Summary
Although biosynthetic gene clusters (BGCs) have been discovered for hundreds of bacterial metabolites, our knowledge of their diversity remains limited. Here, we used a novel algorithm to systematically identify BGCs in the extensive extant microbial sequencing data. Network analysis of the predicted BGCs revealed large gene cluster families, the vast majority uncharacterized. We experimentally characterized the most prominent family, consisting of two subfamilies of hundreds of BGCs distributed throughout the Proteobacteria; their products are aryl polyenes, lipids with an aryl head group conjugated to a polyene tail. We identified a distant relationship to a third subfamily of aryl polyene BGCs, and together the three subfamilies represent the largest known family of biosynthetic gene clusters, with more than 1,000 members. Although these clusters are widely divergent in sequence, their small molecule products are remarkably conserved, indicating for the first time the important roles these compounds play in Gram-negative cell biology.
Introduction
Microbial natural products are widely used in human and veterinary medicine, agriculture, and manufacturing, and are known to mediate a variety of microbe-host and microbe-microbe interactions. Connecting these natural products to the genes that encode them is revolutionizing their study, enabling genome sequence data to guide the discovery of new molecules (Bergmann et al., 2007; Challis, 2008; Franke et al., 2012; Freeman et al., 2012; Kersten et al., 2011; Laureti et al., 2011; Lautru et al., 2005; Letzel et al., 2012; Nguyen et al., 2008; Oliynyk et al., 2007; Schneiker et al., 2007; Walsh and Fischbach, 2010; Winter et al., 2011). The thousands of prokaryotic genomes in sequence databases provide an opportunity to generalize this approach through the identification of biosynthetic gene clusters (BGCs): sets of physically clustered genes that encode the biosynthetic enzymes for a natural product pathway.
Besides core biosynthetic enzymes, many BGCs also harbor enzymes to synthesize specialized monomers for a pathway. For example, the erythromycin gene cluster encodes a set of enzymes for biosynthesis of two deoxysugars, d-desosamine and l-mycarose, that are appended to the polyketide aglycone (Oliynyk et al., 2007; Staunton and Weissman, 2001), while BGCs for glycopeptide antibiotics contain enzymes to synthesize the nonproteinogenic amino acids β-hydroxytyrosine, 4-hydroxyphenylglycine, and 3,5-dihydroxyphenylglycine that their core nonribosomal peptide synthetases use in the assembly of their peptidic scaffolds (Kahne et al., 2005; Pelzer et al., 1999). In many cases, transporters, regulatory elements, and genes that mediate host resistance are also contained within the BGC (Walsh and Fischbach, 2010). Although some BGCs are so well understood that the biosynthesis of their small molecule product has been reconstituted in heterologous hosts (Pfeifer et al., 2001) or in vitro using purified enzymes (Lowry et al., 2013; Sattely et al., 2008), little is known about the vast majority of BGCs, even those that have been connected to a small molecule product.
Here, we report the results of a systematic effort to identify and categorize BGCs in 1,154 sequenced genomes spanning the prokaryotic tree of life. We envisioned that the resulting ‘global map’ of biosynthesis would enable BGCs to be systematically selected for characterization by searching for, e.g., biosynthetic novelty, presence in undermined taxa, or patterns of phylogenetic distribution that indicate functional importance. Surprisingly, the map revealed large and very widely distributed BGC families of unknown function. We experimentally characterized the most prominent of these families, leading to the unexpected finding that gene clusters responsible for producing aryl polyene carboxylic acids constitute the largest BGC family in the sequence databases.
Results and Discussion
The ClusterFinder algorithm detects BGCs of both known and unknown classes
Several algorithms have been developed for the automated prediction of BGCs in microbial genomes (Khaldi et al., 2010; Li et al., 2009; Medema et al., 2011; Starcevic et al., 2008; Weber et al., 2009), but each of these tools is limited to the detection of one or more well-characterized gene cluster classes. As a more general solution to the gene cluster identification problem, we developed a hidden Markov model-based probabilistic algorithm, ClusterFinder, that aims to identify gene clusters of both known and unknown classes. ClusterFinder is based on a training set of 732 BGCs with known small molecule products that we compiled and manually curated (SI Table I). To scan a genome for BGCs, it converts a nucleotide sequence into a string of contiguous Pfam domains and assigns each domain a probability of being part of a gene cluster, based on the frequencies at which these domains occur in the BGC and nonBGC training sets, and the identities of neighboring domains (Figure 1a, Experimental Procedures). Since ClusterFinder is based solely on Pfam domain frequencies, and Nature uses distinct assemblages of the same enzyme superfamilies to construct unrelated natural product classes, ClusterFinder exhibits relatively little training set bias and is capable of identifying new classes of gene clusters effectively (See Experimental Procedures for a detailed description of how we validated ClusterFinder).
