comparing the activity of both N and C-terminal GFP fusions to PARP1. Both incorporated similar amounts of 32P-ADPr, suggesting that the N-terminal location of the tag does not affect PARP enzymatic activity, similar to published results for PARP10 using the similarly sized N-terminal TAP tag (Supplementary Fig. 1a)14. We examined the possibility that the presence of a tag itself affects PARP enzymatic activity by comparing NAD+ incorporation of GFP-PARP10, SBP-PARP10 and untagged PARP10 (Supplementary Fig. 1b). These results show that the presence of the GFP fusion had no appreciable affect on PARP activity. Finally bead bound GFP-PARP10 and soluble PARP10 exhibited similar enzymatic activity suggesting that performing the analysis on beads does not affect PARP activity (Supplementary Fig. 1b).
Kleine et al. predicted PAR synthesis activity for H-Y-E containing PARPs. Consistent with this prediction and previously reported results, PARP1, 2, 5a and 5b incorporated ADPr in a manner consistent with PAR synthesis15,40,41,43 (Fig. 1). PARP1 and 2 automodification reactions contained additional signal that did not resolve in the SDS-PAGE gel and instead remained in the well, potentially due to branching, or very long polymer length (Fig. 1).
In contrast to predictions by Kleine et al., the H-Y-E containing PARP3 and 4 incorporated ADPr in a pattern indicative of MAR synthesis, consistent with previously published reports for full length PARP345 and bacterially expressed PARP4 catalytic domain42. PARP4 is a component of the cytoplasmic Vault complex, and the ADP-ribosylation activity of purified Vault complexes containing PARP4 suggest that it can generate PAR42. Thus PARP4 could exhibit PAR synthesis activity when bound to Vault proteins, or the PAR activity found in Vault complexes could be due to other co-purifying PARPs. Interestingly, the Coomassie stain for GFP-PARP4 shows a single band at ~200kD, however additional lower molecular weight signal was identified on the autoradiogram. This signal is likely due to a C-terminal cleavage product of GFP-PARP4 since the N-terminal GFP fusion is detectable at that molecular weight via anti-GFP blot, whereas an antibody raised against the very C-terminus of PARP4 does not detect the lower molecular weight band (Supplementary Fig. 2).
PARP6, 7, 8, 10, 11, 12, 14, 15 and 16 each incorporated ADPr in a manner consistent with MAR synthesis (Fig. 1). Identification of MAR activity for PARP7, 10, 12, 14, 15 and 16 is consistent with previously published reports13,14,22,26,47. Interestingly, PARP15 exhibited weak automodification activity, instead strongly modifying a co-precipitating protein. This result suggests that in contrast to other PARPs, PARP15 is not a major target of its own enzymatic activity.
PARP9 failed to incorporate detectable amounts of ADPr, suggesting that it is catalytically inactive, in agreement with previous reports (Fig. 1)44. Very weak NAD+ incorporation was observed for both isoforms of PARP13, predicted to be inactive due to the presence of a Q-Y-V catalytic motif for PARP13.1, and the lack of a catalytic domain for PARP13.214,50. Previous reports on the activity of bacterially purified PARP13 catalytic domain did not detect any NAD+ incorporation14 and our results are consistent with this (Supplementary Fig. 3). Instead the weak incorporation identified in PARP13 purified from 293F cells could be due to activity of a sub-stoichiometric co-purifying PARP.
The majority of PARPs are mono(ADP-ribosyl)ases
To further confirm ADP-ribosylation activity, in vitro automodified PARPs were treated with enzymes specific for PAR or MAR hydrolysis or chemicals known to release ADPr from proteins at the site of protein linkage. The signal that remained attached to the PARP was then examined by resolving on SDS-PAGE followed by autoradiography, and the released reaction products examined by thin layer chromatography (TLC) or high resolution TBE-polyacrylamide sequencing gels capable of resolving single units of ADPr (Fig. 2, 3). Each assay was performed a minimum of two times and the results were highly concordant.
PAR hydrolysis was analyzed using T. curvata poly(ADP-ribose) glycohydrolase (T.c. PARG) treatment, which hydrolyzes PAR chains, releasing ADPr as a product48. There have been conflicting reports on the ability of PARG to hydrolyze proximal ADPr-protein linkages or MAR14,48,51. Under our reaction conditions we did not observe substantial signal release from automodified PARP1E988Q, shown to generate only MAR modifications, suggesting that T.c. PARG does not hydrolyze MAR to a significant extent, although a few exceptions are described below (Supplementary Fig. 4a)29. MAR hydrolysis was analyzed using MacroD1 and terminal (ADP-ribose) glycohydrolase (TARG1)36,52,53. Both enzymes hydrolyze MAR, however whereas MacroD1 has no known activity on PAR51,53,54, TARG1 can release PAR chains through hydrolysis at the proximal ADPr-protein linkage, but cannot hydrolyze released polymers to ADPr36.
We first verified that the hydrolysis activity of T.c. PARG, MacroD1 and TARG is not affected by their substrate being bead bound by comparing hydrolysis of bead bound automodified GFP-PARP10 or SBP-PARP10 to soluble PARP10 (Supplementary Fig. 4b). There was no effect of bead binding on hydrolysis activity. To identify an appropriate enzyme concentration for the reactions, automodified GFP-PARP10 was treated with increasing amounts of TARG1, MacroD1 and T.c. PARG ranging from 50nM to 500nM (Supplementary Fig. 4c). We also directly compared TARG1 and MacroD1 hydrolysis activity on automodified PARP1, 7, 10 and 14 (Supplementary Fig. 4d). Hydrolysis activity of both enzymes was comparable and we chose MacroD1 for our subsequent analyses to minimize the possibility of reactivity to PAR (Supplementary Fig. 4d). For all analyses, treatment with the same concentrations catalytically inactive mutants of T.c. PARG, TARG and MacroD1 did not result in significant hydrolysis of ADPr modifications and were used as controls (Supplementary Fig. 4a-d).
