Summary
We demonstrate that SPOP is a critical component of the DDR. SPOP forms nuclear foci and interacts with the ATM kinase after DNA damage. Depletion of SPOP results in an impaired DDR and hypersensitivity to radiation.
Abstract
Speckle-type POZ protein (SPOP) is an adaptor of the cullin 3-based ubiquitin ligase responsible for the degradation of oncoproteins frequently overexpressed in many tumor cells. Altered expression and somatic mutations of SPOP have been observed in various tumor types with chromosomal aberrations, indicating a role of SPOP in maintaining genome stability, although a detailed mechanism remains unclear. Here, we show that SPOP is a component of the DNA damage response (DDR). SPOP is recruited to DNA double-strand break sites and it forms nuclear foci after DNA damage. SPOP foci colocalize with γ-H2AX foci and are predominantly dependent on the activity of the ataxia-telangiectasia mutated (ATM) kinase. Furthermore, SPOP interacts with ATM in response to DNA damage. Finally, we demonstrate that knocking down of SPOP resulted in an impaired DDR and a hypersensitivity to ionizing irradiation. Together, we highlight a critical role of SPOP in the DDR.
Introduction
An optimal DNA damage response (DDR) is critical for mammalian cells to maintain genome stability (1). Orchestrated by a comprehensive signaling network, the DDR reacts to environmental DNA damaging stress (both from endogenous and exogenous sources) by initiation of cell cycle checkpoints, DNA repair, and programmed cell death mechanisms. The overall functions of the DDR seek to eliminate or reduce the risk of erroneous DNA replication being passed to daughter cells. Suboptimal DDRs have been well documented in a majority of tumor cells, indicating a role for DDR proteins as tumor suppressors (2). It is also believed that activation of optimal DDR plays a critical role as an antitumor barrier during early tumorigenesis (3–5). On the other hand, optimal DDR is also responsible for cell survival in response to DNA damaging agents, such ionizing irradiation (IR) and many of the chemotherapeutic drugs (6). Additionally, recent evidence also suggests that hyperactive DDR might promote tumor invasion and metastasis (7,8). Therefore, elucidating the regulatory pathways of the DDR is critical to the understanding of tumor initiation, progression, and therapeutic responses.
Of the >100 genes, and their products, directly involved in the DDR, there are damage sensors, which recognize DNA strand breaks and recruit downstream proteins to damage sites; signal transducers, which amplify signals by posttranslational modification, and effectors, which typically are negative regulators of the cell cycle control. These proteins are well conserved through evolution, and the functional significance of the signaling transduction pathways has been extensively studied. For example, the ataxia-telangiectasia mutated (ATM) protein, mutation of which causes the autosomal recessive genetic disease ataxia telangiectasia, can be recruited and activated in response to DNA double-strand breaks (DSBs) (9). Activated ATM kinase then phosphorylates a large number of proteins to facilitate an optimal DDR (10). For example, ATM phosphorylation of Brca1 (11,12) inhibitor 2 (13), and deoxycytidine kinase (14) are critical for cells to initiate the G2/M cell cycle checkpoint.
E3 ubiquitin ligases play critical roles in cellular functions, including the DDR, by targeting specific protein substrates for proteasome-mediated degradation (15). Speckle-type poxvirus and zinc finger protein, SPOP, serves as an adaptor for cullin 3 (Cul3)-based ubiquitin ligases, a large family of the E3 ubiquitin ligases (16). SPOP is a nuclear protein belongs to the MATH-BTB protein family and was originally identified by immunoscreening as an autoantigen in a scleroderma patient sample (17). Substrates of SPOP include the apoptosis factor DAXX (16), the breast cancer metastasis suppressor BRMS1 (18) and the Hedgehog signaling transcription factors Gli2 and Gli3 (19–21). In addition, drosophila SPOP mediates degradation of the Jun kinase phosphatase puckered, resulting in tumor necrosis factor/Eiger-dependent apoptosis (22). Recently, SPOP was characterized as a tumor suppresser due to its role in reducing the cellular level of the p160 steroid receptor coactivator SRC-3 oncoprotein (23,24). Additionally, SPOP was identified as a regulator in response to fludarabine treatment in chronic lymphocytic leukemia cells (25). Recent literature has shown that loss of SPOP expression was common in gastric cancer, colorectal cancer as well as prostate cancer (26). More interestingly, exome sequencing studies have identified SPOP as the gene most commonly affected by somatic non-synonymous point mutations in prostate cancer (27–30). Reduced expression, as well as somatic mutations, of SPOP is associated with chromosomal aberrations in the cancers, indicating a role of SPOP in the maintenance of genome stability.
