Abstract
Mechanical properties of the extracellular matrix (ECM) have profound effects on cellular functions. Here, we applied novel photosensitive poly-dimethylsiloxane (photoPDMS) chemistry to create photosensitive, biocompatible photoPDMS as a rigidity-tunable material for study of mechanoresponsive cellular behaviors. By modulating the PDMS crosslinker to monomer ratio, UV light exposure time, and post-exposure baking time, we achieved a broad range of bulk Young’s modulus for photoPDMS from 0.027 - 2.48 MPa. Biocompatibility of photoPDMS was assayed and no significant cytotoxic effect was detected as compared to conventional PDMS. We demonstrated that the bulk Young’s modulus of photoPDMS could impact cell morphology, adhesion formation, cytoskeletal structure, and cell proliferation. We further fabricated photoPDMS micropost arrays for multiscale study of mechanoresponsive cellular behaviors. Our results suggested that adherent cells could sense and respond to changes of substrate rigidity at a sub-focal adhesion resolution. Together, we demonstrated the potential of photoPDMS as a photosensitive and rigidity-tunable material for mechanobiology studies.
Keywords: Rigidity-tunable materials, photosensitive elastomer, biointerfaces, rigidity-sensing, mechanotransduction, microfabrication
INTRODUCTION
Recently, increasing lines of evidence have suggested that in addition to soluble factors in the local cellular microenvironment, mechanical properties of the extracellular matrix (ECM) (e.g., matrix rigidity) can also mediate cellular functions including proliferation, migration, and differentiation1, 2. Several cellular mechanosensory machineries including focal adhesions (FAs) and actin cytoskeletal structure have been implicated to sense and interpret ECM mechanical signals through a mechano-chemical conversion process known as mechanotransduction3, 4. Generating synthetic rigidity-tunable polymer systems is of great current interest to studies of cellular mechano-sensitive and -responsive behaviors5, 6.
In conventional mechanotransduction studies, natural ECM proteins or synthetic ECM analogs such as polyacrylamide or polyethylene glycol gels have been used extensively as rigidity-tunable materials7–9. Rigidities of these hydrogel materials are modulated by changing the ECM protein concentration or the cross-linker density. Photosensitive polymers have also been used in cellular mechanics studies. For example, Burton and Taylor modulated the bulk rigidity of a trimethyl-terminated phenylmethyl polysiloxane silicone film by exposing the film with high-energy UV irradiation for different durations10. Photosensitive hydrogel systems have also been recently developed for in situ spatiotemporal controls of rigidity as well as generating rigidity gradients11–13. Such dynamically controlled matrix rigidity or patterned rigidity or rigidity gradient has been proved powerful for studying mechanoresponsive cellular behaviors, such as mechanics-mediated directional cell migration and stem cell differentiation12, 14–18.
Poly-dimethylsiloxane (PDMS) is a well-established thermal-curable, gas permeable and biocompatible material19. It has been broadly used in mechanobiology studies owing to its capability to generate various microscale structures using microfabrication and soft lithography20. Thus, synthesizing PDMS-based materials with photosensitive properties (such as its bulk rigidity)is of particular interest for mechanobiology research.
Here, we modified a reported fabrication method to add benzophenone as a photoinhibitor in PDMS to generate photosensitive PDMS (photoPDMS)21, 22. The bulk Young’s modulus of photoPDMS could be conveniently controlled within a broad range by modulating the PDMS crosslinker to base monomer ratio, UV light exposure time, and post-exposure baking time. To illustrate general applications of photoPDMS for mechanobiology studies, we performed comparative studies to investigate the effect of the bulk Young’s modulus of photoPDMS on mechanoresponsive cellular behaviors, including cell morphology, FA formation, cytoskeletal structure, and proliferation. In addition, we fabricated photoPDMS micropost arrays to investigate the spatial resolution of cellular rigidity sensing23, 24. The photoPDMS micropost array provided an ideal platform to study rigidity sensing at the micrometer scale by varying the micropost array geometry and the sub-micrometer scale (or the sub-FA scale) by varying the bulk Young’s modulus of photoPDMS. Although cellular rigidity sensing has been suspected to involve dynamics and signaling of FAs, whether cells can sense matrix rigidity at the sub-FA scale, i.e. within individual FAs, remains undetermined. By taking advantage of the photoPDMS micropost array and its multiscale mechanical properties (i.e., structural rigidity at the micrometer scale and local material rigidity at the sub-micrometer scale), we demonstrated that adherent cells might sense and respond to local matrix rigidity at the sub-FA scale.
EXPERIMENTAL SECTION
Fabrication of flat photoPDMS substrates
Briefly, PDMS prepolymer with different weight ratios of PDMS curing agent and base monomer was prepared (Sylgard 184, Dow-Corning, Midland, MI). Benzophenone dissolved in xylene (with the w/w ratio of 3:5) was then added to PDMS prepolymer at a 1% wt. concentration to generate UV-sensitive photoPDMS prepolymer. photoPDMS prepolymer was degassed under vacuum for 30 min to remove trapped air before poured into a petri dish and exposed to UV light (~365 nm) for different durations using a portable UV lamp (Black-Ray UV lamp, Ted Pella Inc, Redding, CA). UV light intensity was about 15 mW/cm2. After exposure, photoPDMS was placed immediately in an oven and cured at 110°C for a designated period of time.