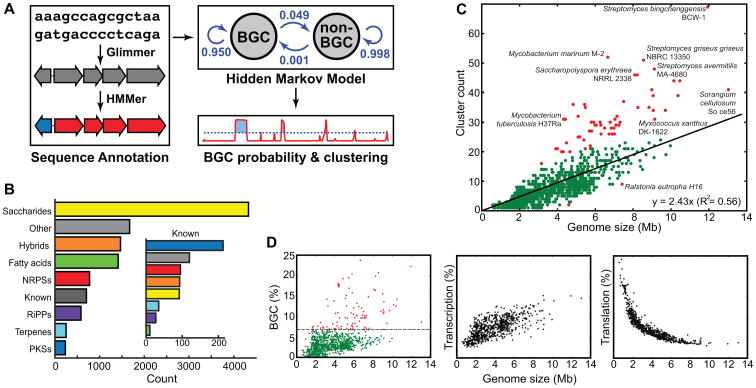
Figure 1. ClusterFinder flowchart and distribution of BGC classes and counts.
a, Flowchart of the four-step BGC prediction pipeline: (i) annotation of a genome sequence and compression to a string of Pfam domains, (ii) calculation of posterior probabilities of a BGC hidden state, (iii) clustering of genes that contain Pfam domain(s) with posterior probabilities of BGC hidden state above the threshold, and (iv) annotation of the predicted BGCs using an expanded version of the antiSMASH algorithm. b, Distribution of BGC classes for known (inset) and predicted BGCs. “Other” gene clusters include gene clusters from other known classes as well as a manually curated set of 1,024 putative gene clusters that fall outside known biosynthetic classes. Unexpectedly, 40% of all predicted BGCs encode saccharides, more than twice the size of the next largest class. c, Number of predicted BGCs by genome size. Most bacterial species follow a linear trend (the equation in the bottom-right corner); outliers (defined as having residuals >8) are colored red. d, The proportions of bacterial genomes devoted to secondary metabolite biosynthesis (left panel; 6.7% of species that devote >7.5% of their genome to biosynthesis are marked red), transcription (middle panel), and translation (right panel).
A global phylogenomic analysis of BGCs provides a quantitative perspective on bacterial secondary metabolite biosynthesis
Our method predicted a total of 33,351 putative BCGs (with an estimated false-positive rate of 5%) in 1,154 genomes of organisms throughout the prokaryotic tree of life (Figure 1c-d, SI Text 1), which we subjected to an extensive phylogenomic analysis (SI Text 2-3, SI Figures 1a-d, 2, 3, SI Tables I-II). We divided the predicted BGCs into two categories – high-confidence (10,724; used in all subsequent analyses) and low-confidence (22,627) – based on assignment to one of ∼20 well-validated BGC classes or on manual inspection for clusters that could not be assigned to any known class. Within the high-confidence set, 7,377 of the predicted gene clusters (69%) were not detected by antiSMASH (Blin et al., 2013; Medema et al., 2011); the difference is due primarily to the fact that antiSMASH does not detect certain BGC classes (including many oligosaccharides), highlighting the need for a tool that identifies BGCs independent of class (Figure 1b).