Here, we report that SPOP is an essential element of the DDR regulatory network. In response to DNA damage, SPOP interacts with ATM and forms nuclear foci. Depletion of SPOP in cells results in an impaired DDR and an increased radiosensitivity.
Materials and methods
Cell culture
The human cervical cancer cell line HeLa, the human breast cancer cell line MDA-MB-231 and the human osteosarcoma cell line U2OS, all obtained from The American Type Culture Collection (Manassas, VA), were cultured in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT) with 10% fetal bovine serum (Hyclone). All cell lines were grown in a 5% CO2 incubator at 37°C. Ionizing radiation was delivered by an X-Rad 320 X-ray irradiator (Precision X-Ray, North Branford, CT).
Antibodies and siRNA oligos
The SPOP antibody was purchased from Santa Cruz Biotechnology (Dallas, TX), and the dilution used was 1:1000. The γ-H2AX antibody was purchased from Cell Signaling Technology (Danvers, MA), and the dilution used was 1:5000. The ATM antibody and phosphor-Serine 1981 ATM antibody were purchased from Abcam (Cambridge, MA), and the dilutions used were 1:3000 for both. The control small interfering RNA (siRNA) and two independent SPOP siRNA oligos were purchased from Santa Cruz Biotechnology and Dharmcon (Waltham, MA), and 20 nM oligos were used in each experiment. The ATM siRNA oligo (5′-CACCUUUUUCUUGGGUUUUGGCUCCUU-3′) was developed by our lab, and 20 nM oligo was used in this study. siRNAs were transfected into cells by oligofectamine reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction.
Western blotting
Cells were incubated with the lysis buffer (Fisher, Pittsburgh, PA) containing the protease inhibitor cocktail (Roche, Indianapolis, IN), and the protein concentration was determined using the DC Protein Assay kit (Bio-Rad, Hercules, CA). The 4–12% Bis–Tris precast gels (Bio-Rad) were used for electrophoresis. Equal volumes of cell total protein were loaded and subsequently transferred from gels to the nitrocellulose membrane. The membrane was blotted with 5% of non-fat milk (Lab Scientific, Livingston, NJ), followed by the incubation with primary and horseradish peroxidase-conjugated secondary antibodies for overnight or 2 h, respectively. Signals were detected by adding chemiluminescence reagents (GE, Buckinghamshire, UK).
RNA isolation and real-time PCR
Total RNA was isolated from cells using RNAfast200 (Pioneer Biotechnology, Shaanxi, China). Five hundred nanograms of RNA was used to synthesize complementary DNA by using PrimeScript™ RT Master Mix (Takara Biotechnology, Dalian, China). The SPOP primer/probe sets (SPOP-F: 5′-ATTCCAGGCTCACAAGGCTATCTTA-3′, SPOP-R: 5′-CAGCTGCCAG CAAATCATCA-3′) and the SYBR® Premix Ex Taq™ II for real-time PCR were purchased from Takara Biotechnology. All real-time PCR experiments were performed in triplicate.