Mechanical characterization of photoPDMS
We performed standard tensile testing to quantify the Young’s modulus E of cured photoPDMS as a function of benzophenone concentration, PDMS curing agent to base monomer ratio, UV exposure time, and post-exposure baking time. Briefly, cured photoPDMS specimens were cut into 20 mm × 40 mm × 2 mm cuboids. Strain-stress curves of photoPDMS specimens were measured at room temperature by an Instron tensile test machine (Norwood, MA) controlled by a computer following the standard ASTM D638-ISO 527 protocol over a stain range from 0.1 to 10%. Before tensile testing, a tare load was applied to hold photoPDMS samples firmly in the Instron machine. An extensometer was used to determine precisely the sample displacement. The specimens were tested at a rate of 5 mm per min. The bulk Young’s modulus E was calculated by fitting linearly the stress-strain curves. For photoPDMS prepared under different conditions, three identical samples were prepared with each tested three times.
Fabrication of photoPDMS micropost arrays
photoPDMS micropost arrays were fabricated using silicon micropost array masters as previously reported25, 26. Briefly, silicon micropost array masters were first fabricated using photolithography and deep reactive ion etching (DRIE) and then silanized with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane (United Chemical Technologies, Bristol, PA) for 4 hr under vacuum to aid subsequent releases of negative PDMS templates from the silicon masters. To make a negative mold containing an array of holes, PDMS prepolymer (with a 1:10 w/w ratio of curing agent to base monomer) was poured over the silicon micropost array master, cured at 110°C for 20 min, peeled off, oxidized with an oxygen plasma for 1 min (~200 mTorr; Plasma Prep II, SPI Supplies, West Chester, PA), and silanized with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane vapor overnight under vacuum. To generate the final photoPDMS micropost array, photoPDMS prepolymer was first prepared, poured over the negative PDMS micropost array mold and degassed under vacuum for 10 min. A 25 cm × 25 cm cover glass, which served as the substrate for the photoPDMS micropost array, was then placed on top of the negative PDMS mold. After curing at 110°C for a designated period of time, the photoPDMS micropost array was peeled off from the negative PDMS mold to release the final photoPDMS micropost array. When peeling induced collapse of the photoPDMS micropost array, we regenerated the micropost array by sonication in 100% ethanol for 60 sec followed by dry-release with liquid CO2 using a critical point dryer (Samdri®-PVT-3D, Tousimis, Rockville, MD).
Cell culture and reagents
NIH/3T3 mouse embryonic fibroblasts (ATCC, Manassas, VA) were maintained in the growth medium consisting of high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% bovine serum (Atlanta Biological, Atlanta, GA), 100 μg mL−1 L-glutamine, 100 units mL−1 penicillin, and 100 μg mL−1 streptomycin. Culture media were refreshed every 3 days. Fresh 0.25% trypsin-EDTA in phosphate buffered saline (PBS) was used to re-suspend cells. Cells were seeded at a low density of 3,000 cells/cm2in growth media onto PDMS or photoPDMS surfaces.
Surface functionalization of photoPDMS surfaces and cell seeding
Fibronectin was used to functionalize surfaces of cured PDMS and photoPDMS substrates to promote cell adhesion. Cured PDMS and photoPDMS substrates were washed with 100% ethanol 3 times and then rinsed with distilled water before blown dried under nitrogen. PDMS and photoPDMS substrates were then treated with UV ozone (ozone cleaner; Jelight, Irvine, CA) for 7 min to activate their surfaces before soaked in fibronectin solutions (50 μg/mL; BD Biosciences, San Jose, CA) prepared using distilled water for at least 1 hr followed by rinsing three times with PBS.
To promote cell adhesion on the photoPDMS micropost array, microcontact printing was used as previously described25. Briefly, a flat PDMS stamp was fabricated and coated with a saturating concentration of fibronectin (50 μg/mL) in distilled water for 1 hr, washed in distilled water, and then blown dried under nitrogen. The fibronectin-coated PDMS stamp was placed in conformal contact with UV ozone-treated, surface-oxidized photoPDMS micropost array, to facilitate fibronectin transfer from the stamp to the photoPDMS micropost array. Following microcontact printing, protein adsorption to all photoPDMS surfaces not coated with fibronectin was prevented by incubating the photoPDMS micropost array in 0.2% Pluronics F127 NF (BASF, Ludwigshafen, Germany) in distilled water for 30 min at room temperature. Cells were then seeded at a density of 3,000 cells/cm2 in the growth medium onto the photoPDMS micropost array and then allowed to spread overnight before other assays.
Immunocytochemistry
For F-actin visualization, cells were fixed with 4% paraformaldehyde (Electron Microscopy Science, Hatfield, PA) in PBS. F-actin was detected with fluorophore-conjugated phalloidin (Invitrogen). For immunofluorescence staining of FAs, cells were first incubated in an ice-cold cytoskeleton buffer (50 mM NaCl, 150 mM sucrose, 3 mM MgCl2, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin, and 2 mM PMSF) for 1 min, followed by another incubation for 1 min in the cytoskeleton buffer supplemented with 0.5% Triton X-100 (Roche Applied Science, Indianapolis, IN). Detergent-extracted cells were fixed in 4% paraformaldehyde in PBS, washed with PBS, incubated with a primary antibody to vinculin (Sigma-Aldrich, St. Louis, MO), and detected with fluorophore–conjugated, isotype-specific, anti-IgG secondary antibodies (Invitrogen).
