Summary
The objective of the study was to characterize the mechanism(s) of resveratrol-mediated protection against estrogen-induced breast carcinogenesis. This study provides evidences that resveratrol epigenetically induces NRF2 and its downstream protective pathways in mammary tissues and inhibits estrogen-induced breast carcinogenesis.
Abstract
The importance of estrogens in the etiology of breast cancer is widely recognized. Estrogen-induced oxidative stress has been implicated in this carcinogenic process. Resveratrol (Res), a natural antioxidant phytoestrogen has chemopreventive effects against a variety of illnesses including cancer. The objective of the present study was to characterize the mechanism(s) of Res-mediated protection against estrogen-induced breast carcinogenesis. Female August Copenhagen Irish rats were treated with 17β-estradiol (E2), Res and Res + E2 for 8 months. Cotreatment of rats with Res and E2 inhibited E2-mediated proliferative changes in mammary tissues and significantly increased tumor latency and reduced E2-induced breast tumor development. Resveratrol treatment alone or in combination with E2 significantly upregulated expression of nuclear factor erythroid 2-related factor 2 (NRF2) in mammary tissues. Expression of NRF2-regulated antioxidant genes NQO1, SOD3 and OGG1 that are involved in protection against oxidative DNA damage was increased in Res- and Res + E2-treated mammary tissues. Resveratrol also prevented E2-mediated inhibition of detoxification genes AOX1 and FMO1. Inhibition of E2-mediated alterations in NRF2 promoter methylation and expression of NRF2 targeting miR-93 after Res treatment indicated Res-mediated epigenetic regulation of NRF2 during E2-induced breast carcinogenesis. Resveratrol treatment also induced apoptosis and inhibited E2-mediated increase in DNA damage in mammary tissues. Increased apoptosis and decreased DNA damage, cell migration, colony and mammosphere formation in Res- and Res + E2-treated MCF-10A cells suggested a protective role of Res against E2-induced mammary carcinogenesis. Small-interfering RNA-mediated silencing of NRF2 inhibited Res-mediated preventive effects on the colony and mammosphere formation. Taken together, these results suggest that Res inhibits E2-induced breast carcinogenesis via induction of NRF2-mediated protective pathways.
Introduction
Breast cancer remains one of the leading causes of mortality among women worldwide. In the USA, breast cancer represents the most common neoplasm and the second leading cause of cancer death in women (1). The importance of endogenous estrogens in the etiology of breast cancer is widely recognized and in 2001, the United States government added steroidal estrogens to the list of known human carcinogens (2–4). Estrogens are suggested to cause breast cancer by stimulating cell growth and proliferation through receptor-mediated processes and by way of their toxic metabolites (3,5).
It is widely accepted that environmental and dietary factors play a role in determining one’s breast cancer risk. Epidemiologic evidence suggests that dietary phytoestrogens may inhibit carcinogenesis. Phytoestrogens are biologically active phenolic compounds of plant origin that structurally mimic the mammalian steroid hormone 17β-estradiol (E2) (6,7). Among the phytoestrogens that have been investigated to treat breast cancer, resveratrol (Res) stands out as a unique compound, because its chemical structure resembles mammalian E2 and hence is anticipated to interfere with the functions of E2 (7).
Resveratrol is a naturally occurring stilbene phytoestrogen that is present in various foods and beverages and is known to possess wide variety of biological properties including antioxidant, detoxification, anti-inflammatory and anticancer activities. This compound is known to modulate several signaling kinases (Raf, Src, MAPK, PKD, PKCd), transcription factors (p53/p21, IkB kinase/NF-kB) and other targets (TRAIL/DR4/DR5, Fas/CD95, PI3K/Akt) leading to cancer cell death (6,8). Resveratrol has also been found to induce apoptosis in T47D breast cancer cell line through activation of p53 (9). Further, it has been suggested that Res-induced caspase-3 activation is required for poly adenosine diphosphate ribose polymerase (PARP) degradation and induction of apoptosis in MDA-MB-231 cells (10).
Resveratrol has also been shown to prevent chemically induced mammary carcinogenesis and alter breast development and morphology in animal models of breast cancer (11–13). Resveratrol blocked the formation of preneoplastic lesions, suppressed mammary carcinogenesis, reduced tumor incidence and increased tumor latency in Sprague Dawley rats treated with dimethylbenz[a]anthracene (13). In Sprague Dawley rats, Res inhibited N-methyl-N-nitrosourea and dimethylbenz[a]anthracene-induced mammary carcinogenesis and decreased N-methyl-N-nitrosourea- and dimethylbenz[a]anthracene-induced tumor incidence and multiplicity by 50% (11,12). The combination of Res with quercetin and catechin has been found to reduce primary tumor growth of breast cancer xenografts in a nude mouse model (14). Significant lowering of tumor growth, decreased angiogenesis and increased apoptotic index in MDA-MB-231 tumors have also been reported in Res-treated nude mice (15). However, doubts persist whether the promising effects of Res seen in these preclinical studies can be translated to humans as Res is rapidly metabolized and have very poor bioavailability (16). In a recent study, Patel et al. observed that Res metabolites are converted back to the parent Res compound in target tissues, and this regenerated Res is of greater importance and give rise to the beneficial effects in vivo (17). In fact, Res metabolites provide a reservoir for the regeneration of parent Res compound in situ (17).
Although there have been a number of investigations regarding the effects of Res on different cancers, there are no in vivo reports on the effect of Res on inhibition of estrogen-induced breast cancer. Moreover, many studies report conflicting and inconsistent results (12,18–21). Thus, it is hard to establish the preventive effects of Res on human breast cancer specifically estrogen-induced breast cancer. Therefore, in the current study, we have examined the preventive effects of Res and mechanism(s) of Res-mediated prevention of estrogen-induced breast cancer in the female August Copenhagen Irish (ACI) rat model, a well-established rodent model of human breast carcinogenesis (22–27).
Materials and methods
Treatment of animals and histopathologic analysis
Female ACI rats (4 weeks of age; Harlan Sprague Dawley, Indianapolis, IN) were housed under controlled temperature, humidity and lighting conditions. After a 1-week acclimatization period, rats were divided into the following groups: Control, E2, Res and Res + E2. Rats were implanted subcutaneously with 3mg E2 pellets prepared in 17mg cholesterol as a binder. Resveratrol was given as a 50mg subcutaneous pellet every other month. The control group received a 17mg cholesterol pellet only. All the animals were fed an AIN-76A phytoestrogen-free diet (Dyets, Bethlehem, PA), and water was given ad libitum. At the end of the experimental time period (8 months), animals were anesthetized using isoflurane and euthanized. Mammary (both tumor and normal) tissues were removed and snap frozen in liquid nitrogen for future analyses. A portion of the excised tissues was stored in 10% buffered formalin for histopathologic analyses. Tumor incidence and the number of tumor nodules were counted at the time of dissection. The formalin-fixed tissues were embedded in paraffin, and sections of 4 to 5 μm thickness were cut. Paraffin-embedded sections of the mammary tumors and mammary tissues were stained with hematoxylin and eosin for histopathologic evaluation by a board-certified pathologist. Tumors in the breast fat pads were identified grossly at the time of animal dissection. Tumor size was measured in millimeters, and the volume (length × breadth × height) of tumor in cubic millimeters was obtained. Animal protocols used in the current study were approved by the Institutional Animal Care and Use Committee.