Strikingly, 40% of all predicted BGCs encode saccharides, more than twice the size of the next largest class. Notably, only 13% of previously reported BGCs encode the biosynthesis of saccharides (SI Text 4). 93% of species harbor saccharide gene clusters, and in 33% of species, more than half of the predicted gene clusters encode saccharides. Cell-associated saccharides such as lipopolysaccharides (Park et al., 2009), capsular polysaccharides (Kadioglu et al., 2008), and polysaccharide A (Mazmanian et al., 2005; Mazmanian et al., 2008) are known to play key roles in microbe-host and microbe-microbe interactions, while diffusible saccharides have a range of biological activities, most notably antibacterial (Flatt and Mahmud, 2007; Weitnauer et al., 2001). The functions of many of the putative saccharide BGCs are still a mystery: 32%, including BGCs from entirely unexplored genera, are not closely related to any known gene cluster (SI Figure 1e). Saccharide BGC repertoires are also surprisingly diverse: only 37% occur in the genomes of two species chosen at random from the same genus (compared to 43% for polyketides, 60% for terpenoids and 74% for fatty acids, SI Figure 1f). The abundance of novel oligosaccharide BGC families raises the possibility that more clinically relevant saccharides such as the antidiabetic drug acarbose and the antibiotics gentamicin and avilamycin will be discovered (Kersten et al., 2013). Another BGC class of unexpectedly large size is the ribosomally synthesized and posttranslationally modified peptides (RiPPs (Arnison et al., 2013)). Notably, RiPP BGCs are as prevalent in our data set as those encoding nonribosomal peptides (Figure 1b).
A BGC distance network reveals unexplored regions of the biosynthetic universe
We next sought to study the relationships among BGCs systematically, with the ultimate goal of creating a global BGC map that could be searched systematically to identify clusters of biosynthetic or taxonomic interest. We adapted a measure of the evolutionary distance between multi-domain proteins (Lin et al., 2006) to calculate an all-by-all distance matrix for the 10,724 BGCs in our high confidence set along with the 732 members of our training set. Using MCL clustering to identify groups of related nodes, we define 905 BGC families with distinct core genetic components. The resulting BGC distance network (Figure 2, SI Text 5, SI Figure 4) revealed an unexpected finding: the presence of large cliques that represent very widely distributed BGC families without any experimentally characterized members.
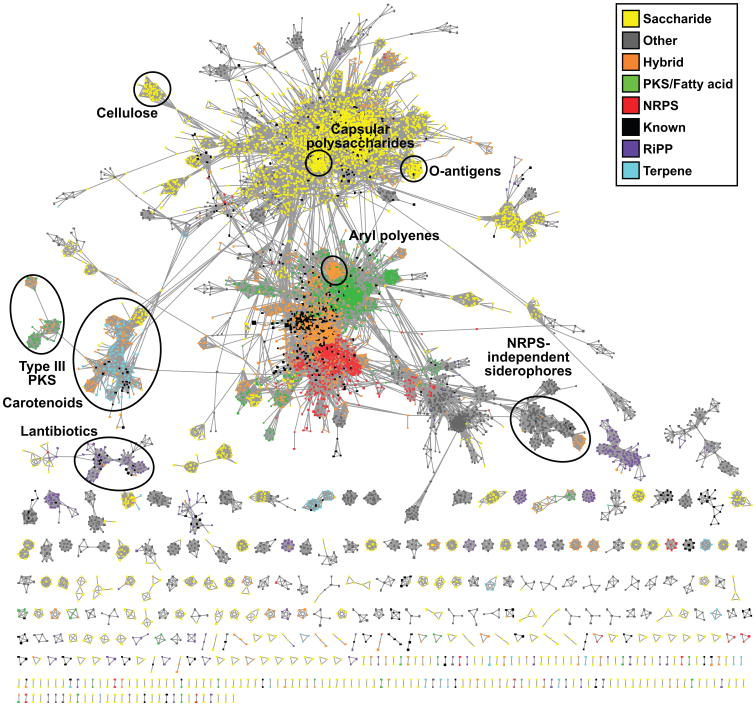
Figure 2. A systematic analysis of bacterial BGCs.
Similarity network of known and putative BGCs, with the BGC similarity metric threshold at 0.5 (See also SI Figure 4). The topology of the network is robust to changes in the distance threshold, as described in the Extended Experimental Procedures. One connected component harbors most of the gene clusters (72%), and is largely composed of two linked subgraphs: one dominated by oligosaccharides and the other a mixture of nonribosomal peptides (NRPs) and polyketides/lipids, indicating that BGCs from these classes share a significant number of gene families with one another. Smaller BGC families with more unique compositions are represented at the bottom of the figure; only 812 BGCs (7.6%) do not have any connections with other BGCs at the chosen cutoff. A selection of node clusters within the network has been highlighted to show how gene cluster families form cliques within the network. The highlighted groups include widely distributed gene cluster families for O-antigens, capsular polysaccharides, carotenoids, and NRPS-independent siderophores, along with one of the lantibiotic BGC families and an unknown family of BGCs with type III polyketide synthases. The aryl polyene family that we characterized further in this study is shown in the middle of the network.