Cell proliferation assay and clonogenic survival assay
The cell proliferation assay was performed on the RNA oligos-transfected HeLa cells seeded in 96-well plates. Following the treatment with mock or irradiation, cell proliferation was detected by Cell Titer 96@ AQueous One Solution Cell Proliferation Assay system (Promega, Madison, WI) at 0, 24 and 48 h after the treatment. Generally, 20 μl of the One Solution reagent was loaded to each well and the mixture of the reagent and cells were put into incubator for 2h. Cell proliferation was then measured using a microplate reader (BioRad Model 680) with the absorbance of 490nm. The clonogenic survival assay was performed on the RNA oligos-transfected HeLa cells growing in 6-well plates. The plates were subjected to irradiation with indicated doses and, 10–14 days later, cells were stained with crystal violet (0.1%) for 30min. The colonies with 50 or more cells were then scored. The survival fractions were calculated as described previously (31).
Immunofluorescence and immunoprecipitation
Cells plated on glass slides (Fisher) were fixed with 4% paraformaldehyde for 15min and permeabilized in 0.5% Triton X-100 for 10min. Cells were rinsed with phosphate-buffered saline (PBS) after the indicated treatment and incubated with primary antibody diluted in 1% bovine serum albumin 1 h at room temperature or overnight at 4°C. Cells were washed three times with 0.1% PBS-T and incubated with fluorescent-conjugated secondary antibody diluted in 1% bovine serum albumin for 1h at room temperature in the dark. Cells were washed three times with 0.1% PBS-T. 4′,6-diamidino-2-phenylindole (1mg/ml) diluted in 1× PBS was used to stain DNA for nuclear visibility. Slides were mounted using microscope immersion oil and visualized using a fluorescence microscope. For immunoprecipitation, cells were lysed with lysis buffer following harvest. The cell lysates were incubated with Protein G Sepharose (Sigma–Aldrich, St Louis, MO) and the SPOP antibody, the ATM antibody, goat IgG or rabbit IgG, respectively, overnight at 4°C. The immunoprecipitates were washed three times with the IP Washing Buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol), then subjected to western blotting.
Single cell gel electrophoresis assay (comet assay)
Comet Assay kits, purchased from Trevigen (Gaithersburg, MD), were used in this study. The neutral comet assay was performed according to the manufacturer’s instructions. Generally, cells were transfected with siRNA oligos and irradiated with mock or IR. Cells were then harvested 0, 4 and 8 h after IR and mixed with low-melting agarose for the subsequent electrophoresis. The nuclear DNA was stained with SYBR green dye for 10 min and visualized using a fluorescent microscope. Olive tail moment was measured using CASP software.
Statistics
Data were determined by Student’s t-test and P values ≤0.05 were considered significant.
Results
SPOP forms nuclear foci in response to DNA damage
To characterize SPOP’s functions in the DDR, we first examined if SPOP is recruited to DNA damage-induced DSBs. In unperturbed HeLa cells, SPOP expression is diffused with weak speckles. However, when cells were treated with IR (4 Gy), we observed nuclear focus formation of SPOP (Figure 1) 1 h after IR. The SPOP foci apparently colocalized with the foci of γ-H2AX, a phosphorylated form of the histone variant H2AX and a marker of DNA DSBs (32). Therefore, these observations indicate that SPOP is recruited to DSBs in response to DNA damage. Similar patterns of focus formation and colocalization were observed in HeLa cells treated with camptothecin, a DNA topoisomerase I inhibitor. This phenotype is also observed in other cell lines including the breast cancer cell line MDA-MB-231 and the osteosarcoma cell line U2OS (Supplementary Figure S1, available at Carcinogenesis Online).
Fig. 1.
SPOP forms nuclear foci in response to DNA damage. Exponentially growing HeLa cells were treated with mock, IR (4 Gy) or camptothecin (CPT, 10 μM). One hour after IR and 2h after CPT, cells were fixed and immunostained with the anti-SPOP (red) or anti-γ-H2AX (green) antibody. 4′,6-diamidino-2-phenylindole (DAPI) was used for DNA staining (blue).
To assess if SPOP messenger RNA (mRNA) levels changes after IR, we conducted reverse transcription–polymerase chain reaction to assess SPOP mRNA changes 1, 6, 12, 24, 48 h after IR (4 Gy) in HeLa cells. We have also assessed the changes of the protein level. As shown in Supplementary Figure S2, available at Carcinogenesis Online, SPOP mRNA did not show significant changes in the 1 and 6 h time points, but it displayed temporary increases 12–24 h after IR. The increase of mRNA was correlated with the protein level.