EdU cell proliferation assay
For the EdU cell proliferation assay, NIH/3T3 cells were first starved at confluence in the growth medium supplemented with 0.5% bovine serum for 48 hr to synchronize cell cycle before trypsinization. Synchronized cells were re-plated on the photoPDMS substrates at 3,000 cells/cm2 and recovered in the complete growth medium for 12 to 24 hr, and were then exposed to 4 μM 5-ethynyl-2′-deoxyuridine (EdU; Invitrogen) in the growth medium for 9 hr. Cells were then fixed with 3.7% formaldehyde in PBS (Electron Microscopy Science), permeabilized with 0.3% Triton X-100 in PBS, blocked with 10% goat serum, and stained with Alexa Fluor 488 conjugated azide targeting alkyne groups in the EdU incorporated in the newly synthesized DNA. Cells were co-stained with Hoechst 33342 (Invitrogen) to visualize cell nucleus. Cell proliferation rate was quantified as the percentage of EdU positive cells among the whole cell population.
Cell adhesion assay
For cell adhesion assay, NIH/3T3 cells were seeded on PDMS and photoPDMS substrates at a density of 4,500 cells/cm2. One, two, and four hours after cell seeding, the PDMS or photoPDMS substrates were transferred into a new six-well plate filled with PBS. After rinsing briefly with PBS, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) for 15 min. The number of attached cells on the PDMS and photoPDMS substrates was manually counted using epi-fluorescence microscopy (Carl Zeiss Axio Observer Z1; Carl Zeiss MicroImaging, Thornwood, NY). Cell adhesion rate was quantified as the ratio between the number of attached cells and the total number of cells seeded.
Quantitative analysis of cell spreading area and FAs
Cell spread area and FA formation were quantified as previously described25. In brief, immunofluorescence images of actin microfilaments and vinculin were obtained using epi-fluorescence microscopy (Carl Zeiss Axio Observer Z1; Carl Zeiss MicroImaging) equipped with a thermoelectrically-cooled monochrome CCD camera (AxioCam camera; Carl Zeiss MicroImaging) and a 40× objective (0.75 NA; EC Plan NEOFLUAR®; Carl Zeiss MicroImaging). Images were captured using the Axiovision Software (Carl Zeiss MicroImaging) and processed using custom-developed MATLAB programs (Mathworks, Natick, MA). To determine cell spread area, the Canny edge detection method was used to binarize actin fibers and vinculin-expressing FAs in the images, and then image dilation, erosion, and fill operations were used to fill in the gaps between white pixels in the images. The resultant white pixels were summed to quantify cell spread area. To quantify FA number and area, the grayscale vinculin image was thresholded to produce a black and white FA image from which the white pixels, representing FAs, were counted and summed.
Statistics
p-value was calculated using the student’s t-test function in Excel (Microsoft, Seattle, WA).
RESULTS AND DISCUSSIONS
Conventional PDMS consists of repeating –OSi(CH3)2– units. The PDMS base monomer is vinyl terminated, while the crosslinkers are methyl terminated and contain silicon hydride – OSiHCH3– units. During curing, PDMS base monomers cross-link via a reaction between the monomer vinyl groups and the silicon hydride groups of the crosslinkers to form Si–CH2–CH2–Si linkages27, 28.
For photoPDMS, benzophenone is added into PDMS to act as a photoinhibitor, which generates free radicals under UV light exposure. Benzophenone radicals can abstract a hydrogen atom from a suitable hydrogen donor and thus react with both the silicon hydride units in the PDMS crosslinkers and the vinyl groups in the PDMS monomers (Scheme S1). This reaction prevents crosslinking reactions between PDMS monomers and crosslinkers and thus decreases the degree of PDMS crosslinking during the post-exposure bake. A high percentage of benzophenone (5% wt concentration) has been added into PDMS to create a positive-acting photodefinable elastomer21, 22, 29. After UV exposure, unexposed PDMS gets cured and cross-linked during the post-exposure bake, while exposed PDMS remains uncrosslinked and can be washed away in toluene. Thus, benzophenone, which is commonly used as a photoinitiator in free radical polymerizations, acts as a photoinhibitor in photoPDMS.
In this work, we decreased the amount of benzophenone in photoPDMS to moderately reduce the extent of PDMS crosslinking so that the bulk modulus of photoPDMS could be tuned by UV light exposure. We performed a systematic study using standard tensile testing to characterize the bulk Young’s modulus of photoPDMS containing a low concentration benzophenone (1% wt) as a function of the PDMS crosslinker to base monomer ratio, UV light exposure time, and post-exposure baking time. photoPDMS samples were prepared using different UV light exposure time ranging from 0 - 3 min, different post-exposure baking time ranging from 20 min to 40 hr, and different PDMS crosslinker to monomer ratio (1:10 or 1:30 by wt). Benzophenone concentration in photoPDMS was fixed at 1% wt as described, and the UV light intensity was about 15 mW/cm2. All photoPDMS samples were baked in an oven at 110°C immediately after UV light exposure.