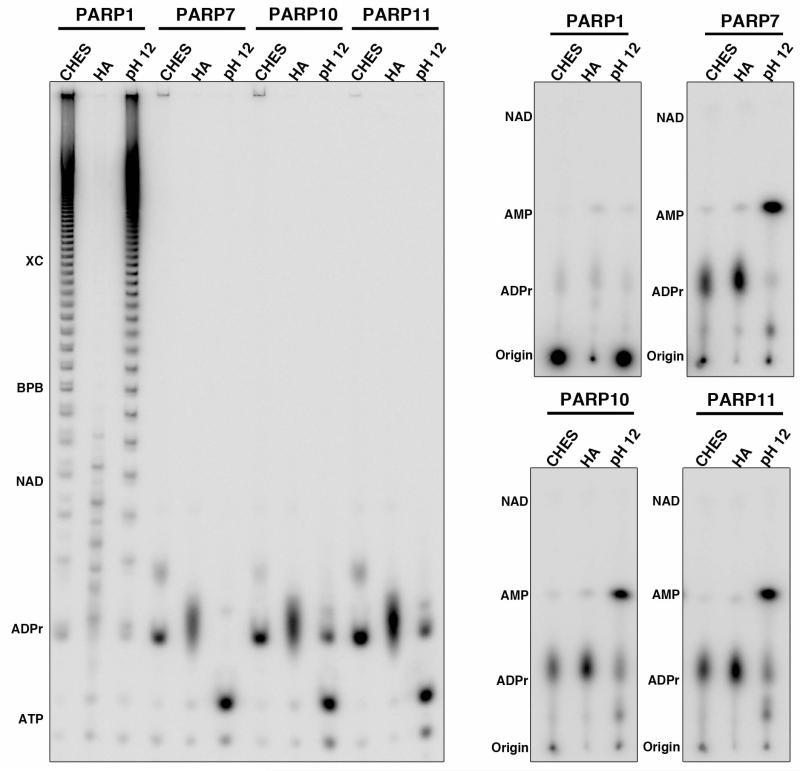
Several chemicals have been identified that specifically cleave ADPr-protein linkages, releasing intact ADPr modifications55. Three were tested: 2-(cyclohexylamino)ethanesulfonic acid (CHES), pH9, Hydroxylamine, pH 7.5 and sodium hydroxide, pH12 (Fig. 4). Although CHES and NaOH treatment showed similar amounts of PAR release from in vitro automodified PARP1, NaOH resulted in degradation of free ADPr to AMP, consistent with previous reports56. We were unable to obtain substantial release of intact PAR upon neutral hydroxylamine treatment of PARP1, previously shown to result in robust release of carboxylate ADPr linkages55 (Fig. 4). Therefore, CHES treatment was utilized for subsequent assays due to its ability to release ADPr from both acidic and lysine residues, allowing for the identification of ADPr modifications from both chemical linkages55.
Figure 4. Comparison of chemical treatments to release ADPr linkages.
Automodified PARP1, 7, 10 and 11 were treated with 100mM CHES, pH9, 2mM EDTA or 0.4M Hydroxylamine, pH 7.5, 2mM EDTA or 100mM Tris, pH12, 1mM EDTA. Released products were analyzed both on 20% TBE-polyacrylamide sequencing gels (left) and TLC (right). Hydroxylamine treatment does not release PAR ladders from PARP1 and high pH treatment results in the degradation of ADPr to AMP.
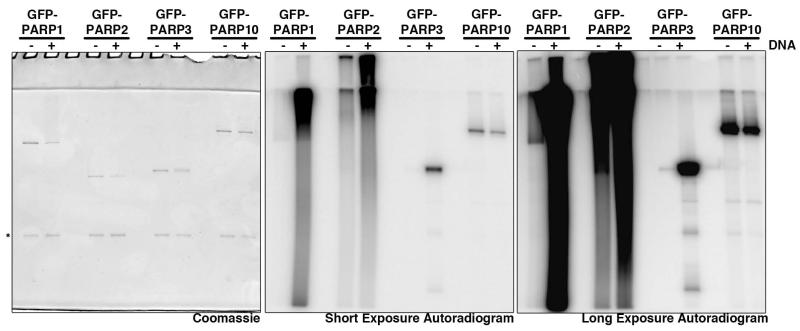
PARP1 and 2 contain DNA binding domains, and their enzymatic activity is highly upregulated upon addition of DNA41,57. PARP3 does not contain a defined DNA binding domain, however it was recently shown to bind and to be activated by DNA46,58. Our initial analyses of PARP3 activity identified low enzymatic activity in the absence of DNA to a point where the enzymatic activity of PARP3 was difficult to discern above background. Therefore, we compared the activity of GFP-PARP1-3 in the absence or presence of DNA (Fig. 5). Automodification activity of all three PARPs was highly increased in the presence of DNA, consistent with previous results. In contrast, addition of DNA to PARP10 automodification reactions resulted in a slight decrease in activity (Fig. 5). Addition of DNA to the PARP3 reactions did not change its reaction product-PARP3 still resolved as a distinct band consistent with MAR synthesis, indicating that ADPr synthesis activity is not altered upon upregulation of PARP enzymatic activity. Because addition of DNA to incorporation reactions increases activity without altering the pattern of NAD+ incorporation, PARPs1-3 were automodified in the presence of DNA for TLC and sequencing gel analysis.
Figure 5. DNA dependence of PARP enzymatic activity.
GFP-PARPs 1, 2, 3 and 10 were immunoprecipitated from 293F cells and subjected to NAD+ incorporation reactions in vitro with 10μM cold NAD+ supplemented with a constant ratio of 32P-NAD+, with or without the addition of activated DNA. Automodified PARPs were resolved on SDS-PAGE gels and subjected to coomassie staining and autoradiography. Addition of DNA to PARPs 1, 2, and 3 results in upregulation of enzymatic activity whereas PARP10 activity is slightly decreased.
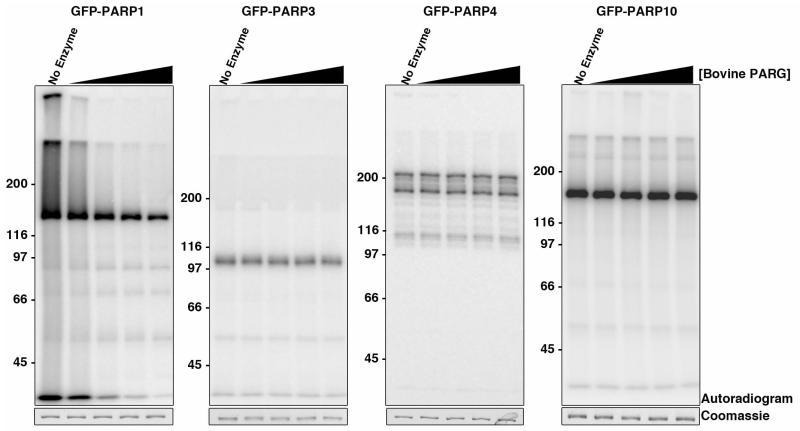
Results for H-Y-E PARPs are shown in Figure 2 and summarized in Table 1. CHES treatment of the H-Y-E PARPs 1, 2, 5a and 5b resulted in release of PAR, seen as ladders on high-resolution DNA sequencing gels (Fig. 2). T.c. PARG treatment hydrolyzed these PAR chains, causing the SDS-PAGE signal for each to collapse to distinct bands at their respective molecular weight, releasing ADPr as the major product as detected by TLC (Fig. 2; see Supplementary Fig. 4e for migration patterns of cold and hot standards on TLC). Sequencing gel analysis of the released T.c. PARG product showed that some short ADPr oligomers remained (Fig. 2), consistent with previous data demonstrating that T.c. PARG has lower activity on short PAR chains59. Finally, although MacroD1 did not cause a similar collapse of automodification signal to distinct band, treatment resulted in some release of PAR chains for PARPs 1, 2, 5a and 5b (Fig. 2). Treatment with MacroD1 and T.c. PARG catalytic dead mutants had no effect on the ADPr modifications. In contrast, CHES, MacroD1 and T.c. PARG treatment of H-Y-E PARPs 3 and 4 resulted in the release of ADPr seen on both TLC and sequencing gel (Fig. 2), similar to what was observed for H-Y-(I/L/Y) PARPs (below, Fig. 3). Since T.c PARG treatment resulted in minor ADPr release for PARP3 and 4, we confirmed that they contain attached MAR by treating with increasing amounts of bovine PARG using automodified PARP1 and PARP10 as controls (Fig. 6). Bovine PARG treatment of PARP3, 4 and 10 had no effect on the ADPr signal attached to the protein whereas a dose-dependent loss of ADPr signal was identified for PARP1. Together these results identify MAR synthesis activity for PARPs 3 and 4 (Fig. 6).