While most known families of secondary metabolites are unique to a small set of organisms, a few are taxonomically widespread. These include the O-antigens, capsular polysaccharides, carotenoids and NRPS-independent siderophores, which can all be clearly distinguished as prominent cliques within our distance network. From a fundamental microbiological perspective, these are among the most important families of molecules produced by microbes and, as such, they have been very intensively studied (Challis, 2005; Rehm, 2010; Samuel and Reeves, 2003; Walter and Strack, 2011). Although we had anticipated finding small gene cluster families of unknown function, we were surprised to discover families harboring hundreds of uncharacterized clusters, distributed widely throughout entire bacterial phyla.
We selected the most prominent of these families for experimental characterization: a set of 811 BGCs, distributed between two subfamilies (hereafter, subfamily 1 and 2), that were not detected by any of the existing BGC identification tools (e.g., antiSMASH), likely because the ketosynthase and adenylation domains they harbor are from uncharacterized, evolutionarily distant clades. BGCs in this family are ∼20 kb in size and harbor a core set of genes that include adenylation, ketosynthase, acyl/glycosyltransferase, ketoreductase, dehydratase, thiolation, and thioesterase domains, as well as an outer membrane lipoprotein carrier protein and an MMPL family transporter (Figure 3a, SI Figures 5 and 8). These clusters are found in a wide variety of Gammaproteobacteria (Acinetobacter, Aggregatibacter, Escherichia, Klebsiella, Pantoea, Pseudoalteromonas, Pseudomonas, Serratia, Shewanella, Vibrio, and Yersinia), as well as a broader set of Beta- (Burkholderia, Neisseria) and Epsilonproteobacteria (Campylobacter) (Figure 3a).
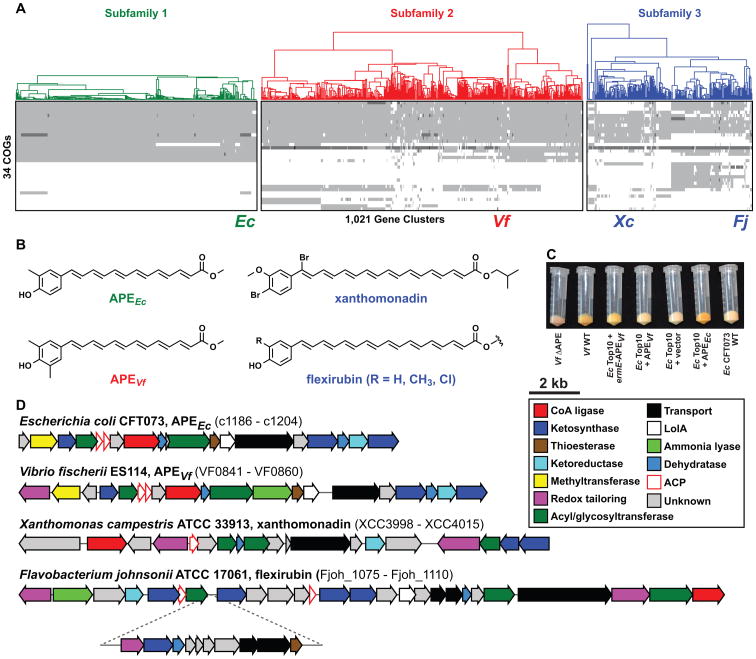
Figure 3. APE gene clusters comprise the largest known BGC family.