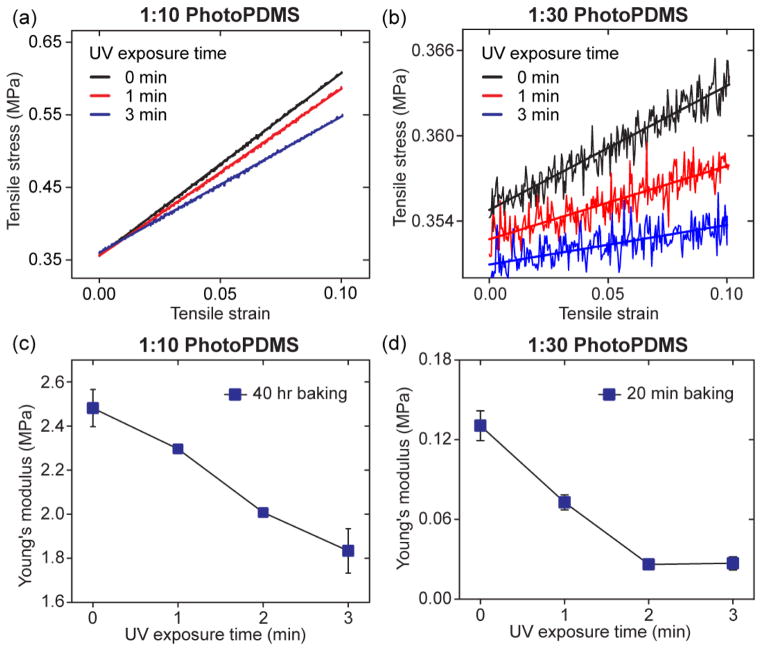
Figure 1a&b plot the strain-stress data measured for 1:10 (Fig. 1a) and 1:30 (Fig. 1b) photoPDMS treated with different periods of UV light exposure (0 - 3 min). 1:10 and 1:30 photoPDMS was post-exposure baked for 40 hr and 20 min, respectively. Slopes of the strain-stress data were quantified as the bulk Young’s modulus E of photoPDMS, with the results plotted in Fig. 1c&d. From Fig. 1c&d, a general trend was observed that the bulk Young’s modulus E of photoPDMS would decrease monotonically with increasing UV light exposure time, regardless of the PDMS crosslinker to base monomer ratio or post-exposure baking time. For example, for 1:10 photoPDMS, the bulk Young’s modulus E decreased about 27% from 2.48 MPa to 1.82 MPa when UV light exposure time increased from 0 min to 3 min. Similarly, for 1:30 photoPDMS, E decreased about 79% from 0.13 MPa to 0.027 MPa when UV light exposure time increased from 0 min to 3 min. Interestingly, the bulk Young’s modulus E of photoPDMS could be stable over a long term at room temperature, in distinct contrast to conventional PDMS. Figure S1 compared the Young’s modulus E of 1:30 photoPDMS measured either right after sample fabrication or after a 6-month storage at room temperature. 1:30 photoPDMS exposed to UV light for more than 2 min were stable without any significant change in E after a 6-month storage at room temperature, probably because benzophenone might have capped and thus protected reactive groups in uncured PDMS. Together, by modulating the PDMS crosslinker to base monomer ratio, UV light exposure time, and post-exposure baking time, we achieved a 100-fold range of bulk rigidity in photoPDMS from 0.027 - 2.48 MPa (Table 1), broader than is achievable with conventional PDMS30.
Figure 1.
Mechanical characterization of photoPDMS. (a&b) Strain-stress curves generated from tensile testing of 1:10 (a) and 1:30 (b) photoPDMS prepared after different UV light exposure time, as indicated. 1:10 and 1:30 photoPDMS was baked at 110°C for 40 hr and 20 min, respectively. Slopes of linear fittings of the strain-stress raw data were quantified as the bulk Young’s Modulus E of photoPDMS. (c&d) Young’s Modulus E of 1:10 (c) and 1:30 (d) photoPDMS as a function of UV light exposure time. 1:10 and 1:30 photoPDMS was baked at 110°C for 40 hr and 20 min, respectively.
Table 1.
Bulk Young’s modulus E of photoPDMS (kPa) as a function of the PDMS crosslinker to monomer ratio, UV light exposure time, and post-exposure baking time.
| UV exposure time (min) | 0 | 1 | 2 | 3 |
|---|---|---|---|---|
| 1:10 photoPDMS 40 hr baking | 2,480 | 2,300 | 2,010 | 1,830 |
| 1:10 photoPDMS 20 min baking | 1,610 | 1,150 | 720 | 630 |
| 1:30 photoPDMS 20 min baking | 130 | 73 | 26 | 27 |
We next evaluated biocompatibility and potential cytotoxic effect of photoPDMS. We compared proliferation rates of NIH/3T3 fibroblasts plated on conventional 1:10 and 1:30 PDMS and 1:10 and 1:30 photoPDMS substrates (without any UV light exposure). Both PDMS and photoPDMS substrates were baked for 40 hr and functionalized with fibronectin to promote cell adhesion. NIH/3T3 cells were seeded at a density of 3,000 cells/cm2. Cell densities after 24, 48, and 96 hr of culture were quantified (Fig. S2). No significant difference in cell density was observed between PDMS and photoPDMS substrates with comparable bulk Young’s modulus for all the three different time points (Fig. S2), suggesting a negligible effect of benzophenone in photoPDMS on cell viability and growth.
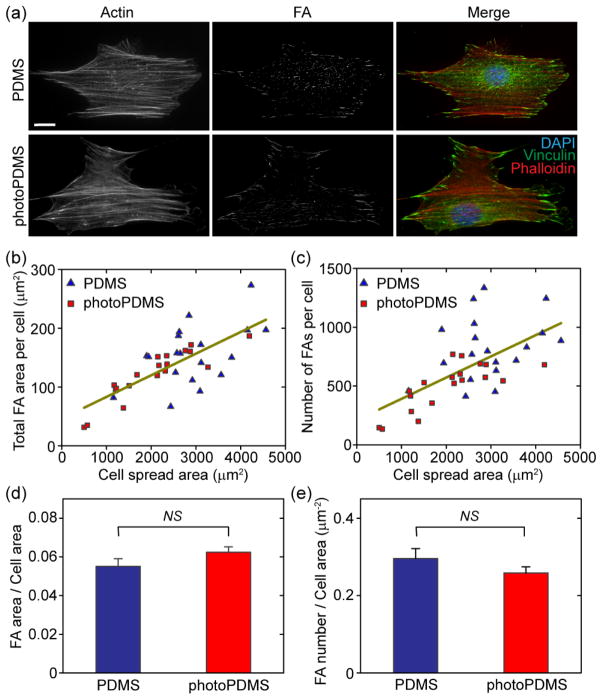
In addition to cell proliferation, we further examined FA formation and cytoskeletal structures for single NIH/3T3 cells plated on PDMS and photoPDMS substrates with comparable bulk Young’s modulus. No visible difference could be observed for actin cytoskeletal structures or FAs of NIH/3T3 cells plated on PDMS and photoPDMS substrates (Fig. 2a). Quantitative morphometric analysis of cell populations on both PDMS and photoPDMS substrates revealed similar correlations between FAs and cell spreading (Fig. 2b&c). We further quantified the total FA area per cell area and the number of FAs per cell area, and no significant difference was found for NIH/3T3 cells plated on PDMS and photoPDMS substrates. Together, our comparative studies in Fig. S2 and Fig. 2 suggested that photoPDMS did not induce detectable cytotoxic effect and benzophenone in photoPDMS did not significantly affect normal behaviors of NIH/3T3 cells.