Table 1. Summary of PARP enzymatic activity.
| Activity | PARP | Other Names | Catalyt ic Motif |
|---|---|---|---|
| PAR | 1 | PARP ARTD1 |
H-Y-E |
| 2 | ARTD2 | H-Y-E | |
| 5a | TNKS1 ARTD5 |
H-Y-E | |
| 5b | TNKS2 ARTD6 |
H-Y-E | |
| MAR | 3 | ARTD3 | H-Y-E |
| 4 | vPARP ARTD4 |
H-Y-E | |
| 6 | ARTD17 | H-Y-I | |
| 7 | tiPARP ARTD14 |
H-Y-I | |
| 8 | ARTD16 | H-Y-I | |
| 10 | ARTD10 | H-Y-I | |
| 11 | ARTD11 | H-Y-I | |
| 12 | ARTD12 | H-Y-I | |
| 14 | BAL2 ARTD8 |
H-Y-L | |
| 15 | BAL3 ARTD7 |
H-Y-L | |
| 16 | ARTD15 | H-Y-Y | |
| Inactive | 9 | BAL1 ARTD9 |
Q-Y-T |
| 13 | ZC3HAV1 ARTD13 |
Y-Y-V |
Figure 6. Bovine PARG treatment of poly and mono(ADP-ribosyl)ating PARPs.
Automodified GFP-PARPs 1, 3, 4 and 10 were treated with a titration of bovine PARG ranging from 1-10ng/mL. GFP-PARP1, which produces PAR, is sensitive to bovine PARG treatment whereas GFP-PARPs 3, 4 and 10, which produce MAR, do not show any decrease in signal upon bovine PARG treatment. Assay was repeated twice.
Results for non H-Y-E PARPs are shown in Figure 3 and summarized in Table 1. H-Y-(I/L/Y) PARPs exhibited release of ADPr as the major product of CHES and MacroD1 treatment (Fig. 3). Treatment with T.c PARG also resulted in release of ADPr for some of these PARPs (albeit to a lesser extent than MacroD1 treatment), suggesting that some of the mono(ADP-ribosyl)ated sites might be sterically accessible for cleavage with the bacterial enzyme. (Fig. 3 and Supplementary Fig. 5). The minor signal present at 2 units of ADPr for CHES treatment for many of the non-H-Y-E PARPs might be due to artifact as it was also identified for PARP1E988Q, although it is possible that this mutant could also synthesize short oligomers not previously detected (Supplementary Fig. 5). Additionally, increased background signal was observed for PARP4 and 16, possibly due to low incorporation activity under these conditions. Together these results suggest that the primary activity of the H-Y-(I/L/Y) PARPs is mono(ADP-ribosyl)ation.
The Donor and Acceptor loops impact enzymatic activity.
Our results identifying PARP3 and 4 as MARylating enzymes suggested that the simple presence of the H-Y-E motif is not sufficient to determine PAR synthesis- other structural elements must also be important. Since it has been postulated that the D-loop is an important structural component of the NAD+ binding pocket30, we examined the D-loops of the H-Y-E PARPs. In addition we examined their acceptor loops since they are thought to bind to elongating ADPr and protein targets and could therefore help determine the type of ADPr modification generated31.
It is possible that the lack of PAR activity for PARP3 is due to the structure of its D-loop since this loop is a major structural difference between the PARP3 and PARP1 and 2 catalytic domains. The PARP1 D-loop contains 3 proline residues to contribute to loop rigidity whereas PARP3 only has one (Supplementary Fig. 6). Additionally, PARP3 lacks a tyrosine residue present in PARP1 that interacts with substrate, potentially affecting its ability to bind NAD+ (Supplementary Fig. 6)60,61. Together this suggests that structural features of the D-loop can impact catalytic activity and that the presence of a H-Y-E is not the sole indicator of PAR activity. The acceptor site is partially lined by the loop between β sheets 4 and 5 of the PARP catalytic domain and is thought to be important for substrate recognition14,31,32. Differences in amino acids lining the acceptor pocket are suggested to effect the type of ADPr that can be generated61 and the acceptor loop length is specifically implicated in determining PARP catalytic activity14. Therefore we tested its impact on enzymatic activity or the type of modification that can be generated.
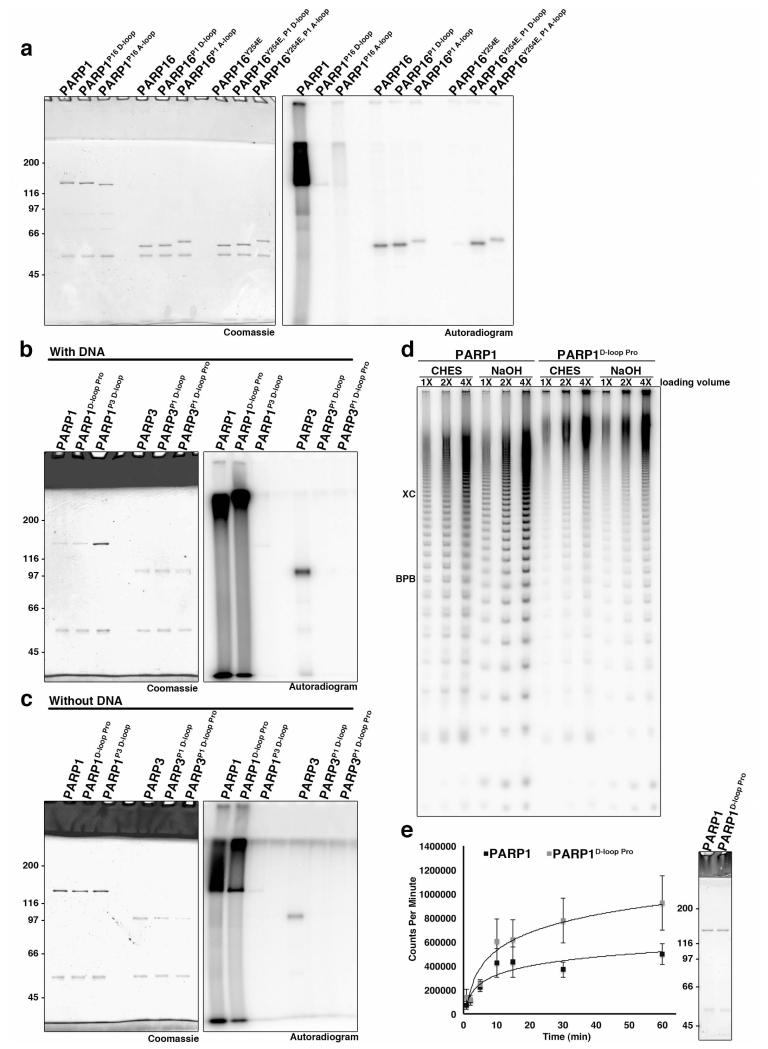
We tested the importance of the D-loop and acceptor loop on PARP1 enzymatic activity by assaying NAD+ incorporation activity of PARP1/PARP16 chimeras in which either the donor or acceptor loops were swapped (Fig. 7a). The PARP16 D-loop was specifically chosen since it lacks prolines and is presumed to be relatively flexible, is not homologous to PARP1-3 D-loop sequence, and comes from a PARP with low MAR generating activity (see discussion). PARP1P16 D-loop no longer generated PAR and instead exhibited MAR synthesis activity, similar to what was previously shown for PARP1 catalytic glutamate point mutants29, confirming the importance of the D-loop on enzymatic activity (Fig. 7a). In contrast PARP1P16 A-loop synthesized polymer, although incorporation activity was greatly reduced (Fig. 7a). This highlights the importance of the acceptor loop to PARP enzymatic activity and is consistent with a function for the acceptor pocket in binding of the terminal ADPr unit to be elongated by PARP1.