ATM activity is required for SPOP nuclear focus formation in the DDR
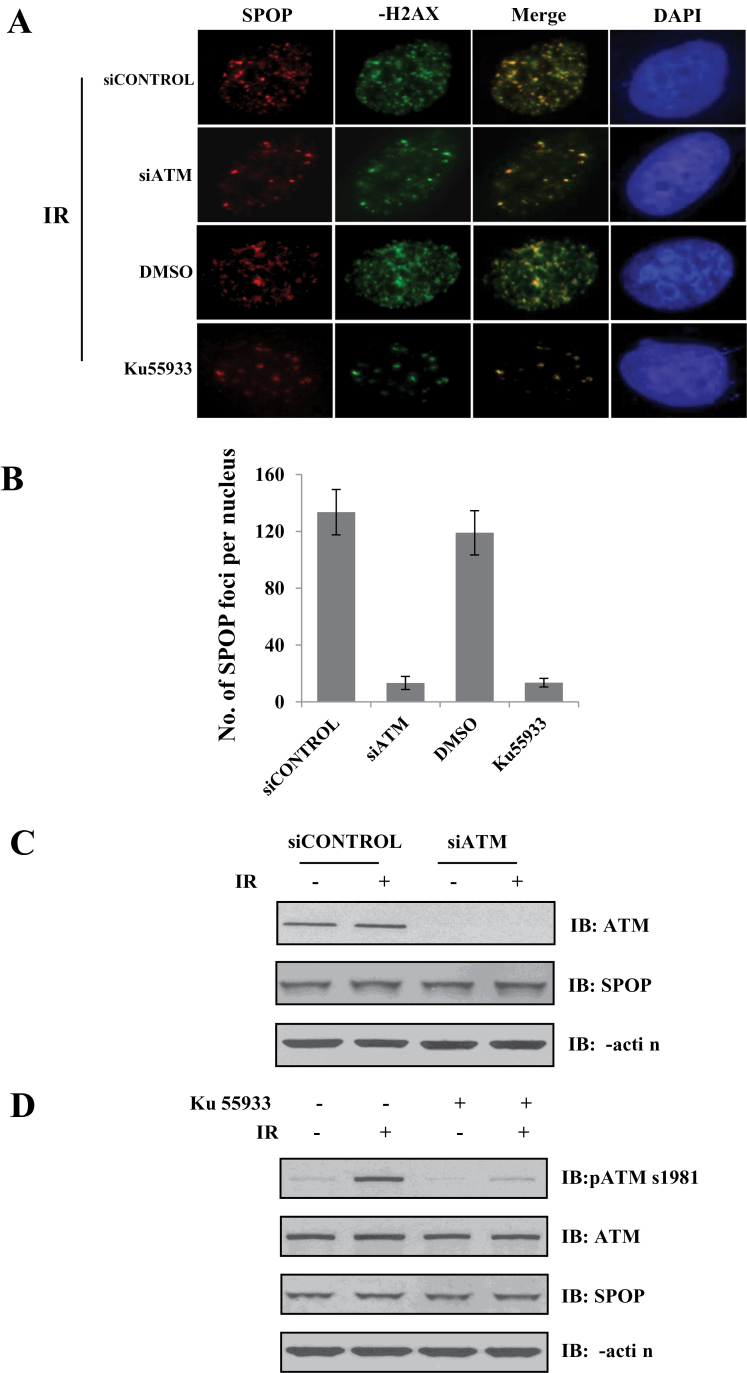
Owing to the central role of the ATM kinase in the DDR, we tested whether DNA damage-induced SPOP focus formation is dependent on ATM. To achieve this goal, we knocked down endogenous ATM by transient transfection of ATM siRNA into HeLa cells. We found that ATM depletion significantly reduced the formation of SPOP foci (Figure 2A and B). Inhibition of ATM by a specific ATM inhibitor Ku55933 also achieved a similar effect. It is noted that ATM depletion or inhibition did not alter the whole protein level of SPOP (Figure 2C).