Figure 2.
Evaluation of cytotoxic effect of photoPDMS on cell morphology and FA formation of NIH/3T3 fibroblasts. (a) Representative immunofluorescence images showing single NIH/3T3 fibroblasts plated on 1:10 PDMS (top) and 1:10 photoPDMS (bottom; UV light exposure time: 0 min) surfaces. Both PDMS and photoPDMS were baked at 110°C for 40 hr. Scale bar, 20 μm. Cells were co-stained for nuclei (DAPI; blue), actin (red), and vinculin (green). (b&c) Quantitative and correlative analysis of cell morphology and FA formation of NIH/3T3 fibroblasts. b & c show the total FA area per cell (b) and the number of FAs per cell (c) as a function of cell spread area. Each data point in b & c represents an individual cell plated on either PDMS or photoPDMS substrates as indicated. Data trends in b & c were plotted using linear least square fitting. Cell number n = 19, for both PDMS and photoPDMS substrates. (d&e) Bar plots showing the total FA area per cell area (d) and the FA number per cell area for single NIH/3T3 fibroblasts plated on the PDMS and photoPDMS substrates. Data represents the means ± standard error of mean (s.e.m) from 3 independent experiments. NS, statistically not significant (p > 0.05).
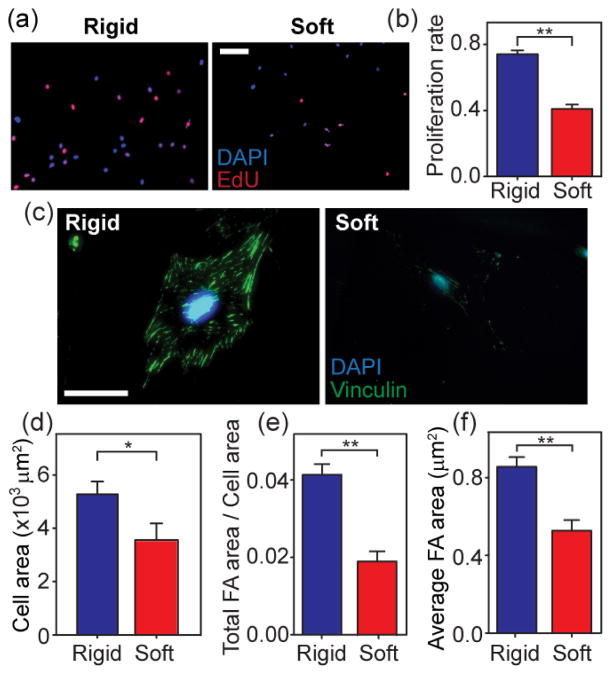
NIH/3T3 cells are mechano-sensitive and -responsive. It has been reported that substrate rigidity can impact cell morphology, FA formation, cytoskeletal contractility, and proliferation of NIH/3T3 cells9, 31. Thus, we examined whether changes of the bulk Young’s modulus E of photoPDMS could mediate mechanoresponsive behaviors of NIH/3T3 cells. We selected the most rigid and softest photoPDMS substrates (Rigid: 1:10 photoPDMS with 40 hr baking and 0 min UV light exposure, and E = 2.48 MPa; Soft: 1:30 photoPDMS with 20 min baking and 3 min UV light exposure, and E = 0.027 MPa). Both photoPDMS substrates were coated with fibronectin before seeding single NIH/3T3 cells. Figure 3 plotted the data for proliferation rate, cell spread area, and FA formation of NIH/3T3 cells as a function of E. Proliferation rate (Fig. 3a&b), cell spread area (Fig. 3c&d), total FA area per cell area (Fig. 3e), and average FA size (Fig. 3f) were all significantly greater for NIH/3T3 cells plated on the rigid photoPDMS substrate as compared to the cells plated on the soft one. Our observations here were consistent with results reported for NIH/3T3 cells plated on polyacrylamide gels of different rigidities9, 31. Thus, our results in Fig. 3 suggested that photoPDMS could serve as a photosensitive, rigidity-tunable biocompatible material to study mechano -sensitive and -responsive cellular behaviors.
Figure 3.