a, Heat map and dendrogram of all 1,021 detected APE family gene clusters, based on Clusters of Orthologous Groups generated by OrthoMCL (Li et al., 2003) using our adapted version of the Lin distance metric (Lin et al., 2006) that includes sequence similarity. Light grey indicates the presence of one gene from a COG, whereas darker grey tones indicate the presence of two or three genes from a COG. The two BGC subfamilies that functioned as the starting point of our analysis (subfamilies 1 and 2) are shown in green and red, respectively, while the smaller BGC subfamily that includes the xanthomonadin and flexirubin gene clusters (subfamily 3) is shown in blue. The positions of the two experimentally targeted gene clusters (Ec for Escherichia coli CFT073 and Vf for Vibrio fischeri ES114) as well as the Xanthomonas campestris ATCC 33913 xanthomonadin (Xc) and Flavobacterium johnsonii ATCC 17061 flexirubin (Fj) gene clusters are indicated below the heat map. See SI Figure 5 for a version with more detailed annotations. b, Chemical structures obtained for the APE compounds from E. coli and V. fischeri, and the previously determined chemical structures of xanthomonadin and flexirubin. Note the difference in polyene acyl chain length as well as the distinct tailoring patterns on the aryl head groups. c, Bacterial pellets from strains harboring APE gene clusters showing the pigmentation conferred by aryl polyenes. d, Genetic architecture of the four characterized aryl polyene gene clusters. The inset in the Flavobacterium johnsonii flexirubin gene cluster is a sub-cluster putatively involved in the biosynthesis of dialkylresorcinol (Fuchs et al., 2013), which is acylated to an APE to form flexirubin. See SI Data File 1 for schematics of all 1,021 APE gene clusters from panel A.
The unexplored BGC family encodes the biosynthesis of aryl polyene carboxylic acids
We set out to identify the small molecule product of two clusters in the family, one each from subfamilies 1 and 2. We used circular polymerase extension cloning (CPEC) (Quan and Tian, 2009) to amplify and assemble the 18 gene, 15.5 kb cluster from E. coli CFT073 (c1186-c1204), and we transferred a plasmid harboring the cluster into E. coli Top10. The transformants exhibited a strong yellow pigmentation that was absent in the empty vector control strain and not observed in the native host strain (Figure 3c), but the pigment did not appear to diffuse into liquid or solid culture medium. We liberated the pigment from an organic extract of the cell mass by mild base hydrolysis and purified it by HPLC. Comparative HPLC analysis of extracts from the cluster+ and cluster- strains revealed the presence of a compound unique to the cluster+ strain with an absorption maximum of 425 nm, consistent with a yellow chromophore (SI Figure 7e). Purification of milligram quantities of the compound for structural characterization required the development of an isolation procedure that rigorously excluded exposure to light. A combination of 1D- and 2D-NMR experiments and high-resolution MS on the purified compound revealed that it was an aryl polyene (APE) carboxylic acid consisting of a 4-hydroxy-3-methylphenyl head group conjugated to a hexaenoic acid (Figure 3b, SI Figure 7c, e-f and SI Data File 2).
To study the 20 gene, 18.9 kb cluster from Vibrio fischeri ES114 (VF0841-VF0860), we first deleted the cluster from its native producer. The yellow pigmentation that is observed in wild type V. fischeri under normal laboratory growth conditions was absent in the V. fischeri knockout strain (Figure 3c). We then proceeded to amplify, assemble, and introduce the V. fischeri cluster into E. coli Top10, but the native cluster failed to confer yellow pigmentation on its heterologous host. We then constructed a modified variant of the cluster in which the ermE* promoter was inserted upstream of the operon starting with VF0844. Introduction of this construct into E. coli resulted in a yellow-pigmented strain that produced a new compound with an absorption maximum at 425 nm (Figure 3c). Purification of the V. fischeri compound and analysis by a combination of 1D- and 2D-NMR experiments and high-resolution MS revealed a structure with a similar scaffold to the E. coli APE but a 4-hydroxy-3,5-dimethylphenyl head group (Figure 3b, SI Figure 7d-f and SI Data File 2). Taken together, these data suggest that the cluster representatives from this family encode APE carboxylic acids.
The aryl polyene BGCs are the largest family in the sequence databases
To our surprise, the E. coli and V. fischeri APEs are similar in structure to flexirubin (Fuchs et al., 2013; McBride et al., 2009), a pigment that was previously isolated from the CFB group bacterium Flexibacter elegans, and xanthomonadin (Goel et al., 2002), the compound that gives Xanthomonas spp. their characteristic yellow color. The biosynthetic genes for flexirubin and xanthomonadin are known (Fuchs et al., 2013; Goel et al., 2002; McBride et al., 2009); both are part of a smaller, distinct subfamily in the ClusterFinder results set (subfamily 3 in Figure 3a), but little else is known about the genes in either cluster. Intriguingly, although the clusters in subfamily 3 share similar Pfam domain content to those in subfamilies 1 and 2, the percent identities of their constituent proteins are very low (<20% for some amino acid sequences, see SI Figure 6a). When we turned to a more sensitive approach in which we used MultiGeneBlast (Medema et al., 2013) to look for sequence similarity at the level of the entire gene cluster, we observed distant but recognizable homology between multiple gene pairs from BGCs from subfamily 3 and subfamilies 1 and 2, indicating that the APE clusters might share a common ancestor. Indeed, when we performed a maximum-likelihood phylogenetic analysis of the ketosynthase and adenylation enzyme superfamilies based on structure-guided multiple sequence alignments (SI Text 6, SI Figure 6b-d), we found that the APE KS and A enzymes cluster together in separate uncharacterized clades that are only distantly related to all other known members of these enzyme superfamilies. Based on this evidence, we conclude that the three subfamilies together comprise a single BGC family of >1000 gene clusters (Figure 3a). Notably, the APE family is, to our knowledge, the largest family of gene clusters in the database, even exceeding the size of the well-known carotenoids (870 clusters, as detected using the same methods, see SI Table III).