Figure 7. Impact of donor and acceptor loops on enzymatic activity.
a) Wild type and chimeric GFP-PARP1 and 16 constructs were expressed in 293F cells, immunoprecipitated and subject to in vitro NAD+ incorporation assays. Assays were repeated twice. Replacement of PARP1 donor loop with PARP16’s (PARP1P16 D-loop) inhibited PAR synthesis activity instead resulting in MAR synthesis. In contrast replacement with the acceptor loop of PARP16 (PARP1P16 A-loop) retained PAR synthesis, but decreased total PAR activity. PARP16 activity was unchanged after replacement with either the PARP1 donor or acceptor loop. In contrast, PARP16Y254E inhibits the endogenous MAR synthesis activity that is restored by replacement with either the donor or acceptor loops of PARP1. b, c) Wild type, PARP1D-loop Pro and chimeric GFP-PARP1 and 3 constructs were expressed in 293F cells, immunoprecipitated and subject to in vitro NAD+ incorporation assays either with (b) or without (c) DNA. Assays were repeated twice. Mutation of the 3 proline residues in the PARP1 D-loop results in an enrichment for PAR that resolves higher on the SDS-PAGE gel whereas replacement with the PARP-3 donor loop results in MAR synthesis activity in the absence and presence of DNA. The GFP-PARP3P1 D-loop construct is not active. d) Wild type GFP-PARP1 and GFP-PARP1D-loop Pro were automodified and attached PAR chains were released with either CHES or NaOH and analyzed by sequencing gel. Assay was repeated twice. GFP-PARP1D-loop Pro is enriched for longer polymer. e) NAD+ incorporation kinetics of wild type GFP-PARP1 and GFP-PARP1D-loop Pro indicate that, although both proteins have similar initial reaction rates, the PARP1D-loop Pro incorporation reaction does not plateau with similar kinetics, n≥3, error bars represent standard deviation, data fit to logarithmic regression. Representative coomassie stain of input shown to right. See also Supplementary Figures 6 and 7.
Replacement of the PARP16 donor or acceptor loops with those from PARP1 did not change the MAR activity of PARP16, although PARP16P1 A-loop exhibited decreased enzymatic activity (Fig. 7a). Replacement of the tyrosine from the catalytic residues with glutamate completely abolished activity, similar to what was previously shown for PARP1014. Interestingly, substituting either the donor or acceptor loop of PARP1 in PARP16Y254E restores MAR synthesis activity, indicating the primary sequence of the catalytic residues are not the sole determinant of enzymatic activity (Fig. 7a).
Because the D-loop appeared to have the greatest impact on enzymatic activity, altering the nature of the modification generated by PARP1, we further investigated its contribution to ADPr synthesis. To do so, we generated PARP1/PARP3 chimeras in which the donor loops were swapped, and a PARP1 mutant in which the 3 proline residues of the D-loop (P881, P882, P885) were mutated to alanines (PARP1D-loop Pro) (Fig. 7b, c). We first examined enzymatic activity using fixed concentrations of 32PNAD+ (10 μM) in the presence and absence of DNA. PARP1D-loop Pro still had PAR synthesis activity in both conditions, although the pattern of incorporation differed from wild type PARP1. Higher molecular weight product appeared to dominate the reaction product in PARP1D-loop Pro reactions regardless of the reaction conditions (Fig. 7b, c and Supplementary Fig. 7a, see below). In contrast, the PARP1P3 D-loop no longer generated PAR and instead exhibited MAR synthesis activity consistent with results from the PARP1P16 D-loop chimera (Fig. 7a-c). Because of the high level of PAR synthesis for wild type PARP1 and PARP1D-loop Pro in the presence of DNA, the coomassie signal appears weaker due to smearing of the signal (Fig. 7b). In contrast, since PARP1P3 D-loop resolves as a distinct band, the coomassie signal is much stronger (Fig. 7b). Unlike the PARP16 chimera, the PARP3P1 D-loop was inactive. These results suggest that simple replacement with a PAR competent D-loop is not sufficient for PAR synthesis activity (Fig. 7a-c).
To determine if a similar pattern of PAR synthesis occurs for PARP1D-loop Pro at physiological NAD+ levels, we assayed product formation at increasing concentrations of NAD+ substrate that include higher, more physiological concentrations of NAD+ (Supplementary Fig. 7b). This analysis was done in the absence of DNA since our initial results indicated that the lower activity of the both PARP1 and PARP1D-loop Pro highlighted the difference in the pattern of incorporation. At all NAD+ concentrations PARP1D-loop Pro reactions were enriched for PAR resolving at higher MW relative to wild type, confirming that this enrichment is due to the inherent enzymatic activity of the PARP1D-loop Pro (Supplementary Fig. 7b).
Two possibilities could result in the higher MW PAR found in the PARP1D-loop Pro: an increase in branching, which has been suggested to contribute to high molecular weight PAR41, and the synthesis of longer polymer. To test for differences in the levels of branching, we treated automodified wild type and PARP1D-loop Pro with phosphodiesterase I and analyzed the released products via 2D-TLC (Supplementary Fig. 7c). The (PR)2AMP to PRAMP signal ratio found in wild type and PARP1D-loop Pro were similar, suggesting that there was no difference in branching between the two products.
To test for differences in polymer length, PAR synthesized by wild type PARP1 and PARP1D-loop Pro was analyzed by resolving CHES and NaOH released product on TBE sequencing gels (Fig. 7d). CHES release was incomplete, with the unreleased product for the PARP1D-loop Pro samples highly enriched for high molecular weight PAR (Supplementary Fig. 7d). PAR released from PARP1D-loop Pro reactions exhibited a higher molecular weight distribution relative to wild type for both CHES and NaOH as analyzed by TBE gel analysis suggesting that they contained longer polymers (Fig. 7d). To better understand the reaction product generated by PARP1 and PARP1D-loop Pro, kinetic analysis was performed (Fig. 7e). Wild type PARP1 and PARP1D-loop Pro incorporated NAD+ with similar kinetics at early time points (1-5 minutes), however at later time points (including 30 min. when samples shown on Fig. 7d were obtained) when the incorporation kinetics of PARP1 plateaued, PARP1D-loop Pro incorporation continued to increase, consistent with the hypothesis that the higher molecular weight product results from generation of longer PAR polymers (Fig. 7e).
Amino acid specificity of PARP ADP-ribosylation reactions
PARP1 has been shown to modify acidic residues and lysines34-39. Because differences in the structures of the acceptor pockets or other structural or enzymatic differences that exist between MAR versus PAR generating PARPs could impact which amino acids are targeted for modification, we determined if MAR generating PARPs exhibit similar a.a. target selectivity. To do so we examined PARP3, 6, 9, 10, 11, 12, and 16 automodification reactions using mass spectrometry (Table 2 and Supplementary Table 2). PARP3 was specifically examined since it contains H-Y-E but only makes MAR. Our data identify glutamate and aspartate residues as the most commonly targeted amino acids for modification, especially for PARP3. Many preparations also contained modifications on lysine (Table 2) consistent with previously published results for PARP1, although we cannot rule out the possibility that non-enzymatic chemical ligation is responsible for modifications at these sites, a known phenomenon62.