Cell culture
Non-tumorigenic human breast epithelial cell line MCF-10A was purchased from American Type Culture Collection (ATCC, Manassas, VA). MCF-10A cells were grown in Dulbecco’s modified Eagle’s medium/F12 (50:50) media (Mediatech, Herndon, VA). Twenty-four hours before treatment, cells were washed twice with phosphate-buffered saline (PBS) and then grown in phenol red-free Dulbecco’s modified Eagle’s medium/F12 (50:50) media supplemented with 5% charcoal-dextran stripped horse serum. Cells were treated with E2 (10 and 50nM) and Res (50 µM) for up to 48h.
Real-time PCR analysis
Total RNA was isolated from ACI rat tissues and cells in culture using Tri Reagent (Molecular Research Center, Cincinnati, OH) according to the supplier’s protocols. Five microgram total RNA was reverse transcribed using random hexanucleotide primers and superscript II reverse transcription system (Invitrogen, Carlsbad, CA). Real-time PCR by SYBR Green chemistry was performed using iCycler iQ5 system (Bio-Rad Laboratories, Hercules, CA) with rat-specific NRF2, NQO1, SOD3, OGG1, FMO1, AOX1 QuantiTect primers (Qiagen, Valencia, CA) and pri-miR-93 specific primers, which can detect this pri-miR in both MCF-10A cell line and rat samples. The expression of U6 snRNA was used for normalization and quantification of miR-93, whereas expression of cyclophilin was used to normalize and quantitate the mRNA expression of NRF2, NQO1, SOD3, OGG1, FMO1 and AOX1. Data were analyzed from at least four different animals/cell line samples from each group. Primers used were as follows: pri-U6: 5′-CTCGCTTCGGCAGCACA-3′ and 5′-AACGCTTCACGAATTTGCGT-3′, Pri-miR-93: 5′-AAGTGCTGTTCGTGCAGGT-3′ and 5′-CTCGGGAAGT GCTAGCTCA-3′, cyclophilin: 5′-CCCACCGTGTTCTTCGACAT-3′ and 5′-CCAGTGCTCAGAGCACGAAA-3′.
RNA interference
Small-interfering RNA for NRF2 and scrambled small-interfering RNA were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). MCF-10A cells were transfected with siNRF2 (20 nmol/l) using Lipofectamine 2000 transfection reagent (Invitrogen). Scrambled small-interfering RNA- (20 nmol/l) transfected MCF-10A cells were used as a negative control as described recently (28).
Western blot analysis
Approximately 50mg of ACI rat tissues were homogenized in a tissue protein extraction buffer (T-PER, Thermo Scientific, Rockford, IL). MCF-10A cell lysates were prepared in RIPA buffer with protease inhibitor cocktail (Sigma–Aldrich, St Louis, MO). The Pierce BCA Protein Assay kit was used to determine protein concentrations (Pierce, Rockford, IL). Eighty microgram total protein from ACI rat tissues or 30 µg protein from MCF-10A cells was size fractionated on a 12% sodium dodecyl sulfate–polyacrylamide gel, and transferred onto a polyvinylidene difluoride membrane (Millipore Corp., Billerica, MA) under standard conditions (28,29). NRF2, NQO1, SOD3, OGG1, P53 (Santa Cruz Biotechnology) and cleaved PARP (Cell signaling Technology, Danvers, MA) primary antibodies were used individually for immunodetection. Chemiluminescent detection was performed using the BM Chemiluminescence Detection kit (Roche, Indianapolis, IN) and Alpha Innotech FluorChem HD2 (Alpha Innotech, San Leandro, CA) gel documentation system. Membranes were reprobed with α-tubulin antibody (Santa Cruz Biotechnology) using the methods described above. Intensities of the bands were quantified and normalized using AlphaEase FC StandAlone software (version 6.0.0.14; Alpha Innotech).
Methylation-specific PCR
DNA methylation patterns in the CpG islands of the promoter region of the human NRF2 gene were determined by methylation-specific PCR (30). Sodium bisulfite modification of MCF-10A cells’ genomic DNA was performed using the EZ DNA methylation Kit (Zymo Research, Irvine, CA) according to the manufacture’s protocol. Briefly, 500ng of isolated DNA from MCF-10A cells treated with Res (50 µM) in the presence or absence of E2 (50nM) for 24h was used for the sodium bisulfate modification. Freshly prepared CT Conversion Reagent was added and incubated at 50°C for 16h. The modified DNA was purified using Zymo-Spin I Column and eluted in M-Wash Buffer according to manufacturer’s protocol. Primer sequences used for methylation-specific PCR were designed using MethPrimer v1.1 beta software (www.urogene.org/methprimer/index1.html). The primer sequences were as follows: 5′-GGTGGGTAATATTGATTATTTTTTGA-3′ (sense) and 5′-AATATAAACAACTCCAACAACTCATA-3′ (antisense) for unmethylated reaction (PCR product 147bp), and 5′-GGGTGGGT AATATTGATTATTTTTC-3′ (sense) and 5′-ATATAAACAACTCCGACAA CTCGTA-3′ (antisense) for methylated reaction (PCR product 147bp). PCR was carried out in a 25 µl reaction mixture, containing 10× reaction buffer, 2.5mM dNTP mix, primers (0.5 µl each 10 µmol/µl solution), Taq DNA polymerase (Thermo Scientific, Rockford, IL) and 5ng of bisulfite-modified DNA in a GeneAmp PCR System 2400 thermal cycler (Perkin Elmer, San Jose, CA). The cycling conditions consisted of an initial denaturation at 95°C for 5min, 35 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1min, followed by final extension for 10min at 72°C. Amplified products were electrophoresed on 2% agarose gels and visualized under ultraviolet light after staining with ethidium bromide.
Casapase-3/7 activity assay
Caspase-3/7 activities were measured using the Apo-ONE Homogeneous Caspase-3/7 Assay kit (Promega, Madison, WI) according to the manufacturer’s protocol. MCF-10A cells were seeded in triplicate in 96-well white flat bottom cell culture plates (Corning, Lowell, MA) and treated with Res (50 µM) in the presence or absence of E2 (10nM) for 24h. After 24h of treatment, cells were lysed with lysis buffer containing caspase substrate Z-DEVD-R110 and incubated at room temperature for 6h. Fluorescence was measured at an excitation/emission wavelength of 485/535nm.
8-Hydroxydeoxyguanosine estimation
8-Hydroxydeoxyguanosine (8-OHdG), an accepted marker of oxidative-stress-mediated DNA damage, was estimated in E2-, Res- and Res + E2-exposed ACI rat mammary tissues, mammary tumors and MCF-10A cells treated with E2 in the presence or absence of Res using Oxiselect Oxidative DNA damage ELISA kit (Cell Biolabs, San Diego, CA) as described previously (28).
Colony formation assay
Five-hundred MCF-10A cells were seeded in 6-well plates and allowed to grow for 24h in phenol red-free complete media. The cells were then incubated with Res (25 µM) in the presence or absence of E2 (10nM) for 72h, washed in PBS and incubated for an additional 8 days in complete medium. The colonies obtained were washed with PBS and fixed in 10% formalin for 10min and again washed twice with PBS followed by staining with crystal violet (0.1% wt/vol solution in 10% ethanol). The colonies were photographed, counted and compared with respective untreated cells. Each treatment was done in triplicate.