Fig. 2.
ATM is involved in the formation of SPOP nuclear foci in response to DNA damage. HeLa cells transfected with control or ATM siRNA or treated with Ku55933 (10nM) were subjected to immunofluorescence microscopy (A) or immunoblotting (C) using indicated antibodies. (A) The localization of SPOP (red) and γ-H2AX (green) 1h after IR (4 Gy). (B) Quantitative data of foci/nuclear of SPOP. (C and D) Endogenous expression levels of SPOP in HeLa cells transfected with control or ATM siRNA (C) or treated with Ku55933 (10 µM, 2h) (D) in the presence or absence of IR (4 Gy, 1h).
SPOP interacts with ATM in response to IR
We further investigated if there is an interaction between SPOP and ATM by coimmunoprecipitation. We did not observe an interaction of SPOP and ATM in mock-treated cells; however, we observed that ATM was present in the SPOP immunoprecipitates in response to IR (Figure 3A). A reciprocal experiment showed that SPOP was detectable in the ATM immunoprecipitates only after cells were irradiated (Figure 3B). These results indicate that SPOP interacts with ATM in response to IR-induced DNA damage.
Fig. 3.
SPOP interacts with ATM in response to DNA damage. Endogenous SPOP (in A) or ATM (in B) was immunoprecipitated in cells 1h after treating with mock or IR (4 Gy). The immunoprecipitates were subjected to immunoblotting using indicated antibodies. Total cell lysates were included as loading controls.
SPOP knockdown resulted in suboptimal DNA damage repair
Recruitment of SPOP to DNA DSB sites shown in Figure 1 indicates that SPOP might be required for DNA damage repair. Therefore, we examined if depletion of endogenous SPOP by siRNA impacted DNA damage repair processes. Two independent siRNA oligos were used to knockdown SPOP in HeLa cells, and both oligos were efficient in reducing the SPOP level (Figure 4A). We first investigated the existence of γ-H2AX foci in control or SPOP-depleted cells in response to IR. In control cells, the number of γ-H2AX foci per nucleus increased at 1 h after treatment and returned to a slightly above normal level at 24 h posttreatment because of the repair of the DSBs. However, SPOP knockdown cells showed a prolonged presence of γ-H2AX foci after IR (Figure 4B). This strongly suggests that SPOP is required for efficient DSB repair. We also conducted a neutral gel comet assay and the results are consistent with the γ-H2AX foci (Figure 4C), as significantly higher olive tail moment in SPOP knockdown cells were observed, indicating a delay of DNA strand break repair in the absence of SPOP.
Fig. 4.
SPOP knockdown leads to defects in DNA repair. Two independent siRNA oligos were used to inhibit endogenous expression of SPOP in HeLa cells. Twenty-four hours after transfection of control or SPOP siRNA, cells were subjected to IR (4 Gy). One hour after IR, cells were harvested and subjected to the following assays: (A) Immunoblotting of endogenous SPOP after siRNA knockdown. (B) Fluorescence microscopy staining and quantification of the existence of γ-H2AX foci. (C) The signal cell electrophoresis assay (the comet assay) and quantification of olive tail moment.
SPOP depletion results in a hypersensitivity to IR
We further investigated the effect of SPOP knockdown on cellular radiosensitivity. Using a cell proliferation assay, we found that, although SPOP knockdown in the absence of DNA damage caused mild cell death, it greatly sensitized HeLa cells to IR (Figure 5A). We also conducted the clonogenic survival assay assessing radiosensitivity. Compared with control cells, SPOP knockdown cells showed a significant reduction in cell survival after IR (Figure 5B). Together, our data demonstrate a critical role of SPOP in regulation of radiosensitivity.
Fig. 5.