Table 2. ADP-ribosylation sites identified by mass spectrometry analysis of automodified PARPs.
For the peptides highlighted in grey the MS/MS spectra do not contain enough information to distinguish between ADP-ribosylation on the internal residue from the modification on the peptide N-terminus
| Protein | Amino acid | Position | Peptide sequence |
|---|---|---|---|
| PARP3 (540 aa) | Glu | 19 | _PKPWVQTE(ad)GPEK_ |
| Glu | 22 | _PKPWVQTEGPE(ad)K_ | |
| Glu | 33 | _QAGREE(ad)DPFR_ | |
| Glu | 41 | _STAE(ad)ALK_ | |
| Glu | 170 | _YTLIEVQAEDEAQE(ad)AVVK_ | |
| Glu | 238 | _GFE(ad)ALEALEEALK_ | |
| Glu | 316 | _TVE(ad)EVPHPLDR_ | |
| Glu | 317 | _TVEE(ad)VPHPLDR_ | |
| Glu | 351 | _VIQTYLE(ad)QTGSNHR_ | |
| Glu | 456 | _E(ad)HHINTDNPSLK_ | |
| Asp | 148 | _D(ad)HFVSHPGK_ | |
| Asp | 217 | _NTMALMD(ad)LDVK_ | |
| Lys | 13 | _PK(ad)PWVQTEGPEK_ | |
| Lys | 44 | _STAEALK(ad)AIPAEK_ | |
| PARP6 (630 aa) | Asp | 600 | _FFFVYED(ad)GQVGDANINTQDPK_ |
| Cys | 237 | _VEVFGYPPSPQAGLLC(ad)PQHVGLPPPAR_ | |
| PARP8 (854 aa) | Cys | 332 | _TDDVC(ad)VTK_ |
| Cys | 367 | _LLNRPC(ad)PAAVK_ | |
| Cys | 376 | _SEEC(ad)LTLK_ | |
| Cys | 395 | _C(ad)EHNTNLKPHK_ | |
| PARP10 (1025 aa) | Glu | 106 | _LE(ad)QHVQALLR_ |
| Lys | 140 | _ALVQLPK(ad)PLSEADVR_ | |
| Lys | 916 | _NATVYGK(ad)_ | |
| PARP11 (331 aa) | Glu | 6 | _AE(ad)ELFSK_ |
| Asp | 80 | _ID(ad)FAEMK_ | |
| Cys | 49 | _WHMFQPDTNSQC(ad)SVSSEDIEK_ | |
| Cys | 65 | _TNPC(ad)GSISFTTSK_ | |
| Lys | 11 | _AEELFSK(ad)_ | |
| PARP12 (701 aa) | Asp | 600 | _D(ad)AAYSHHYSK_ |
| Asp | 611 | _SD(ad)TQTHTMFLAR_ | |
| Cys | 474 | _YVSPQDVTTMQTC(ad)NTK_ | |
| Cys | 584 | _VC(ad)GVHGTSYGK_ | |
| PARP16 (322 aa) | Glu | 77 | _E(ad)LLQSSGDNHKR_ |
| Asp | 37 | _D(ad)SVLRPFPASYAR_ | |
| Lys | 110 | _IQK(ad)LTGAPHTPVPAPDFLFEIEYFDPANAK | |
| Lys | 137 | _LTGAPHTPVPAPDFLFEIEYFDPANAK(ad)_ |
Surprisingly, we also identified ADP-ribosylated cysteines in PARP 6, 8, 11 and 12 samples. Interestingly, although multiple clustered ADP-ribosylated cysteine residues were identified in PARP8, no modifications of acidic residues or lysines were detected, although this does not imply that they are not present. Furthermore, these modifications occurred in the PARP6/8-specific cysteine rich domain that precedes the catalytic domain. Altogether, these observations may suggest that cysteine could be modified by some of the MAR generating PARPs. However, as cysteine modifications were identified prior to addition of exogenous NAD+, it is also possible that MAR-cysteine modification is due to the activity of other enzymes present in the human cell extracts (Supplementary Table 2).
To determine if the cysteine modifications were dependent on PARP activity, all four cysteines identified as ADP-ribosylated in PARP8 were mutated to alanines and automodification reactions were performed for wild type and cysteine mutant GFP-PARP8 (Supplementary Fig. 8a). A ~10-20% decrease in the amount of NAD+ incorporated was identified for the cysteine mutant depending on the concentration of NAD+ tested, suggesting that they are genuine sites of PARP8 automodification (Supplementary Fig. 8a). Additional unidentified modification sites are present in PARP8 as NAD+ incorporation signal remained in the mutant (Supplementary Fig. 8a). NAD+ incorporation was not observed using a catalytic dead point mutant of PARP8 (PARP8H697A), suggesting that the cysteine modifications are dependent on catalytic activity and are not due to non-enzymatic chemical ligation (Supplementary Fig. 8b).
A large number of modification sites, mainly in PARP10 samples, occurred on the N-terminus of peptides suggesting that they could be experimental artifact (Supplementary Table 3). Although all the ADP-ribosylation peptides reported here have been identified very confidently, we are less confident about the precise localization of ADPr modifications at the sites reported in grey in Table 2 (the peptide N-terminus of these peptides might be modified instead of the side chain of the internal residue) and they are therefore shown provisionally. In addition to cysteine, we detected ADP-ribosylation of three arginine residues on PARP10 (Supplementary Table 3). As all three ADP-ribosylated arginines are the first residues of the peptide, the modification could occur either on the guanidine group (arginine side chain) or on the amino group (peptide N-terminus).
No sites were identified for PARP9 protein consistent with the known lack of catalytic activity of this protein.
Discussion
To systematically identify the enzymatic activity of each PARP protein, we performed multiple assays to accurately identify the reaction products for the entire family. Our data indicate that the major activity for most of the PARPs is mono(ADP-ribosyl)ation. We can now confidently ascribe MAR activity to the newly identified functions for PARPs in actin cytoskeletal regulation, membrane organelle regulation, signal transduction, the unfolded protein response, and the cytoplasmic stress response. This suggests that the mechanisms of PARP functions in these pathways are mediated by MAR modifications of target proteins, unless these PARPs function in an ADPr synthesis independent manner.
Although the use of recombinant protein is standard in the PARP field, it is important to note that the use of recombinant proteins expressed as GFP-fusion proteins has potential limitations compared to analysis of endogenous protein. For instance, although the GFP tag had no effect on the enzymatic activity of PARP1 or PARP10 (Supplementary Fig. 1), we cannot completely rule out the possibility that it does not affect the activity of other PARPs. However this systematic analysis would not have been possible without the use of recombinant protein as it enabled purification of large amounts of pure protein necessary for such analysis. Studies of MARylating PARPs have shown that trans substrates are also MAR-modified suggesting that automodification is a useful assay of activity26,47, however it is possible that some PARPs may have substrate specific enzymatic activity not detected using automodification. Identifying the protein targets of MAR activity and identifying mechanisms of MAR function will be critical.