Mammosphere formation assay
Mammosphere formation assay was carried out in ultra low attachment plates (Corning) as described earlier (29). Briefly, 5000 viable MCF-10A cells were seeded into a 24-well plate. Cells were grown in serum-free Dulbecco’s modified Eagle’s medium/F12 (50:50) medium supplemented with 1× B27 (Invitrogen), 20ng/ml epidermal growth factor (Invitrogen), 20ng/ml basic fibroblast growth factor (Invitrogen), 1 µg/ml hydrocortisone (BD Biosciences, Bedford, MA), 5 µg/ml insulin (Invitrogen), penicillin/streptomycin (Lonza, Walkersville, MD) and 4 µg/ml heparin calcium salt (Thermo Scientific) at 37°C under 5% CO2 and incubated with Res (50 µM) in the presence or absence of E2 (10nM). After 6–8 days of incubation, mammospheres were viewed under the microscope and photographed. Three replicate wells from a 24-well plate were used for each experimental condition.
Cell migration assay
Cell migration assay was used to study the metastatic potential of the cells (31). Briefly, a confluent monolayer of MCF-10A cells was established and then a scratch was made through the monolayer, using a standard 200 µl plastic pipet tip, washed twice with PBS and replaced in phenol red-free complete media. The cells were then incubated with Res (50 µM) in the presence or absence of 10nM of E2. Cells migrate into the scratch area as single cells from the confluent sides. After 24h of incubation, the width of the scratch gap was viewed under the microscope and photographed. Three replicate wells from a 6-well plate were used for each experimental condition.
Statistical analyses
Statistical analyses were performed by using Sigma Plot 11.0 (Systat Software, San Jose, CA) and IBM SPSS Statistics 19 software (IBM, Armonk, NY). The average number of tumor nodules per tumor-bearing animal was calculated by dividing the sum of the tumor nodules in all tumor-bearing animals by the total number of tumor-bearing animals. The average number of tumor nodules per rat is expressed as the mean ± SEM. After calculating Kaplan–Meier survival curves of tumor occurrence for latency, we used log-rank test to detect differences in the survival curves between two treatment groups. The unpaired t-test analysis was used to calculate P values for comparisons of mRNA/protein expression levels of NRF2, NQO1, SOD3, OGG1, FMO1 and AOX1 between treated animals and respective age-matched controls. The unpaired t-test was also used to calculate P values for comparisons of miR-93 expression levels between treated animals and respective age-matched controls as well as in MCF-10A cell line. The P values for comparisons of colony formation, caspase-3/7 activity and 8-OHdG levels were also calculated using unpaired t-test. A P < 0.05 was considered significant.
Results
Resveratrol decreases tumor incidence and increases latency of E2-induced mammary tumors
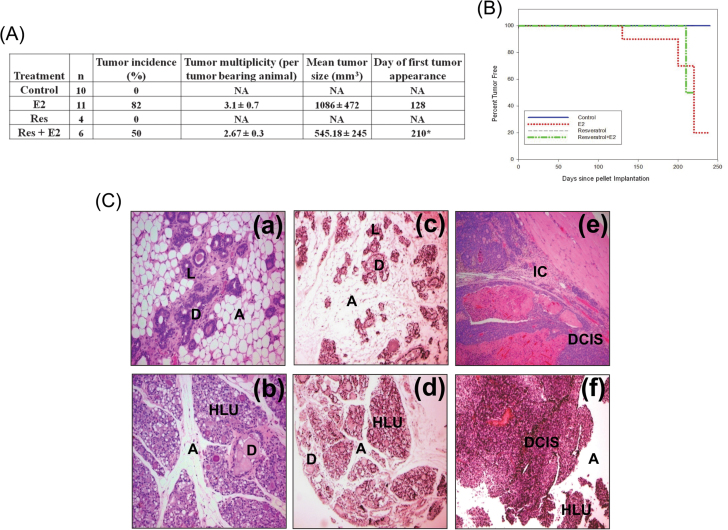
Neither control nor Res-only-treated rats developed mammary tumors. Mammary tumor incidence in rats cotreated with E2 and Res was 50% after 8 months of treatment. The first palpable tumor in this group did not appear until day 210 (Figure 1A). In contrast, in the E2-treated group, the first palpable breast tumors appeared after 128 days of treatment, and mammary tumor incidence was 82% after 8 months of E2 exposure (Figure 1A) (23). Resveratrol treatment significantly increased tumor latency (Figure 1B). In the Res + E2 group, 2.67±0.3 tumor nodule per tumor-bearing animal (tumor multiplicity) was observed, whereas in E2 treatment group, an average of 3.1±0.7 tumor nodules were present in tumor-bearing rats. Although treatment with Res + E2 showed a trend toward decrease in tumor multiplicity compared with the E2 group (Figure 1A), these changes were not statistically significant. Histopathologic examination of mammary tissues from the control and Res groups revealed normal lobular architecture with branched ducts and normal distribution of fat tissue (Figure 1C). Mammary tissues in Res + E2-treated rats showed ductal hyperplasia with increased expansion of terminal lobular units accompanied by compression of and expansion into the surrounding fat tissue (Figure 1C). However, the extent of proliferative/hyperplastic changes in Res + E2-treated mammary tissues was less than the hyperplastic changes observed in rats treated with E2 only (Figure 1C). Although both mammary ductal carcinoma in situ and microinvasive carcinoma were observed in E2-treated rats (Figure 1C), only mammary ductal carcinoma in situ was evident in the rats from Res + E2-treated group (Figure 1C). In most cases, in both E2 and Res + E2 groups, the carcinomas showed mixed morphology, consisting of solid patterns with or without comedo-necrosis. Cribriform and cystic-papillary patterns were also present in some cases. Microscopically, there were no differences in tumor morphology between E2 and Res + E2-treated animals except for invasiveness, which was present only in E2-treated mammary tumors (Figure 1C). Mammary tumors in E2-treated rats showed evidence of invasion of surrounding tissues by breakdown of the basal membrane of the affected tubule-lobular units or ductal units with atypical clusters and poorly formed glands invading the surrounding stroma, whereas animals cotreated with Res + E2 harbored mammary tumors that were confined within the myoepithelium and basement membrane, and non-invasive in nature (Figure 1C).
Fig. 1.
Resveratrol decreases cellular proliferation and tumor incidence and increases latency of E2-induced mammary tumors. Female ACI rats were treated with E2, Res and Res + E2 as described in the Materials and methods. Control animals received cholesterol pellet only. (A) Breast tumor incidence and multiplicity in the rats treated with E2, Res or Res + E2 for 240 days are shown in a tabular form; (B) Kaplan–Meier survival curves for tumor occurrence were plotted for each treatment group, and the log-rank test was used to detect differences in tumor latency curves between groups. Animals in the control or Res-only groups did not develop any tumors; and (C) Histopathologic analyses were performed on mammary tissue of all the rats treated with E2-, Res- or Res + E2. (a) The mammary gland of a representative control ACI rat shows normal lobular architecture (L) with branched ducts (D) and normal distribution of fat tissue/adipocytes (A); (b) E2-treated mammary gland shows increased proliferation with dilated ducts containing inspissated secretions (D) and increased proliferation and expansion of terminal lobular units (HLU) accompanied by compression of and expansion into the surrounding fat tissue/adipocytes (A); (c) the mammary gland of Res-only-treated rats shows normal lobular architecture (L) with branched ducts (D) and normal distribution of fat tissue (A); (d) mammary gland from Res + E2-treated rat displays increased proliferation with dilated ducts containing inspissated secretions (D) and increased proliferation and expansion of terminal lobular units (HLU) accompanied by compression of and expansion into the surrounding fat tissue (A), but the extent of proliferation in this group was lower than the proliferation in E2-treated mammary glands; (e) histomorphologic analysis of mammary tumors from E2-treated rats revealed carcinomas, both in situ (ductal carcinoma in situ) and microinvasive (IC). The carcinomas showed a mixed morphology, with solid patterns with or without comedo-necrosis predominating, but cribriform and cystic-papillary patterns were also present. Microinvasive foci were characterized by breakdown of the basal membrane of the in situ lesion by proliferation of atypical small glands that invaded the surrounding stroma surrounded by a prominent desmoplastic reaction; (f) mammary tumors from animals treated with Res + E2 show ductal carcinoma in situ only. Hyperplastic lobules (HLU) and compressed surrounding fat tissue (A) were also present.