Effect of the bulk Young’s Modulus E of photoPDMS on cell morphology, FA formation, and proliferation of NIH/3T3 fibroblasts. (a) Representative immunofluorescence images of NIH/3T3 fibroblasts co-stained with DAPI (blue) and EdU (pink) after 8 hr of culture on flat photoPDMS substrates of different rigidities. Rigid photoPDMS substrate, E = 2.48 MPa. Soft photoPDMS substrate, E = 0.027 MPa. (b) Proliferation rate of NIH/3T3 cells after 8 hr of culture as a function of E of photoPDMS substrates. Data represents the means ± s.e.m from 3 independent experiments. **, p < 0.01. (c) Representative immunofluorescence images of single NIH/3T3 fibroblasts on flat photoPDMS substrates of different rigidities after 24 hr of culture. Cells were co-stained for nuclei (DAPI; blue) and vinculin (green). (d–f) Bar plots showing cell spread area (d), total FA area per cell area (e), and average FA area for single NIH/3T3 fibroblasts plated on flat photoPDMS substrates of different rigidities after 24 hr of culture. Data represents the means ± s.e.m from 3 independent experiments. n = 32 for 1:10 photoPDMS, and n =30 for 1:30 photoPDM S. *, p < 0.05. **, p < 0.01.
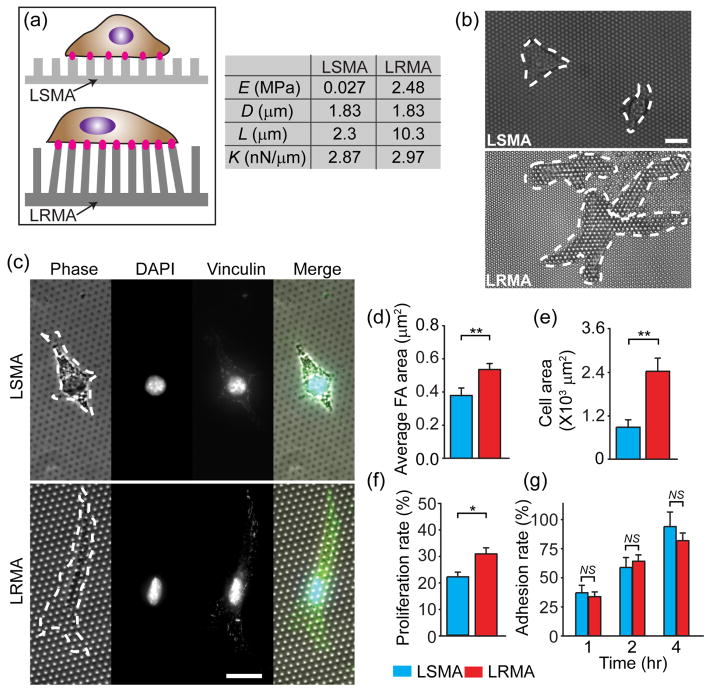
To further illustrate general applications of photoPDMS, we fabricated photoPDMS micropost arrays using photolithography and replica molding (Fig. S3). Mechanical properties of the photoPDMS micropost array were characterized by both the nanoscale bulk Young’s modulus E of photoPDMS and the microscale spring constant K of the photoPDMS micropost. K can be approximately calculated using the Euler-Bernoulli beam theory as K = 3πED4/(64L3), where D and L are the micropost diameter and height, respectively. It should be noted that the Euler-Bernoulli beam theory might not be accurate enough to calculate K values for PDMS microposts with L/D < 2. For such short PDMS microposts, the finite-element method (FEM) could be applied for calculations of their theoretical K values25. Recently, we reported that the PDMS micropost array could be used to decouple microscale substrate rigidity from adhesive and surface properties. By using PDMS micropost arrays with the same surface geometry but different post heights, we showed that the microscale spring constant K of the PDMS micropost could impact cell morphology, FA formation, cytoskeletal contractility and stem cell differentiation25, 32.Yet, it is still unclear whether this microscale spring constant K of the PDMS micropost can dominate and surpass their nanoscale bulk Young’s modulus for regulating mechanoresponsive cellular behaviors. In other words, it is unclear whether rigidity sensing by adherent cells takes place at a nanoscale resolution underneath single FAs or at a microscale resolution across the length of multiple FAs25, 32.
In this study, we fabricated two types of hexagonally spaced photoPDMS micropost arrays, one using the most rigid photoPDMS (“local rigid micropost array” or “LRMA”) while the other using the softest one (“local soft micropost array”, or “LSMA”) (Fig. 4a). The two photoPDMS micropost arrays had the same surface geometry (with the micropost diameter D of 1.83 μm and the post center-to-center spacing of 4 μm) but different post heights, so that their spring constant K calculated from FEM and the Euler-Bernoulli beam theory were comparable (Fig. 4a). The tops of both photoPDMS micropost arrays were functionalized with the same amount of fibronectin by microcontact printing, which was confirmed using fluorescence-conjugated proteins26. As shown in Fig. 4b, 24 hr after cell seeding, NIH/3T3 cells plated on LRMA could spread out and generate significant traction forces to deflect underlying microposts, while the cells on LSMA had more confined morphology and did not generate detectable traction forces and no micropost deflection could be observed. We further examined FA formation of NIH/3T3 cells by immunostaining of vinculin, a FA protein (Fig. 4c). NIH/3T3 cells displayed prominent and mature vinculin-expressing FAs on LRMA but only diffusive and punctate vinculin-expressing FAs on LSMA. Quantified results for FA formation and cell morphology further confirmed that the average FA size (Fig. 4d) and cell spread area (Fig. 4e) were all significantly greater for NIH/3T3 cells plated on LRMA as compared to the cells on LSMA. We further examined cell proliferation and adhesion rates for NIH/3T3 cells plated on LRMA and LSMA. Cell proliferation rate was significantly greater for NIH/3T3 cells plated on LRMA as compared to the cells plated on LSMA (Fig. 4f). Cell adhesion rate during the first 4 hr after cell seeding appeared insensitive to the local nanoscale rigidity difference between LRMA and LSMA (Fig. 4g), suggesting that the local nanoscale rigidity of photoPDMS microposts might exert their mechano-regulatory effects on cellular functions after initial cell attachment and spreading. Together, our data in Fig. 4 suggested that mechanosensitive adherent cells could sense and respond to changes of substrate rigidity at a sub-FA resolution. It is highly likely that rigidity sensing can involve the molecular arrangement, dynamic organization, and signaling of the cellular adhesion machinery at a sub-FA scale.