The MAR activity exhibited by PARP3 and 4, coupled with the analysis of the PARP1/PARP3 and PARP1/PARP16 chimeras demonstrate the importance of the D-loop in the function of the catalytic domain. Its importance is further illustrated by the ability of the PARP1 D-loop chimera to restore PARP16Y254E synthesis activity, suggesting that D-loop function can overcome deficiencies of H-Y-E motif mutants. The proline residues of the PARP1 D-loop have been proposed to provide structural rigidity30 but the requirement for this rigidity in enzymatic activity had not been investigated. The mutation of the D-loop prolines to alanines resulted in an enzyme that generates longer polymer and has increased enzyme kinetics. This increase in polymer length produced by PARP1D-loop Pro could be due to several factors including decreased self-regulation- automodification normally downregulates PARP1 enzymatic activity and this mechanism of self-regulation could be deficient in the PARP1D-loop Pro mutant. Another possibility is an increase in the processivity of the enzyme since communication between the D-loop and the A-loop is possible. Regardless, our results suggest that the D-loop confers important regulatory information to the catalytic domain. Our results also suggest that the acceptor loop is important for the enzymatic activity of PARPs, but not the type of modification generated, since the PARP1P16 A-loop chimera continued to generate PAR, albeit at lower levels. This is consistent with a function for the A-loop in the binding to substrate, either elongating polymer, or protein target.
Consistent with results for PARP1, both acidic residues and lysines were identified as targets of MAR generating PARPs suggesting that, in general, MAR and PAR generating PARPs modify similar amino acids. These results also argue that the sequence and structural constraints limiting PARPs to MAR synthesis, including the H-Y-E containing PARP3, do not affect their ability to modify canonical amino acid targets. In contrast our identification of cysteine modifications suggest that MARylating PARPs may have the ability to modify a larger diversity of amino acids although it remains to be determined if PARylating PARPs can also generate cysteine modifications.
Why do cells need both types of ADPr modifications? MAR and PAR synthesis activities are both evolutionarily conserved, indicating that both have important functions in cellular physiology63. PAR functions during stress responses and physiological pathways that require the rapid assembly of multiprotein complexes, acting as a protein binding scaffold. The consequences of MAR modifications on target protein are less understood. Recent work showing that ADPr-binding Macro domain-containing proteins can specifically bind MARylated targets suggests that one function could be to regulate specific protein-protein interactions, similar to SH2 domains binding to phosphoproteins17,23,64. MAR modifications are also especially interesting because they could serve as primers for further elongation to PAR, with PARPs functioning cooperatively to synthesize polymer, allowing a cell to tightly regulate each step of PAR generation. This possibility is supported by in vitro data showing heterodimerization and activation of PARPs with distinct activities, by the fact that PARPs with each activity are localized in both the cytoplasm and nucleus and by the presence of multiple physiological protein complexes containing MAR and PAR generating PARPs13,27,45,58,65. A closer examination of MAR function, and a closer examination of the ability of MAR to initiate PAR polymerization will be important.
Methods
Cell culture and reagents
293F cells (from ATCC) were grown at 37°/5% CO2 in F17 media supplemented with 2% glutamax (Life Technologies). 32P-NAD+ was from Perkin Elmer. Bovine PARG and 10× Activated DNA was from Trevigen.
NAD+ Incorporation Reaction
GFP-PARPs or SBP-PARPs were expressed in 293F cells. 24-48hrs after transfection, cells were washed 3× in ice-cold PBS and lysed for 20min on ice in cell lysis buffer (CLB, 50mM HEPES, pH7.4, 150mM NaCl, 1mM MgCl2, 1mM EGTA, 1mM DTT, 1% Triton-X 100, 1 μg/mL leupeptin, aprotinin, pepstatin, PMSF). Lysates were subject to ultracentrifugation at 100K*g for 30min. Cleared lysates were incubated for 1hr at 4°C either with anti-GFP antibody (3E6, Life Technologies) pre-bound protein A magnetic beads (Millipore) or streptavidin sepharose (GE Healthcare Life Sciences). Beads were than washed 1×5min in CLB, followed by 3×10min washes in CLB containing 1M NaCl, and 1×5min wash in PARP reaction buffer (PRB; 50mM Tris, pH7.5, 50mM NaCl, 0.5mM DTT, 0.1% Triton-X 100, 1 μg/mL leupeptin, aprotinin, pepstatin). Soluble PARP10 was produced by cleaving the SBP tag from immunoprecipitated SBP-PARP10 at Prescission protease sequence using HRV3C (Sigma-Aldrich).
NAD+ incorporation reactions were performed in PRB containing 10μM NAD+ (unless otherwise indicated) supplemented with 32P-NAD at a 1:20 ratio for 30min at 25°C. For PARPs with low incorporation signals (PARP4, 5a and 16), NAD+ incorporation was performed using a 1:5 ratio for 1hr at 25°C for TLC and sequencing gel analysis of released products. For PARPs 1-3, activated DNA was added to NAD+ incorporation reaction for TLC and sequencing gel analysis of released products. Following NAD+ incorporation, beads were washed 6×5min in PRB supplemented with 1M NaCl, 100μM NAD+ and 10μM ATP and 2×5min in PRB containing 100μM NAD+ and 10μM ATP. Beads were then resuspended in Laemmli sample buffer, heated to 65°C for 10 minutes and run on 8% SDS-PAGE gels followed by autoradiography.
For enzymatic and chemical treatments, beads were treated with indicated enzyme or chemical following NAD+ incorporation and washes. MacroD1 was used at 0.5μM and T.c. PARG was used at 0.1μM for 1 hour at 25°C. CHES release was performed for 2 hours at 37°C. Beads were then resuspended in Laemmli sample buffer and reaction supernatants were collected and either spotted on PEI-cellulose thin layer chromatography plates (Macheray-Nagel) and resolved in 0.15M LiCl/0.15M formic acid or diluted 1:1 in PAR loading buffer (50% urea, 25mM NaCl, 2mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue) and resolved on 20% polyacrylamide-TBE sequencing gels.
For analysis of PARP1D-loop Pro, NAD+ titration was performed using a ratio of 0.75μCi hot NAD+:10μM cold NAD+. For kinetic analysis, reactions were stopped using 20% TCA at indicated time points, incubated on ice for a minimum of 30 minutes, and spun at 16.1K*g for 30 minutes at 4°C. TCA pellets were washed with 5% TCA 3 times and treated with 0.5mL 0.1N NaOH, 2% SDS at 37°C overnight. Resuspended pellets were transferred to scintillation vials containing 5mL of Emulsifier-Safe scintillation fluid (Perkin Elmer) and analyzed by liquid scintillation counting. High pH release of PAR chains was performed in 0.1N NaOH, 20mM EDTA at 60°C for 2 hours. Samples were neutralized with 0.1N HCl and analyzed by sequencing gels as described.
PARP Chimera Generation
Geneart strings (Life Technologies) or G-blocks (IDT) were generated in which either the donor and acceptor loops of PARP’s 1, 3 and 16 were switched. See Supplementary Methods for amino acid sequences that were switched. EcoRV (internal site)/SalI were used to clone PARP1 fragments into PARP1/eGFPC1, SbfI (internal site)/SalI were used to clone PARP3 fragments into PARP3/eGFPC1, and PstI (internal site)/SalI were used to clone PARP16 fragments into PARP16/eGFPC1. Constructs were expressed in 293F cells and used for NAD+ incorporation assays as described above.