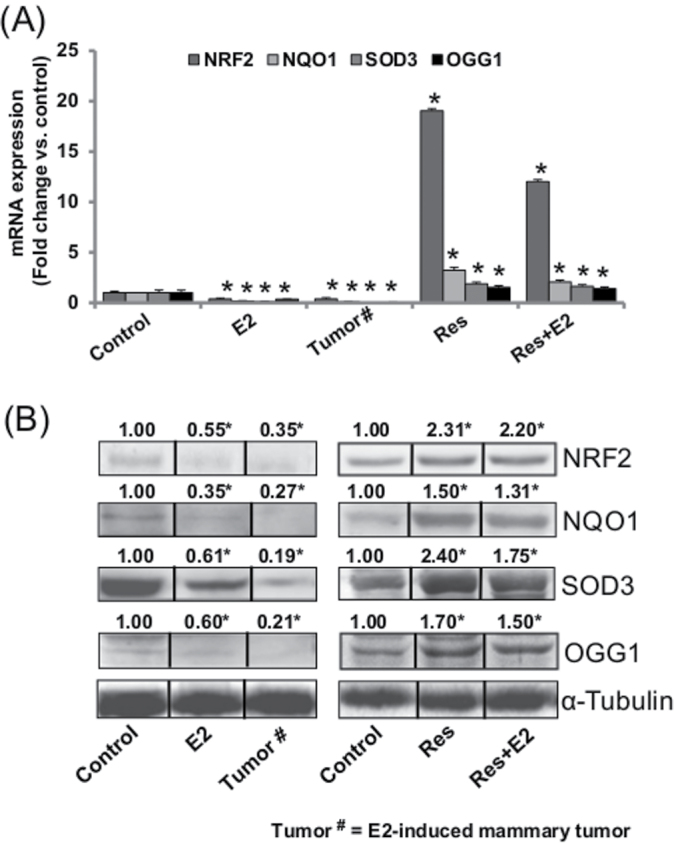
Resveratrol induces NRF2 and NRF2-regulated antioxidant genes and DNA damage repair gene OGG1 in mammary tissues
Nuclear factor erythroid 2-related factor 2 (NRF2) is a known regulator of the antioxidant response (32,33). We examined the mRNA and protein expression levels of NRF2 in E2- and Res + E2-treated mammary tissues and compared it with NRF2 expression in their respective age-matched control mammary tissues. Significant decreases in NRF2 mRNA and protein expression levels in mammary tumors and in mammary tissues of rats treated with E2 for 240 days were determined (Figure 2). Resveratrol alone or in combination with E2 significantly increased mRNA and protein expression of NRF2 compared with its expression in age-matched control mammary tissues (Figure 2). We have earlier reported NRF2-mediated regulation of cancer-protective phase II enzymes like NAD(P)H quinone oxidoreductase 1 (NQO1), superoxide dismutase 3 (SOD3) and oxidative DNA damage repair gene 8-oxoguanine DNA glycosylase 1 (OGG1) in the same animal model (28,29,34). In the present study, we examined the expression of NQO1, SOD3 and OGG1 in E2-induced mammary tumors and in mammary tissues exposed to E2, Res or Res + E2 for 240 days. Similar to NRF2 expression, NQO1, SOD3 and OGG1 mRNA and protein expression levels were significantly decreased in E2-induced mammary tumors and in E2-exposed mammary tissues compared with age-matched mammary tissues from control rats (Figure 2). However, mRNA and protein expression of these genes were significantly increased in Res and Res + E2-treated mammary tissues compared with age-matched mammary tissues from control rats (Figure 2)
Fig. 2.
Resveratrol induces NRF2 and NRF2-regulated antioxidant genes, and DNA damage repair gene OGG1 in mammary tissues. (A and B) NRF2, NQO1, SOD3 and OGG1 mRNA and protein expression in E2-induced mammary tumors and in mammary tissues of rats treated with E2, Res or Res + E2 for 240 days. (A) The bar graph represents a fold change in NRF2, NQO1, SOD3 and OGG1 mRNA expression (mean ± SEM) in mammary tumors and in mammary tissues from at least four different animals compared to age-matched control mammary tissues. (B) Representative western blots are presented to show NRF2, NQO1, SOD3 and OGG1 protein expression in mammary tumors and in mammary tissues of rats treated with E2, Res or Res + E2 for 240 days. The mean fold change from at least three different animals from each treatment group compared to age-matched control mammary tissues is given at the top of each blot. Vertical black lines indicate that intervening lanes have been removed. Each individual western blot panel compares signals from protein samples loaded on the same gel. ‘*’ indicates P value <0.05 compared with respective controls.
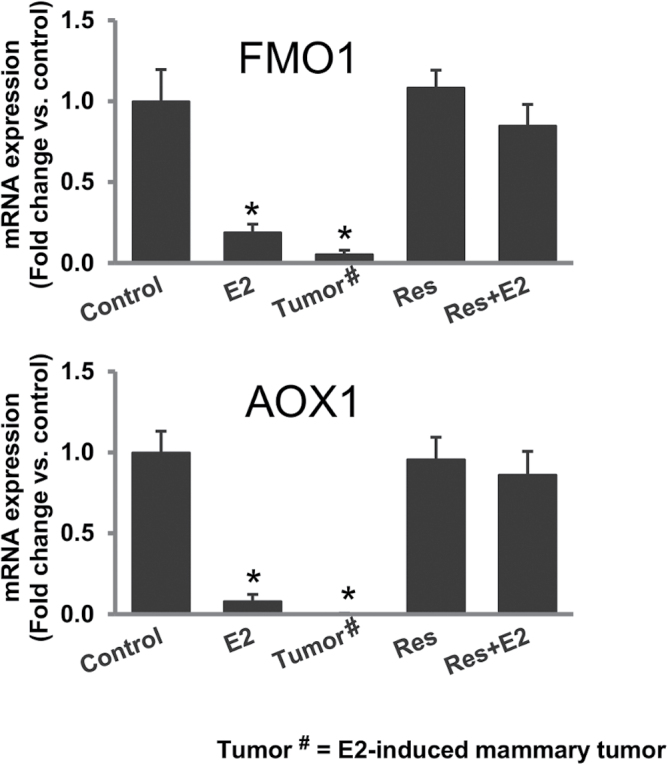
Resveratrol protects against estrogen-mediated inhibition of detoxification genes
NRF2 is known as an important regulator of genes involved in detoxification mechanisms in the biological system (28,32,35). Estrogen itself alters the expression of genes involved in detoxification of xenobiotics (28,36). We examined the effect of E2 treatment on the expression of detoxification genes flavin monooxygenase 1 (FMO1) and aldehyde oxidase 1 (AOX1) in mammary tissues and the role, if any, Res treatment can play in reversal of E2-mediated changes in expression of these detoxification genes. We measured the mRNA expression of two NRF2-regulated detoxification genes FMO1 and AOX1 by real-time PCR. Estrogen treatment significantly decreased mRNA expression of FMO1 and AOX1 in mammary tissues and in mammary tumors compared with age-matched mammary tissues from control rats (Figure 3), whereas Res treatment alone or in combination with E2 reverted the mRNA expression levels of these genes to the control levels (Figure 3).