SPOP inhibition sensitizes HeLa cells to IR. HeLa cells were transfected with control or SPOP siRNA oligos (siSPOP-1 or siSPOP-2). Twenty-four hours after transfection, cells were irradiated with mock (0 Gy) or IR (4 Gy). At indicated time points after IR, cells were subjected to: (A) the 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide proliferation assay (*indicates statistical significance, P < 0.05), and (B) the colony formation assay.
Discussion
Characterized as an adaptor protein in the Cul3 E3 ubiquitin ligase family, SPOP has recently been shown as a tumor suppressor for the proteolysis of the oncogenic coactivator SRC-3/AIB1 (23,24). Somatic mutations as well as altered expressions of SPOP have been observed in many tumor types, including gastric cancer, colorectal cancer as well as prostate cancer (26,29), indicating a potential role of SPOP in genome stability. In this manuscript, we reported, for the first time, that SPOP is an essential element of the DDR. SPOP forms nuclear foci and interacts with ATM in response to DNA damage. Functionally, SPOP is required for DNA damage repair and the maintenance of radioresistance. These observations provide insights into the functional significance of SPOP in genome stability.
As a speckle-type protein, a rough speckled pattern of endogenous SPOP was originally shown in the nuclei of COS7 cell line (17). However, there were very few speckles detected for the endogenous SPOP in HeLa cells (33). Consistent with the previous data, few bright spots of endogenous SPOP were visualized. However, in response to DNA damage, a significant number of nuclear foci were formed. Colocalization of SPOP foci with γ-H2AX foci supports DSB-dependent nuclear recruitment of SPOP in the event of DNA damage. Although at later time points after IR, SPOP mRNA and protein levels increases temporarily, the SPOP focus formation appears to be mediated by posttranslational modification.
Ubiquitination plays a critical role in the DDR as evidenced by that fact that many E3 ubiquitin ligases such as RNF8 and RNF168 catalyze polyubiquitylation of histones H2A and H2AX at DSB sites (34,35). Recent reports have shown that ATM-dependent phosphorylation of the E3 ubiquitin ligase, RNF20-RNF40 heterodimer, is responsible for histone H2B monoubiquitylation, which facilitates the two major DSB repair pathways: non-homologous end-joining and homologous recombination repair (36). A link between SPOP and ATM might provide clues on how SPOP is involved in the DDR. The requirement of ATM activity in SPOP focus formation indicates that ATM is an upstream of the SPOP pathway. Indeed, SPOP contains three ATM consensus phosphorylation motifs (S/T-Q, threonine 25, serine 119 and threonine 319), and one of them, serine 119, located in the conserved N-terminus MATH domain that recruits substrates (37), is found to be mutated (a substitution of serine to asparagine -S119N) in some prostate cancers (29). In a recent study, it has been shown that induction of S119N mutation in LNCAP-ABL cells abrogated degradation of SRC3 (24), indicating an important role of the residue and the surrounding sequence.
The solved crystal structure of SPOP (PDB 3HQI) demonstrates the heterodimeric assembly of the SPOP–Cul3 interaction. The secondary structure of SPOP contains a conserved α-helical C-terminus which encompasses the BTB domains, also known as the ‘3-box’ due to its facilitating of Cul3 binding, as well as its similarity to F-/SOCS-boxes in other cullin-based E3s (37). The structural flexibility attributed to the region between the MATH and Cul3-binding BTB/3-box domains most likely allows for a SPOP dimer to simultaneously interact with multiple SPOP-binding consensus (SBCs) on a substrate. This allows for high-order hetero-oligmerization that facilitates the ubiquitination of bound substrates (38). The most recent crystal structure of the dimeric SPOP C-terminal domain showed that tyrosine 353 plays a critical role in high-order oligomerization (39). To assess a potential role of ATM-mediated SPOP phosphorylation, we have conducted molecular modeling studies to obtain the structural insight into the ATM consensus motifs on SPOP in order to predict the potential effects of ATM-mediated phosphorylation (Figure 6). The structural models of SPOPMATH and SPOPBTB were built directly from the SPOP crystal structure (PDB ID: 3HQI) using the protein preparation protocol of Schrödinger®. We found that, unlike threonine 25, which belongs to the flexible N-terminus of SPOP, both serine 119 and threonine 319 locate at structurally conserved regions that are important for SPOP functions. The crystal structure of the SBC-SPOPMATH complex indicates that serine 119 locates at the SBC-MATH binding interface and is in close contact with the non-polar residue of the SBC motif (37). Phosphorylation of serine 119 therefore will directly affect SPOP-substrate binding. On the other hand, threonine 319 is a part of the Cul3-interacting box region at the C-end of the SPOP BTB domain (SPOPBTB) (37). Similar as the F-/SOCS-boxes that bind to cullins, the ‘3-box’ is functionally important for SPOP-Cul3 binding. Although structural information regarding the SPOP–Cul3 interaction is still not available, the crystal structure of the SPOPBTB has demonstrated that the ‘3-box’ is a relative small fragment consisting two short helixes; threonine 319 locates in the middle of the helical region and is fully solvent exposed (which is typical for residues at protein–protein interfaces). Thus, it is very likely that phosphorylation of threonine 319 will affect the interactions between SPOP and Cul3.