Mass Spectrometry Analysis
Streptavidin binding peptide tagged-PARP6, 8, 9, 10, 11 and 12 fusions were expressed in 293F cells and immunoprecipitated as described above. Following 3×10 minute washes in 1M NaCl/CLB, streptavidin sepharose beads were washed 2×5 minutes in PRB and proteins were eluted in 4mM biotin/PRB for 1 hour at 25°C. PARP16 (1-273 AA) and PARP3 were purified with N-terminal His-tags in from E. coli as previously described66.
PARPs were incubated with 0.2mM NAD+ and ADP-ribosylated PARPs were digested with trypsin by using a modified FASP protocol67. The samples were solubilized in 8 M urea 100 mM Tris pH 8 and loaded on a centrifugal filter (Vivacon 500, 50 kDa MW cutoff, Sartorius Stedim Biotech). The low-molecular-weight components were removed by washing four times with 8 M urea. Urea was removed by washing four times with 100 mM ammonium bicarbonate and proteins were digested with trypsin (trypsin Gold, Promega, Madison, USA). Following overnight digestion, the resulting peptides were eluted from the filter by centrifugation and further recovered with two additional washes using 100 mM ammonium bicarbonate. The eluted peptides were dried in a vacuum centrifuge system (Vacufuge, Eppendorf, Hamburg, Germany) and resuspended in 40 μl 1% formic acid. The acidified peptide mixtures were analyzed by nanoflow LC-MS/MS using a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany). Following separation on a Thermo Scientific EASY-Spray PepMap C18 column (75 μm inner diameter × 50 cm, 2 μm particle size, 100 Å pore size) using an EASY-nLC 1000 UPLC system (Thermo Scientific, Odense, Denmark), peptides were injected into the mass spectrometer through a Thermo Scientific EASY-Spray ion source. Optimal loading amounts were experimentally determined by injecting 1 μl (1/40) of the sample using a 90-minutes gradient (linear gradient from 0 to 22 % acetonitrile for 60 minutes) and the samples were sequentially analyzed multiple times using 150-minutes gradients (linear gradient from 0 to 22 % acetonitrile for 120 minutes) and different mass spectrometric acquisition parameters (see below) to maximize the quality of MS/MS spectra. The Q Exactive was operated in the data dependent mode to automatically switch between MS and MS/MS acquisition with the following general parameters: survey full MS spectra acquired with a m/z range of 300-1800 Th, the resolution set to as value of 70 000 and AGC (automatic gain control) target value of 1 000 000 ions; dynamic exclusion of 40 seconds. In the “standard” acquisition method up to 10 most intense ions were fragmented by higher-energy collisional dissociation (HCD) with a maximum injection time of 60 ms, resolution of 17 500 and target value of 1 000 000. Additionally, the samples were analyzed by two highly sensitive acquisition methods both with a maximum injection time of 1 000 ms and resolution of 35 000. In the first highly sensitive method up to 5 most intense ions were fragmented and scanned with the target value of 500 000; for the second highly sensitive method the most intense precursor ion was sequenced with the target value of 1 000 000.
Raw data were combined and analyzed with Andromeda-based MaxQuant (version 1.3.0.5)68. The search was performed the against UniProtKB human proteome (canonical and isoform sequences; downloaded in April 2013;). Enzyme specificity was set to trypsin allowing for up to four missed cleavages. Methionine oxidation, protein N-acetylation and ADP-ribosylation (mass shift of 541.0611) on glutamate, aspartate, cysteine, arginine, lysine and on the peptide N-terminus were set as variable modifications. MaxQuant was set up to automatically search for the following diagnostic ions in MS/MS spectra matched to ADP-ribosylated peptides: adenine (mass 135.0545), adenosine –H2O (mass 249.0862), adenosine monophosphate (AMP; mass 347.0631), adenosine diphosphate (ADP)69. All reported fragmentation spectra were manually validated using stringent criteria (supplementary MS/MS spectra).
Raw data (mzML format) and manually annotated spectra have been submitted to Peptide Atlas (http://www.peptideatlas.org/), where they are available under the identifier PASS00485.
Supplementary Material
Acknowledgements
We thank Frank Solomon for his helpful comments and discussion about the manuscript. Particular thanks to Thomas Colby for the help with the analysis of mass spectrometry data. Funding for this project was provided while P.C. was a Rita Allen Foundation Scholar and a Kimmel Foundation for Cancer Research Scholar. This work was partially supported by Cancer Center Support (core; grant P30-CA14051) and RO1GM087465 from the National Institutes of Health to PC, and funds from Curt and Kathy Marble to P.C. Work in I.A. laboratory is funded by the Wellcome Trust and the European Research Council. IM acknowledges funding from the Wellcome Trust (Sir Henry Wellcome Fellowship, 088957/Z/09/Z) and DFG (Cologne Cluster of Excellence in Cellular Stress Responses in Aging-Associated Diseases).
Footnotes
Competing financial interests: The authors declare no competing financial interests.
Accession codes: Raw mass spectrometry data and manually annotated spectra have been deposited in Peptide Atlas (http://www.peptideatlas.org/) under the identifier PASS00485.
References
- 1.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. doi:10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 2.Ha GH, et al. Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation. Cell death and differentiation. 2012;19:321–332. doi: 10.1038/cdd.2011.101. doi:10.1038/cdd.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature cell biology. 2005;7:1133–1139. doi: 10.1038/ncb1322. doi:10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 4.Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Molecular biology of the cell. 2009;20:4575–4585. doi: 10.1091/mbc.E09-06-0477. doi:10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. doi:10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 6.Ozaki Y, et al. Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Molecular cell. 2012;47:694–706. doi: 10.1016/j.molcel.2012.06.033. doi:10.1016/j.molcel.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Current opinion in genetics and development. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. doi:10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang S-MA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. doi:10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nature cell biology. 2011;13:623–629. doi: 10.1038/ncb2222. doi:10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 10.Malanga M, Althaus F. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2005;83:354–364. doi: 10.1139/o05-038. doi:10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 11.Petesch S, Lis J. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Molecular cell. 2012;45:64–74. doi: 10.1016/j.molcel.2011.11.015. doi:10.1016/j.molcel.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Giammartino D, Shi Y, Manley J. PARP1 represses PAP and inhibits polyadenylation during heat shock. Molecular cell. 2013;49:7–17. doi: 10.1016/j.molcel.2012.11.005. doi:10.1016/j.molcel.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung A, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. doi:10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Molecular cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. doi:10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez-Gonzalez R, Jacobson M. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. doi:10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 16.Pleschke J, Kleczkowska H, Strohm M, Althaus F. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. The Journal of biological chemistry. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. doi:10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 17.Karras G, et al. The macro domain is an ADP-ribose binding module. The EMBO journal. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. doi:10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. doi:10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes and development. 2012;26:235–240. doi: 10.1101/gad.182618.111. doi:10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goenka S, Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4210–4215. doi: 10.1073/pnas.0506981103. doi:10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouleau M, et al. A key role for poly(ADP-ribose) polymerase 3 in ectodermal specification and neural crest development. PloS one. 2011;6:e15834. doi: 10.1371/journal.pone.0015834. doi:10.1371/journal.pone.0015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacPherson L, et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic acids research. 2013;41:1604–1621. doi: 10.1093/nar/gks1337. doi:10.1093/nar/gks1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verheugd P, et al. Regulation of NF-κ B signalling by the mono-ADP-ribosyltransferase ARTD10. Nature communications. 2013;4:1683. doi: 10.1038/ncomms2672. doi:10.1038/ncomms2672. [DOI] [PubMed] [Google Scholar]