Fig. 3.
Resveratrol protects against estrogen-mediated inhibition of detoxification genes. FMO1 and AOX1 mRNA expression in E2-exposed mammary tumors and in mammary tissues of rats treated with E2, Res or Res + E2 for 240 days. The bar graphs represent fold changes in FMO1 and AOX1 mRNA expression (mean ± SEM) in mammary tumors and in mammary tissues from at least four different animals compared with age-matched control mammary tissues. * indicates P value of <0.05 compared with respective controls.
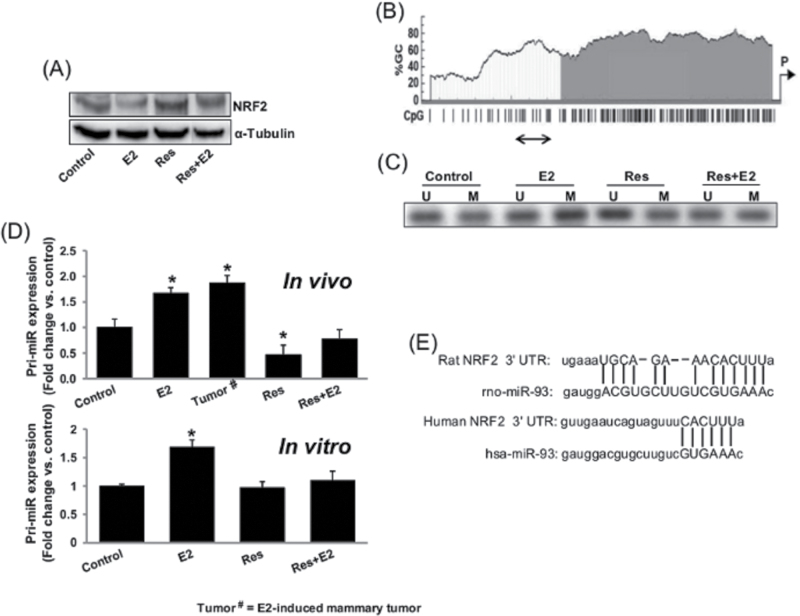
Resveratrol epigenetically regulates NRF2 expression
Canonically estrogen acts through estrogen receptor-mediated signaling pathways; however, estrogen is also known to alter the gene expression epigenetically (37). We demonstrated E2-mediated downregulation of NRF2 and NRF2-responsive genes in female ACI rats (Figure 2), which is an estrogen and progesterone receptor positive breast cancer model (36,38), but the downregulation of NRF2 expression in estrogen receptor negative human non-neoplastic breast epithelial cell line MCF-10A indicated an estrogen receptor independent mechanism of regulation of NRF2 (Figure 4A). Therefore, we examined whether E2 epigenetically regulates expression of NRF2. We treated estrogen receptor negative MCF-10A cells with E2 in the presence or absence of Res for 24h and isolated genomic DNA. After bisulfite modification of genomic DNA from MCF-10A cells, methylation-specific PCR was carried out to determine changes in methylation pattern of human NRF2 promoter. Human NRF2 promoter sequence has CpG-rich islands as determined by MethPrimer software (Figure 4B). Estrogen treatment increased methylation of NRF2 promoter compared with control as revealed by an intense band of PCR product with methylated primer pairs (E2-M; Figure 4C), whereas Res alone or in combination with E2 reversed E2-mediated changes in the methylation status of NRF2 promoter (Figure 4C).
Fig. 4.
Resveratrol epigenetically regulates NRF2 expression. (A) Representative western blot showing NRF2 expression in MCF-10A cells treated with Res (50 µM) in the presence or absence of E2 (10nM) for 24h; (B) CpG-rich islands in NRF2 promoter as determined by MethPrimer software are shown. The promoter region amplified by methylation-specific PCR after bisulphite treatment is shown by a double-headed arrow; (C) Representative picture of agarose gel electrophoresis from methylation-specific PCRs with unmodified and bisulphite-modified genomic DNA from MCF-10A cells treated with Res (50 µM) in the presence or absence of E2 (10nM) for 24h is shown. U = PCR product with unmethylated DNA-specific primer pair; M = PCR product with methylated DNA-specific primer pair; (D) Real-time PCR analyses of pri-miR-93 expression in mammary tumors and in mammary tissues of rats treated with E2, Res and Res + E2 for 240 days and in MCF-10A cells after E2 (10nM), Res (50 µM) and Res + E2 treatment for 24h are carried out. The bar graphs represent a fold change in pri-miR-93 expression (mean ± SEM) in mammary tumors and in mammary tissues from at least four different animals compared with age-matched control mammary tissues (in vivo) and from at least five MCF-10A samples treated with E2 in the presence or absence of Res compared with vehicle-treated control samples (in vitro). * indicates P value of <0.05 compared with respective controls; and (E) Schematic of rat and human NRF2 mRNA 3′-UTR containing potential miR-93 binding site.
MicroRNAs (miRNAs) are small non-coding RNAs that epigenetically regulate the expression of ~60% of all human genes and play important roles in disease processes (39,40). We have recently reported E2-mediated inhibition of NRF2 through upregulation of miR-93 in E2-induced ACI rat mammary tissues and mammary tumors (31). In the present study, we examined the role of Res in E2-mediated changes in miR-93 expression and thus NRF2 expression in mammary tissues. We analyzed expression of miR-93 in rat mammary tumors, mammary tissues and MCF-10A cells by quantitative real-time PCR. Real-time PCR data demonstrated that E2 treatment significantly increased expression of miR-93 in rat mammary tissues (~1.7-fold) and in E2-induced mammary tumors (~1.9-fold) compared with age-matched control mammary tissues, whereas Res treatment alone or in combination with E2 did not increase expression levels of this miRNA compared with control levels (Figure 4D). In MCF-10A cells, E2 treatment significantly increased expression of miR-93 (~1.7-fold) compared with controls and its expression remained at control levels in Res alone or Res + E2-treated MCF-10A cells (Figure 4D).