Fig. 6.
Structural representation of the phosphorylation sites of serine 119 and threonine 319. (A) The SBC-SPOPMATH interface. The SBC residues are illustrated in green-colored sticks; the residues of SPOP are shown in gray lines; the serine 119 residue and the non-polar residue (V98) of SBC motif are highlighted in the space-filling model. (B) Carton representation of the SPOP-BTB domain. The threonine 319 residue is shown in the space-filling model. Both (A) and (B) are based on the crystal structure of SPOP (PDB ID: 3HQI).
Despite the evidence that SPOP depletion cause pronged H2AX foci and higher comet tail moment, it is still not known whether SPOP directly participates in the DNA repair processes. Due to its known role as an adaptor protein of Cul-3-mediated ubiquitination, it is likely that SPOP participate in the DDR as an adaptor for multiple signaling in response to DNA damage, thus playing an indirect role in DNA repair.
In summary, we demonstrate that SPOP is recruited to IR-induced DNA double-strand break sites in a predominantly ATM-dependent manner. We also show that SPOP depletion results in defects in the DDR and an increase in radiosensitivity.
Supplementary material
Supplementary Figures S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
D.Z. was supported in part by funding from the China Scholarship Council; NIH grants (R01CA133093, R01ES016354 to Dr B.X.).
Supplementary Material
Acknowledgements
We thank Dr S.Mitra for helpful comments on the manuscript. We thank Dr R.Boohaker for the proofreading of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ATM
ataxia-telangiectasia mutated
- Cul3
cullin 3
- DDR
DNA damage response
- DSB, double-strand break; IR
ionizing irradiation
- mRNA
messenger RNA
- PBS
phosphate-buffered saline
- SBC
SPOP-binding consensus
- siRNA
small interfering RNA
- SPOP
Speckle-type POZ protein.