Figure 4.
Mechanoresponsive cellular behaviors depending on local rigidity sensing on the photoPDMS micropost array. (a) Schematic showing NIH/3T3 cells adhered on the photoPDMS micropost arrays generated using soft (top; E = 0.027 MPa) and rigid (bottom; E = 2.48 MPa) photoPDMS (LSMA: “local soft micropost array”; LRMA: “local rigid micropost array”). The spring constant K were comparable between LSMA and LRMA. (b&c) Representative phase-contrast and immunofluorescence micrographs of NIH/3T3 cells spreading on LSMA and LRMA after 24 hr of culture. White dashed lines in micrographs highlight cell boundaries. Cells in c were co-stained for nuclei (DAPI) and vinculin. Scale bar: 20 μm. (d&e) Bar plots showing the average FA area (d) and the cell spread area (e) of single NIH/3T3 fibroblasts plated on LSMA and LRMA after 24 hr of culture. Data represents the means ± s.e.m from 3 independent experiments. n = 20 for both LSMA and LRMA. **, p < 0.01. (f) Proliferation rate of single NIH/3T3 fibroblasts plated on LSMA and LRMA. Data represents the means ± s.e.m from 3 independent experiments. *, p < 0.05. (g) Adhesion rate of single NIH/3T3 fibroblasts plated on LSMA and LRMA at different time points as indicated. Data represents the means ± s.e.m from 3 independent experiments. NS, statistically not significant (p > 0.05).
As shown in Fig. 1, the bulk Young’s modulus E of photoPDMS depended strongly on UV light exposure. Thus, in principle, it is possible to spatially modulate UV light intensity using gradient photomasks to generate spatially patterned substrate rigidities or rigidity gradients in photoPDMS. Temporal control of local rigidity in photoPDMS is currently not possible, as after thermal curing the bulk Young’s modulus E of photoPDMS became stable. Generating freestanding tall micropost structures with soft photoPDMS (such as the 1:30 photoPDMS with E = 0.027 MPa used in this work) is difficult because of collapsing of the microposts when demolding from the mold due to strong adhesions between photoPDMS posts and the mold as well as the gravity or adhesive forces between the posts. These adhesive forces and gravity could become dominant over the elastic energy of the micropost structure. We have fabricated photoPDMS micropost arrays with different post heights using the softest photoPDMS with E = 0.027 MPa. The tallest photoPDMS micropost array that could be routinely generated was about 3 μmin height.
Recently, Hoffecker et al. developed a micropatterned hydrogel–photoresist composite system to study rigidity sensing at the cellular or micrometer scale33. In their system, an array of 6.5 μm × 6.5 μm square-shaped islands of rigid SU-8 photoresist were patterned on compliant polyacrylamide hydrogels of different rigidities. Using this composite system, Hoffecker et al. concluded that cellular or micrometer scale rigidity sensing was dominant in NIH/3T3 cells, as the cells had smaller spreading area and formed less stress fibers and FAs on soft hydrogels with rigid SU-8 islands as compare with the cells plated on rigid hydrogels with rigid islands. However, the results by Hoffecker et al. still could not rule out the possibility that adherent cells could also be sensitive to local nanoscale substrate rigidity. In this work, photoPDMS micropost arrays with multiscale mechanical properties were fabricated and applied for multiscale study of mechanoresponsive cellular behaviors. Our results suggested that adherent cells could sense and respond to changes of substrate rigidity at a sub-FA resolution. Our results combined together with those reported by Hoffecker et al. demonstrated that cellular sensing of substrate rigidity could be at multiple size scales ranging from local nanoscale material properties to micron or cellular scale structural properties. Future detailed studies of the size scale of rigidity sensing will provide important insights into designs of biomaterials for tissue engineering and studying mechanics-mediated multiscale pathological processes such as atherosclerosis and cancer metastasis4, 33, 34.
CONCLUSION
In this study, we developed photosensitive, biocompatible photoPDMS as a rigidity-tunable material for study of mechanoresponsive cellular behaviors. By modulating the PDMS crosslinker to monomer ratio, UV light exposure time, and post-exposure baking time, we achieved a 100-fold range of rigidity change from 0.027 to 2.48 MPa for photoPDMS, broader than is achievable with conventional PDMS. By assaying cell proliferation and FA formations, we demonstrated that photoPDMS did not induce detectable cytotoxic effect and benzophenone in photoPDMS did not significantly affect normal cell behaviors. We further fabricated photoPDMS micropost arrays to study the spatial resolution of cellular rigidity sensing. Our results suggested that mechanosensitive adherent cells could sense and respond to changes of substrate rigidity at a sub-FA resolution. Since PDMS is one of the most frequently used structural materials in microfluidics20, 35, developing photoPDMS as a photosensitive, rigidity-tunable material and integrating it into conventional PDMS-based microfluidics will extend microfluidic applications for microscale studies of local cellular microenvironment and its regulation of cell functions. Together, we demonstrated the potential of photoPDMS as a photosensitive and rigidity-tunable material for mechanobiology studies.