Resveratrol induces apoptosis and inhibits oxidative DNA damage and mammary carcinogenic process
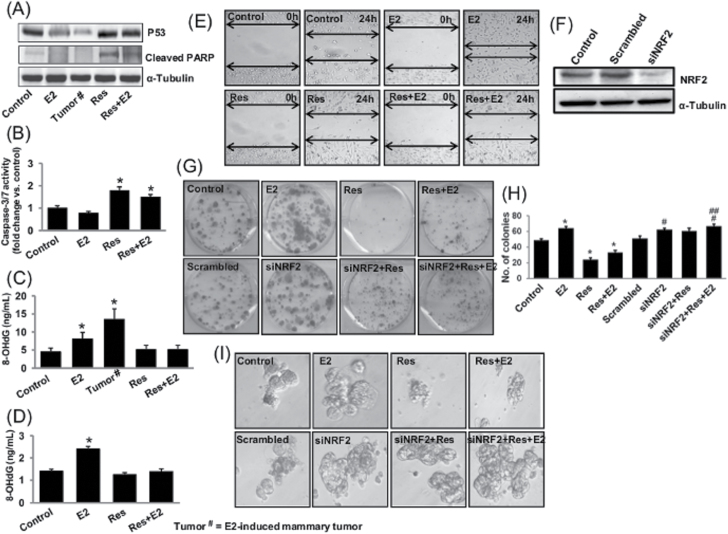
To demonstrate the protective role of Res against E2-mediated carcinogenic process, we examined the effect of Res on apoptosis and oxidative DNA damage in rat mammary tissues and performed colony formation, mammosphere formation and wound healing assays using MCF-10A cells. Resveratrol is known to induce caspases, PARP degradation and induction of apoptosis in p53-dependent fashion (9,10). Expression of p53 and PARP cleavage was decreased in E2-treated mammary tissues and in mammary tumors (Figure 5A), but increased expression of p53 and PARP cleavage in Res- and Res + E2-treated mammary tissues indicated Res-mediated induction of p53-dependent apoptosis (Figure 5A). Similarly, we also observed a decrease in caspase-3/7 activity in E2-treated MCF-10A cells, whereas Res in the presence or absence of E2 significantly increased activities of these caspases compared with their activities in control samples (Figure 5B). DNA 8-OHdG levels were quantified as a marker of oxidative DNA damage in mammary tissues and in MCF-10A cell line. About 2- and 3-fold increase (P < 0.05) in 8-OHdG levels was demonstrated in E2-treated mammary tissues and in E2-induced mammary tumors, respectively, compared with control mammary tissues (Figure 5C). No significant difference in 8-OHdG levels was detected in mammary tissue DNA samples from Res- and Res + E2-treated groups compared with control tissue (Figure 5C). Treatment of MCF-10A cells with E2 for 48h significantly increased 8-OHdG levels compared with vehicle-treated controls (Figure 5D). 8-OHdG levels remained unchanged in Res- or Res + E2-treated MCF-10A cells relative to vehicle-treated cells (Figure 5D). Although E2 treatment increased the number of colonies, mammosphere formation and wound healing potential of MCF-10A cells compared with vehicle-treated MCF-10A cells (Figure 5E, G–I), Res treatment alone or in combination with E2 inhibited all these carcinogenic processes compared with E2- or vehicle-treated MCF-10A cells (Figure 5E, G–I). However, in NRF2-silenced MCF-10A cells, Res treatment could not inhibit colony as well as mammosphere formation (Figure 5G–I). Moreover, silencing of NRF2 itself increased colony and mammosphere formation in MCF-10A Cells (Figure 5G–I). Collectively, silencing of NRF2 in MCF-10A cells abrogated anticarcinogenic effects of Res (Figure 5G–I).
Fig. 5.
Resveratrol induces apoptosis and inhibits oxidative DNA damage and mammary carcinogenesis. Female ACI rats were treated with E2, Res and Res + E2 for 240 days as described in the Materials and methods. (A) Representative western blot showing Res-mediated induction of p53 and cleaved PARP as an indicator of induction of apoptosis in rat mammary tissues; (B) Caspase-3/7 activities in MCF-10A cells; (C) levels of 8-OHdG in mammary tumors and in mammary tissues; and (D) levels of 8-OHdG in MCF-10A cells treated with Res (50 µM) in the presence or absence of E2 (10nM); (E) Representative pictures from wound healing assays with MCF-10A cells treated with Res (50 µM) in the presence or absence of E2 (10nM); (F) western blot showing small-interfering RNA-mediated silencing of NRF2 in MCF-10A cells; (G and H) Representative pictures showing clonogenic potential of the wild-type MCF-10A and siNRF2-transfected MCF-10A cells in the presence or absence of Res (25 µM) and E2 (10nM) (G); Colonies from each treatment group were counted and are presented as bar graph (H); (I) Representative pictures showing mammosphere formation ability of the wild-type MCF-10A and siNRF2-transfected MCF-10A cells in the presence or absence of Res (50 µM) and E2 (10nM). * and # indicate P value of <0.05 compared with respective control and scrambled-transfected MCF-10A cells, respectively. ## indicates P value of <0.05 compared with siNRF2-transfected MCF-10A cells.
Discussion
Estrogens have been implicated in the development of breast cancer (2,23). However, the exact mechanisms by which estrogens exert their carcinogenic effects remain elusive. A growing body of recent literature strongly supports the role of estrogen-metabolism-mediated oxidative stress in estrogen-induced breast carcinogenesis (2,3,23,24,27). Metabolic redox cycling between catechol estrogens and their corresponding quinones generates oxidative stress (3). Induction of antioxidant and/or detoxification enzymes by natural compounds is one of the approaches that can be used to inhibit the overburden of oxidative stress in the estrogen-induced carcinogenic process (28,29).
In light of the chemoprotective properties attributed to Res by recent epidemiological and animal studies, we examined the preventive role of Res and the possible mechanisms by which Res may exert its anticancer effects against estrogen-induced breast cancer. One such mechanism may involve Res’s ability to induce NRF2 and thus its downstream antioxidant genes. Transcription factor NRF2-regulated phase II enzymes protect against the development of cancer by catalyzing reactions that convert highly reactive, carcinogenic chemicals to less reactive products (35,41). In the present study, we investigated the role of Res in prevention against E2-mediated inhibition of antioxidant mechanisms, oxidative DNA damage and subsequent breast carcinogenesis in female ACI rats. The female ACI rat model is an established animal model of hormonal carcinogenesis (22–27). It shares many features of human breast cancer such as genomic instability, increased oxidant stress, DNA damage, etc. (22,23,28,42,43). We have demonstrated that cotreatment with Res inhibited E2-induced breast tumor incidence by 50%, increased tumor latency and decreased tumor size and multiplicity in ACI rats (Figure 1A). We also demonstrated that Res inhibits E2-mediated proliferation of mammary tissues (Figure 1C).
Resveratrol is known to have strong antioxidant activities (44,45), and NRF2 is considered as master regulator of antioxidant response (35,41). Therefore, we examined expression of NRF2 in mammary tumors and mammary tissues of E2-, Res- and Res + E2-treated rats. We demonstrated significant suppression of NRF2 mRNA and protein expression following long-term E2 treatment (240 days), and this suppression was reversed upon cotreatment with Res (Figure 2). NRF2 is a transcription factor that regulates the transcription of cancer-protective phase II enzymes like NQO1, SOD3 and oxidative DNA damage repair gene OGG1 (28,29,34). Decreased mRNA and protein expression of all these genes in E2-treated mammary tumors and mammary tissues and increased expression in Res- and Res + E2-treated mammary tissues confirmed E2-mediated inhibition and Res-mediated upregulation of NRF2 in mammary tissues (Figure 2). We have recently reported the importance of NRF2-regulated NQO1, SOD3 and OGG1 in prevention of E2-induced breast cancer (28,29,34).
AOX1 is a member of the molybd-flavo enzyme family that is involved in the detoxification of a variety of aldehydes and nitrogenous heterocyclic compounds. Recently, AOX1 has emerged as an important diagnostic/prognostic marker for cancer (46,47). Literature evidence also supports cancer preventive role of flavin-containing monooxygenases (48). Flavin-containing monooxygenases are a polymorphic family of enzymes that catalyze the oxidation of nucleophilic heteroatom substances to their respective oxides. The most common substrates for flavin-containing monooxygenase are sulfur- and nitrogen-containing xenobiotics (49). Both FMO1 and AOX1 are NRF2-regulated phase I detoxification genes (50,51). Downregulation of gene expression of FMO1 and AOX1 in E2-treated mammary tissues and in mammary tumors and inhibition of suppression of these genes in Res- and Res + E2-treated mammary tissues point toward the role of these genes in Res-mediated protection against E2-mediated breast carcinogenesis. Thus, prevention of expression of these genes in Res-treated mammary tissues indicates a chemopreventive role of Res during breast carcinogenesis via regulation of NRF2 (Figure 3).