References
- 1. Harper J.W., et al. (2007). The DNA damage response: ten years after. Mol. Cell, 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 2. Motoyama N., et al. (2004). DNA damage tumor suppressor genes and genomic instability. Curr. Opin. Genet. Dev., 14, 11–16 [DOI] [PubMed] [Google Scholar]
- 3. Bartkova J., et al. (2006). Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature, 444, 633–637 [DOI] [PubMed] [Google Scholar]
- 4. Bartkova J., et al. (2005). DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature, 434, 864–870 [DOI] [PubMed] [Google Scholar]
- 5. Gorgoulis V.G., et al. (2005). Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature, 434, 907–913 [DOI] [PubMed] [Google Scholar]
- 6. Zhou B.B., et al. (2003). Targeting DNA checkpoint kinases in cancer therapy. Cancer Biol. Ther., 2 (4 suppl. 1), S16–S22 [PubMed] [Google Scholar]
- 7. Sun M., et al. (2012). Activation of the ATM-Snail pathway promotes breast cancer metastasis. J. Mol. Cell Biol., 4, 304–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boohaker R.J., et al. (2013). ATM-mediated Snail Serine 100 phosphorylation regulates cellular radiosensitivity. Radiother. Oncol., 108, 403–408 [DOI] [PubMed] [Google Scholar]
- 9. Shiloh Y., et al. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol., 14, 197–210 [PubMed] [Google Scholar]
- 10. Matsuoka S., et al. (2007). ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science, 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 11. Xu B., et al. (2001). Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell. Biol., 21, 3445–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu B., et al. (2002). Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res., 62, 4588–4591 [PubMed] [Google Scholar]
- 13. Tang X., et al. (2008). A novel ATM-dependent pathway regulates protein phosphatase 1 in response to DNA damage. Mol. Cell. Biol., 28, 2559–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang C., et al. (2012). Deoxycytidine kinase regulates the G2/M checkpoint through interaction with cyclin-dependent kinase 1 in response to DNA damage. Nucleic Acids Res., 40, 9621–9632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hannah J., et al. (2009). Regulation of DNA damage response pathways by the cullin-RING ubiquitin ligases. DNA Repair (Amst)., 8, 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwon J.E., et al. (2006). BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem., 281, 12664–12672 [DOI] [PubMed] [Google Scholar]
- 17. Nagai Y., et al. (1997). Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett., 418, 23–26 [DOI] [PubMed] [Google Scholar]
- 18. Kim B., et al. (2011). Breast cancer metastasis suppressor 1 (BRMS1) is destabilized by the Cul3-SPOP E3 ubiquitin ligase complex. Biochem. Biophys. Res. Commun., 415, 720–726 [DOI] [PubMed] [Google Scholar]
- 19. Wang C., et al. (2010). Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development, 137, 2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Q., et al. (2009). Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl Acad. Sci. USA, 106, 21191–21196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Q., et al. (2006). A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell, 10, 719–729 [DOI] [PubMed] [Google Scholar]
- 22. Liu J., et al. (2009). Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science, 323, 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li C., et al. (2011). Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene, 30, 4350–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Geng C., et al. (2013). Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc. Natl Acad. Sci. USA, 110, 6997–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moussay E., et al. (2010). Determination of genes and microRNAs involved in the resistance to fludarabine in vivo in chronic lymphocytic leukemia. Mol. Cancer, 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim M.S., et al. (2013). Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS, 121, 626–633 [DOI] [PubMed] [Google Scholar]
- 27. Kan Z., et al. (2010). Diverse somatic mutation patterns and pathway alterations in human cancers. Nature, 466, 869–873 [DOI] [PubMed] [Google Scholar]
- 28. Berger M.F., et al. (2011). The genomic complexity of primary human prostate cancer. Nature, 470, 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barbieri C.E., et al. (2012). Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet., 44, 685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Gallo M., et al. ; NIH Intramural Sequencing Center (NISC) Comparative Sequencing Program. (2012). Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet., 44, 1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cariveau M.J., et al. (2007). Characterization of an NBS1 C-terminal peptide that can inhibit ataxia telangiectasia mutated (ATM)-mediated DNA damage responses and enhance radiosensitivity. Mol. Pharmacol., 72, 320–326 [DOI] [PubMed] [Google Scholar]
- 32. Kuo L.J., et al. (2008). Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo, 22, 305–309 [PubMed] [Google Scholar]
- 33. Hernández-Muñoz I., et al. (2005). Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl Acad. Sci. USA, 102, 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huen M.S., et al. (2007). RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell, 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stewart G.S., et al. (2009). The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell, 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 36. Moyal L., et al. (2011). Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell, 41, 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhuang M., et al. (2009). Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell, 36, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Errington W.J., et al. (2012). Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure, 20, 1141–1153 [DOI] [PubMed] [Google Scholar]
- 39. van Geersdaele L.K., et al. (2013). Structural basis of high-order oligomerization of the cullin-3 adaptor SPOP. Acta Crystallogr. D. Biol. Crystallogr., 69(Pt 9), 1677–1684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.