Supplementary Material
Acknowledgments
We acknowledge financial support from the National Science Foundation (CMMI 1129611 and CBET 1149401), the National Institute of Health (UL1RR024986), and the department of Mechanical Engineering at the University of Michigan, Ann Arbor. We thank Dr. A.M. Sastry for sharing the Instron machine for mechanical characterization of PDMS and photoPDMS samples. We thank P. Mao for his assistance in microfabrication of the silicon micropost array master. We thank M. Yang and C. Chen for sharing the MATLAB program for quantifying cellular contractile forces. The Lurie Nanofabrication Facility at the University of Michigan, a member of the National Nanotechnology Infrastructure Network (NNIN) funded by the National Science Foundation, is acknowledged for support in microfabrication.
Footnotes
SUPPORTING INFORMATION AVAILABLE
A scheme illustrating photoPDMS chemical reaction; Proliferation of NIH/3T3 cells on PDMS and photoPDMS substrates; photoPDMS micropost array fabrication process; Fluorescent intensity of Alexa-488 conjugated proteins coated on PDMS and photoPDMS micropost arrays. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Discher DE, Janmey P, Wang Y-l. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 3.Chen CS. Mechanotransduction - a field pulling together? J Cell Sci. 2008;121(20):3285–3292. doi: 10.1242/jcs.023507. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y, Chen CS, Fu J. Forcing Stem Cells to Behave: A Biophysical Perspective of the Cellular Microenvironment. Annu Rev Biophy. 2012;41(1) doi: 10.1146/annurev-biophys-042910-155306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong JY, Leach JB, Brown XQ. Balance of chemistry, topography, and mechanics at the cell-biomaterial interface: Issues and challenges for assessing the role of substrate mechanics on cell response. Sur Sci. 2004;570(1–2):119–133. [Google Scholar]
- 6.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotech. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 7.Ingber DE. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ Res. 2002;91(10):877–87. doi: 10.1161/01.res.0000039537.73816.e5. [DOI] [PubMed] [Google Scholar]
- 8.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 9.Pelham RJ, Wang YL. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton K, Taylor DL. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature. 1997;385(6615):450–4. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- 11.Frey MT, Wang YL. A photo-modulatable material for probing cellular responses to substrate rigidity. Soft Matter. 2009;5:1918–1924. doi: 10.1039/b818104g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324(5923):59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tse JR, Engler AJ. Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS One. 2011;6(1):e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo CM, Wang HB, Dembo M, Wang Y-l. Cell movement is guided by the rigidity of the substrate. Biophysical J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray DS, Tien J, Chen CS. Repositioning of cells by mechanotaxis on surfaces with micropatterned Young’s modulus. Journal of Biomedical Materials Research Part A. 2003;66A(3):605–614. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- 16.Wong JY, Velasco A, Rajagopalan P, Pham Q. Directed Movement of Vascular Smooth Muscle Cells on Gradient-Compliant Hydrogels†. Langmuir. 2003;19(5):1908–1913. [Google Scholar]
- 17.Ramanan VV, Katz JS, Guvendiren M, Cohen ER, Marklein RA, Burdick JA. Photocleavable side groups to spatially alter hydrogel properties and cellular interactions. J Mater Chem. 2010;20(40):8920–8926. [Google Scholar]
- 18.Young JL, Engler AJ. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials. 2011;32(4):1002–1009. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia Y, Whitesides GM. Soft Lithography. Angew Chem Int Edit. 1998;37(5):550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 20.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–73. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 21.Jothimuthu P, Carroll A, Bhagat AAS, Lin G, Mark JE, Papautsky I. Photodefinable PDMS thin films for microfabrication applications. J Micromech Microeng. 2009;19(4) [Google Scholar]
- 22.Bhagat AA, Jothimuthu P, Papautsky I. Photodefinable polydimethylsiloxane (PDMS) for rapid lab-on-a-chip prototyping. Lab Chip. 2007;7(9):1192–7. doi: 10.1039/b704946c. [DOI] [PubMed] [Google Scholar]
- 23.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100(4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saez A, Buguin A, Silberzan P, Ladoux B. Is the Mechanical Activity of Epithelial Cells Controlled by Deformations or Forces? Biophysical J. 2005;89(6):L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu JP, Wang YK, Yang MT, Desai RA, Yu XA, Liu ZJ, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Meth. 2010;7(9):733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang MT, Fu J, Wang YK, Desai RA, Chen CS. Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity. Nat Protocols. 2011;6(2):187–213. doi: 10.1038/nprot.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sia SK, Whitesides GM. Microfluidic devices fabricated in Poly(dimethylsiloxane) for biological studies. Electrophoresis. 2003;24(21):3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 28.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane) Analytical Chemistry. 1998;70(23):4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 29.Cong H, Pan T. Photopatternable Conductive PDMS Materials for Microfabrication. Adv Fun Mater. 2008;18(13):1912–1921. [Google Scholar]
- 30.Brown XQ, Ookawa K, Wong JY. Evaluation of polydimethylsiloxane scaffolds with physiologically-relevant elastic moduli: interplay of substrate mechanics and surface chemistry effects on vascular smooth muscle cell response. Biomaterials. 2005;26(16):3123–3129. doi: 10.1016/j.biomaterials.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. American Journal of Physiology - Cell Physiology. 2000;279(5):C1345–C1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 32.Weng S, Fu J. Synergistic regulation of cell function by matrix rigidity and adhesive pattern. Biomaterials. 2011;32(36):9584–9593. doi: 10.1016/j.biomaterials.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffecker IT, Guo W-h, Wang Y-l. Assessing the spatial resolution of cellular rigidity sensing using a micropatterned hydrogel-photoresist composite. Lab Chip. 2011;11(20):3538–3544. doi: 10.1039/c1lc20504h. [DOI] [PubMed] [Google Scholar]
- 34.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 35.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJ, Whitesides GM. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21(1):27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.