Furthermore, the mechanism by which Res inhibits proliferation of mammary tissues is most likely through upregulation of NRF2, as redox status of the system plays an important role in the regulation of proliferation of cells including stem cells (52). NRF2 is suggested to regulate redox control mechanism (32,35,41), and dampening of its expression by E2 might be involved in reduced overall redox regulation by impacting the expression of reactive oxygen species scavenging/detoxifying genes NQO1 and SOD3, whose expression is dependent on NRF2 (28,41). Treatment with antioxidants N-acetyl-cysteine and glutathione has been shown to be sufficient to limit the cells’ proliferation potential (53). NRF2 has also been shown to be involved in inhibition of cell growth by inducing cell cycle arrest (54). Therefore, it may be logical to suggest that decreased NRF2 level and corresponding increased oxidative stress in E2-treated mammary tissues and in mammary tumors might be one of the possible mechanisms of E2-mediated increased proliferation, whereas Res treatment prevents mammary cells proliferation potential by inducing expression of NRF2 and thus inhibiting oxidative stress in mammary tissues.
Physical interaction of estrogen receptor alpha (ERα) and NRF2 has been suggested as one of the mechanisms of E2-mediated inhibition of NRF2-responsive genes in ERα positive cell lines (55). Epigenetic regulation of NRF2 has also been previously established (56). Estrogen-mediated downregulation of NRF2 in ER and PR positive ACI rat mammary tissues as well as in ER negative human breast cell line MCF-10A suggests the possibility that an ER-independent mechanism might be involved in the regulation of NRF2 (Figures 2 and 4A). Methylation-specific PCRs with E2-, Res- and Res + E2-treated DNA samples of MCF-10A cells indicate alterations in methylation status of NRF2 promoter region (Figure 4C) that suggest toward epigenetic regulation of NRF2 during E2-induced breast carcinogenesis. Although E2 treatment led to increased promoter methylation, cotreatment with Res reversed E2-mediated changes in methylation pattern of NRF2 promoter (Figure 4C).
miRNAs are another important epigenetic regulators of the expression of human genes and play important roles in disease processes (39,40). Increased expression of NRF2-regulating miRNA miR-93 in E2-treated mammary tissues, mammary tumors, E2-treated MCF-10A cells and reversal of E2-mediated change in the expression of this miRNA in Res- and Res + E2-treated mammary tissues, and in MCF-10A cells indicates yet another epigenetic mechanism of regulation of NRF2 during E2-induced breast carcinogenesis (Figure 4D).
Resveratrol is known to induce apoptosis (9,10). We demonstrated that Res induces activities of caspases and apoptosis in vitro as well in rat mammary tissues in p53-dependent fashion (Figure 5A and B). We also demonstrated a significant increase in 8-OHdG levels in E2-treated mammary tissues, mammary tumors and MCF-10A cells. Further, we demonstrated no change in 8-OHdG levels in Res- and Res + E2-treated mammary tissues and in MCF-10A cells compared with respective controls (Figure 5C and D). The increased level of 8-OHdG is considered a critical marker of oxidative stress and carcinogenesis (57). Increased colony formation and mammosphere formation in E2-treated MCF-10A cells and a decrease in Res and Res + E2-treated MCF-10A cells compared with vehicle-treated MCF-10A cells further confirm the anticarcinogenic role of Res (Figure 5G–I). Resveratrol treatment in the absence of NRF2 (siNRF2-mediated silencing) was unable to inhibit carcinogenic processes (Figure 5G–I). Moreover, cotreatment of Res and E2 in the absence of NRF2 further increased colony as well as mammosphere formation (Figure 5G–I). Because E2 treatment leads to inhibition of NRF2 expression (Figure 2), increase in colony and mammosphere formation might be due to E2-mediated further inhibition of NRF2 in siNRF2-transfected cells. Abrogation of protective effects of Res on the colony as well as mammosphere formation after silencing of NRF2 suggest that Res inhibits carcinogenic processes through induction of NRF2-mediated protective pathways (Figure 5G–I). Overall, these findings support the role of Res in the prevention of mammary carcinogenesis. Our studies provide direct evidences for NRF2-mediated protective role of Res against E2-induced breast carcinogenesis. Thus, Res may serve as a potential chemopreventive agent in the development of therapeutic strategies for the prevention of estrogen-induced breast neoplasia.
Funding
National Institutes of Health Grant (CA 109551); the University of Missouri Research Board Grant (H.K.B.); the School of Pharmacy, University of Missouri-Kansas City.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- 8-OHdG
8-hydroxydeoxyguanosine
- ACI
August Copenhagen Irish
- E2
17β-estradiol
- miRNA
microRNA
- NRF2
nuclear factor erythroid 2-related factor 2
- PARP
poly adenosine diphosphate ribose polymerase
- PBS
phosphate-buffered saline
- Res
resveratrol.
References
- 1. DeSantis C., et al. (2014). Breast cancer statistics, 2013. CA Cancer J. Clin., 64, 52–62 [DOI] [PubMed] [Google Scholar]
- 2. Bhat H.K., et al. (2003). Critical role of oxidative stress in estrogen-induced carcinogenesis. Proc. Natl Acad. Sci. USA, 100, 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavalieri E.L., et al. (1997). Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc. Natl Acad. Sci. USA, 94, 10937–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. NTP. (2002). Report on Carcinogens. National Toxicology Program, Research Triangle Park, NC [Google Scholar]
- 5. Yager J.D., et al. (2006). Estrogen carcinogenesis in breast cancer. N. Engl. J. Med., 354, 270–282 [DOI] [PubMed] [Google Scholar]
- 6. Mense S.M., et al. (2008). Phytoestrogens and breast cancer prevention: possible mechanisms of action. Environ. Health Perspect., 116, 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sirtori C.R., et al. (2005). Phytoestrogens: end of a tale? Ann. Med., 37, 423–438 [DOI] [PubMed] [Google Scholar]
- 8. Saiko P., et al. (2008). Resveratrol and its analogs: defense against cancer, coronary disease and neurodegenerative maladies or just a fad? Mutat. Res., 658, 68–94 [DOI] [PubMed] [Google Scholar]
- 9. Alkhalaf M. (2007). Resveratrol-induced apoptosis is associated with activation of p53 and inhibition of protein translation in T47D human breast cancer cells. Pharmacology, 80, 134–143 [DOI] [PubMed] [Google Scholar]
- 10. Alkhalaf M., et al. (2008). Resveratrol-induced apoptosis in human breast cancer cells is mediated primarily through the caspase-3-dependent pathway. Arch. Med. Res., 39, 162–168 [DOI] [PubMed] [Google Scholar]
- 11. Banerjee S., et al. (2002). Suppression of 7,12-dimethylbenz(a)anthracene-induced mammary carcinogenesis in rats by resveratrol: role of nuclear factor-kappaB, cyclooxygenase 2, and matrix metalloprotease 9. Cancer Res., 62, 4945–4954 [PubMed] [Google Scholar]
- 12. Bhat K.P., et al. (2001). Estrogenic and antiestrogenic properties of resveratrol in mammary tumor models. Cancer Res., 61, 7456–7463 [PubMed] [Google Scholar]
- 13. Whitsett T., et al. (2006). Resveratrol, but not EGCG, in the diet suppresses DMBA-induced mammary cancer in rats. J. Carcinog., 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlachterman A., et al. (2008). Combined resveratrol, quercetin, and catechin treatment reduces breast tumor growth in a nude mouse model. Transl. Oncol., 1, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garvin S., et al. (2006). Resveratrol induces apoptosis and inhibits angiogenesis in human breast cancer xenografts in vivo . Cancer Lett., 231, 113–122 [DOI] [PubMed] [Google Scholar]