- 24.Cho S, et al. PARP-14, a member of the B aggressive lymphoma family, transduces survival signals in primary B cells. Blood. 2009;113:2416–2425. doi: 10.1182/blood-2008-03-144121. doi:10.1182/blood-2008-03-144121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbarulo A, et al. Poly(ADP-ribose) polymerase family member 14 (PARP14) is a novel effector of the JNK2-dependent pro-survival signal in multiple myeloma. Oncogene. 2013;32:4231–4242. doi: 10.1038/onc.2012.448. doi:10.1038/onc.2012.448. [DOI] [PubMed] [Google Scholar]
- 26.Jwa M, Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1alpha-mediated unfolded protein response. Nature cell biology. 2012;14:1223–1230. doi: 10.1038/ncb2593. doi:10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nature communications. 2013;4:2240. doi: 10.1038/ncomms3240. doi:10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papini E, Schiavo G, Sandona D, Rappuoli R, Montecucco C. Histidine 21 is at the NAD+ binding site of diphtheria toxin. The Journal of biological chemistry. 1989;264:12385–12388. [PubMed] [Google Scholar]
- 29.Marsischky GT, Wilson BA, Collier RJ. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. The Journal of biological chemistry. 1995;270:3247–3254. doi: 10.1074/jbc.270.7.3247. [DOI] [PubMed] [Google Scholar]
- 30.Wahlberg E, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nature biotechnology. 2012;30:283–288. doi: 10.1038/nbt.2121. doi:10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 31.Ruf A, Rolli V, de Murcia G, Schulz G. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. Journal of molecular biology. 1998;278:57–65. doi: 10.1006/jmbi.1998.1673. doi:10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
- 32.Han S, Tainer JA. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. International journal of medical microbiology: IJMM. 2002;291:523–529. doi: 10.1078/1438-4221-00162. [DOI] [PubMed] [Google Scholar]
- 33.Otto H, et al. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. doi:10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao Z, Gao P, Liu H.-w. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. Journal of the American Chemical Society. 2009;131:14258–14260. doi: 10.1021/ja906135d. doi:10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 35.Chapman J, Gagné J-P, Poirier G, Goodlett D. Mapping PARP-1 Auto-ADP-ribosylation Sites by Liquid Chromatography-Tandem Mass Spectrometry. Journal of proteome research. 2013 doi: 10.1021/pr301219h. doi:10.1021/pr301219h. [DOI] [PubMed] [Google Scholar]
- 36.Sharifi R, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. The EMBO journal. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. doi:10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nature methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. doi:10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 38.Altmeyer M, Messner S, Hassa P, Fey M, Hottiger M. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic acids research. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. doi:10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messner S, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic acids research. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. doi:10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. doi:10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 41.Amé J, et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. The Journal of biological chemistry. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. doi:10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 42.Kickhoefer VA, et al. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. The Journal of cell biology. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cook B, Dynek J, Chang W, Shostak G, Smith S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Molecular and cellular biology. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. doi:10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguiar R, Takeyama K, He C, Kreinbrink K, Shipp M. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. The Journal of biological chemistry. 2005;280:33756–33765. doi: 10.1074/jbc.M505408200. doi:10.1074/jbc.M505408200. [DOI] [PubMed] [Google Scholar]
- 45.Loseva O, et al. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. The Journal of biological chemistry. 2010;285:8054–8060. doi: 10.1074/jbc.M109.077834. doi:10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rulten S, et al. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Molecular cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. doi:10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ß1. PloS one. 2012;7 doi: 10.1371/journal.pone.0037352. doi:10.1371/journal.pone.0037352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slade D, et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. doi:10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindahl T, Satoh M, Poirier G, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends in biochemical sciences. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. doi:10.1016/S0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 50.Kerns J, Emerman M, Malik H. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS genetics. 2008;4 doi: 10.1371/journal.pgen.0040021. doi:10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barkauskaite E, et al. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nature communications. 2013;4:2164. doi: 10.1038/ncomms3164. doi:10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jankevicius G, et al. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nature structural and molecular biology. 2013;20:508–514. doi: 10.1038/nsmb.2523. doi:10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenthal F, et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nature structural and molecular biology. 2013;20:502–507. doi: 10.1038/nsmb.2521. doi:10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 54.Chen D, et al. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. The Journal of biological chemistry. 2011;286:13261–13271. doi: 10.1074/jbc.M110.206771. doi:10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cervantes-Laurean D, Jacobson E, Jacobson M. Preparation of low molecular weight model conjugates for ADP-ribose linkages to protein. Methods in enzymology. 1997;280:275–287. doi: 10.1016/s0076-6879(97)80119-x. doi:10.1016/S0076-6879(97)80119-X. [DOI] [PubMed] [Google Scholar]
- 56.Bredehorst R, et al. Two different types of bonds linking single ADP-ribose residues covalently to proteins. Quantification in eukaryotic cells. Eur J Biochem. 1978;92:129–135. doi: 10.1111/j.1432-1033.1978.tb12730.x. doi:10.1111/j.1432-1033.1978.tb12730.x. [DOI] [PubMed] [Google Scholar]
- 57.Simonin F, et al. The carboxyl-terminal domain of human poly(ADP-ribose) polymerase. Overproduction in Escherichia coli, large scale purification, and characterization. The Journal of biological chemistry. 1993;268:13454–13461. [PubMed] [Google Scholar]
- 58.Augustin A, et al. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. Journal of cell science. 2003;116:1551–1562. doi: 10.1242/jcs.00341. doi:10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 59.Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose) The Journal of biological chemistry. 1986;261:14902–14911. [PubMed] [Google Scholar]
- 60.Lehtio L, et al. Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J Med Chem. 2009;52:3108–3111. doi: 10.1021/jm900052j. doi:10.1021/jm900052j. [DOI] [PubMed] [Google Scholar]
- 61.Steffen JD, Brody JR, Armen RS, Pascal JM. Structural Implications for Selective Targeting of PARPs. Front Oncol. 2013;3:301. doi: 10.3389/fonc.2013.00301. doi:10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobson E, Cervantes-Laurean D, Jacobson M. Glycation of proteins by ADP-ribose. Molecular and cellular biochemistry. 1994;138:207–212. doi: 10.1007/BF00928463. [DOI] [PubMed] [Google Scholar]
- 63.Citarelli M, Teotia S, Lamb R. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC evolutionary biology. 2010;10:308. doi: 10.1186/1471-2148-10-308. doi:10.1186/1471-2148-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forst A, et al. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure. 2013;21:462–475. doi: 10.1016/j.str.2012.12.019. doi:10.1016/j.str.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 65.Kanai M, et al. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Molecular and cellular biology. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. doi:10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunstan M, et al. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nature communications. 2012;3:878. doi: 10.1038/ncomms1889. doi:10.1038/ncomms1889. [DOI] [PubMed] [Google Scholar]
- 67.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. doi:10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 68.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. doi:10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 69.Matic I, Ahel I, Hay R. Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nature methods. 2012;9:771–772. doi: 10.1038/nmeth.2106. doi:10.1038/nmeth.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.