- 16. Subramanian L., et al. (2010). Resveratrol: challenges in translation to the clinic–a critical discussion. Clin. Cancer Res., 16, 5942–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel K.R., et al. (2013). Sulfate metabolites provide an intracellular pool for resveratrol generation and induce autophagy with senescence. Sci. Transl. Med., 5, 205ra133. [DOI] [PubMed] [Google Scholar]
- 18. Gehm B.D., et al. (1997). Resveratrol, a polyphenolic compound found in grapes and wine, is an agonist for the estrogen receptor. Proc. Natl Acad. Sci. USA, 94, 14138–14143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Corre L., et al. (2005). Resveratrol and breast cancer chemoprevention: molecular mechanisms. Mol. Nutr. Food Res., 49, 462–471 [DOI] [PubMed] [Google Scholar]
- 20. Niles R.M., et al. (2006). Resveratrol is rapidly metabolized in athymic (nu/nu) mice and does not inhibit human melanoma xenograft tumor growth. J. Nutr., 136, 2542–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato M., et al. (2003). Prepubertal resveratrol exposure accelerates N-methyl-N-nitrosourea-induced mammary carcinoma in female Sprague-Dawley rats. Cancer Lett., 202, 137–145 [DOI] [PubMed] [Google Scholar]
- 22. Li S.A., et al. (2002). Prevention of solely estrogen-induced mammary tumors in female aci rats by tamoxifen: evidence for estrogen receptor mediation. J. Endocrinol., 175, 297–305 [DOI] [PubMed] [Google Scholar]
- 23. Mense S.M., et al. (2008). Estrogen-induced breast cancer: alterations in breast morphology and oxidative stress as a function of estrogen exposure. Toxicol. Appl. Pharmacol., 232, 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mense S.M., et al. (2009). Vitamin C and alpha-naphthoflavone prevent estrogen-induced mammary tumors and decrease oxidative stress in female ACI rats. Carcinogenesis, 30, 1202–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shull J.D., et al. (1997). Ovary-intact, but not ovariectomized female ACI rats treated with 17beta-estradiol rapidly develop mammary carcinoma. Carcinogenesis, 18, 1595–1601 [DOI] [PubMed] [Google Scholar]
- 26. Singh B., et al. (2010). Dietary quercetin exacerbates the development of estrogen-induced breast tumors in female ACI rats. Toxicol. Appl. Pharmacol., 247, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh B., et al. (2009). Antioxidant butylated hydroxyanisole inhibits estrogen-induced breast carcinogenesis in female ACI rats. J. Biochem. Mol. Toxicol., 23, 202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh B., et al. (2012). Superoxide dismutase 3 is induced by antioxidants, inhibits oxidative DNA damage and is associated with inhibition of estrogen-induced breast cancer. Carcinogenesis, 33, 2601–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh B., et al. (2012). Induction of NAD(P)H-quinone oxidoreductase 1 by antioxidants in female ACI rats is associated with decrease in oxidative DNA damage and inhibition of estrogen-induced breast cancer. Carcinogenesis, 33, 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chmelarova M., et al. (2013). Methylation in the p53 promoter in epithelial ovarian cancer. Clin. Transl. Oncol., 15, 160–163 [DOI] [PubMed] [Google Scholar]
- 31. Singh B., et al. (2013). MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis, 34, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kensler T.W., et al. (2007). Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol., 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 33. Li W., et al. (2009). Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog., 48, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh B., et al. (2013). Antioxidant-mediated up-regulation of OGG1 via NRF2 induction is associated with inhibition of oxidative DNA damage in estrogen-induced breast cancer. BMC Cancer, 13, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reddy N.M., et al. (2007). Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol. Genomics, 32, 74–81 [DOI] [PubMed] [Google Scholar]
- 36. Singh B., et al. (2011). Partial inhibition of estrogen-induced mammary carcinogenesis in rats by tamoxifen: balance between oxidant stress and estrogen responsiveness. PLoS One, 6, e25125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh K.P., et al. (2012). DNA demethylation by 5-aza-2-deoxycytidine treatment abrogates 17 beta-estradiol-induced cell growth and restores expression of DNA repair genes in human breast cancer cells. Cancer Lett., 316, 62–69 [DOI] [PubMed] [Google Scholar]
- 38. Weroha S.J., et al. (2006). Overexpression of cyclins D1 and D3 during estrogen-induced breast oncogenesis in female ACI rats. Carcinogenesis, 27, 491–498 [DOI] [PubMed] [Google Scholar]
- 39. Friedman R.C., et al. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res., 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kloosterman W.P., et al. (2006). The diverse functions of microRNAs in animal development and disease. Dev. Cell, 11, 441–450 [DOI] [PubMed] [Google Scholar]
- 41. Kensler T.W., et al. (2010). Nrf2: friend or foe for chemoprevention? Carcinogenesis, 31, 90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arnerlöv C., et al. (2001). Intratumoral variations in DNA ploidy and s-phase fraction in human breast cancer. Anal. Cell. Pathol., 23, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li J.J., et al. (2004). Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc. Natl Acad. Sci. USA, 101, 18123–18128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carrizzo A., et al. (2013). Antioxidant effects of resveratrol in cardiovascular, cerebral and metabolic diseases. Food Chem. Toxicol, 61, 215–226 [DOI] [PubMed] [Google Scholar]
- 45. Wood L.G., et al. (2010). Antioxidant and anti-inflammatory effects of resveratrol in airway disease. Antioxid. Redox Signal., 13, 1535–1548 [DOI] [PubMed] [Google Scholar]
- 46. Haldrup C., et al. (2013). DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J. Clin. Oncol., 31, 3250–3258 [DOI] [PubMed] [Google Scholar]
- 47. Varisli L. (2013). Identification of new genes downregulated in prostate cancer and investigation of their effects on prognosis. Genet. Test. Mol. Biomarkers, 17, 562–566 [DOI] [PubMed] [Google Scholar]
- 48. Wojcieszyńska D., et al. (2012). Flavin-dependent enzymes in cancer prevention. Int. J. Mol. Sci., 13, 16751–16768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ziegler D.M. (2002). An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab. Rev., 34, 503–511 [DOI] [PubMed] [Google Scholar]
- 50. Li J., et al. (2005). Dissecting tBHQ induced ARE-driven gene expression through long and short oligonucleotide arrays. Physiol. Genomics, 21, 43–58 [DOI] [PubMed] [Google Scholar]
- 51. Maeda K., et al. (2012). Aldehyde oxidase 1 gene is regulated by Nrf2 pathway. Gene, 505, 374–378 [DOI] [PubMed] [Google Scholar]
- 52. Hochmuth C.E., et al. (2011). Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell, 8, 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Buchon N., et al. (2009). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev., 23, 2333–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Havens C.G., et al. (2006). Regulation of late G1/S phase transition and APC Cdh1 by reactive oxygen species. Mol. Cell. Biol., 26, 4701–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ansell P.J., et al. (2005). Repression of cancer protective genes by 17beta-estradiol: ligand-dependent interaction between human Nrf2 and estrogen receptor alpha. Mol. Cell. Endocrinol., 243, 27–34 [DOI] [PubMed] [Google Scholar]
- 56. Yu S., et al. (2010). Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One, 5, e8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Valavanidis A., et al. (2009). 8-hydroxy-2’ -deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health. C. Environ. Carcinog. Ecotoxicol. Rev., 27, 120–139 [DOI] [PubMed] [Google Scholar]