The lack of homology even between the xanthomonadin and flexirubin biosynthetic genes (both in subfamily 3) is so profound that these pigments have never been connected in the literature: indeed, both previously discovered APEs have been proposed as chemosystematic markers of a genus (Flexibacter and Xanthomonas) because of their “limited distribution among bacteria” (Fautz and Reichenbach, 1979; Jenkins and Starr, 1982; Reichenbach et al., 1980; Starr et al., 1977; Wang et al., 2013). Our results, however, show that APE family BGCs are widely distributed throughout the Gram-negative bacterial tree of life (Figure 4, SI Figure 3). Notably, their pattern of phylogenetic distribution is markedly discontinuous: clusters are present in some strains but not others of most genera (36.4% of the complete genomes in a typical genus harbor the cluster, but note the high standard deviation of 37.9%). The most parsimonious explanations for this distribution pattern are frequent gene cluster loss from the descendants of a cluster-harboring ancestor, or frequent horizontal transfer among the descendants of a cluster-negative ancestor. Two lines of evidence support the possibility of frequent horizontal transfer: The family 1 cluster from E. coli O157:H7 is located on an O-island (Dong and Schellhorn, 2009), and the family 2 cluster from Acinetobacter sp. ADP1 resides on an element that has been identified as horizontally transferred (Barbe et al., 2004). Their broad distribution, and the fact that such widely divergent gene clusters have small molecule products that are so similar in structure, suggests the possibility that aryl polyenes play an important role in Gram-negative cell biology.
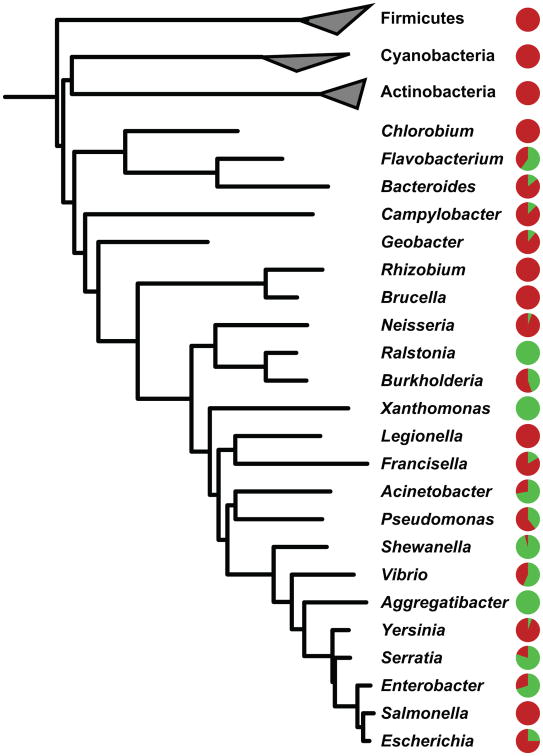
Figure 4. APE gene clusters are widely but discontinuously distributed among Gram-negative bacteria.
Presence/absence pattern of APE gene clusters across all complete genomes from selected bacterial genera, mapped onto the PhyloPhLan high-resolution phylogenetic tree (Segata et al., 2013). For each genus, the pie chart represents the percentage of sequenced genomes in which APE gene clusters are present (green) or absent (red). BGCs from the APE family occur throughout all subphyla of the Proteobacteria, as well as in a range of genera from the CFB group. The discontinuous presence/absence pattern suggests that gene cluster gain and/or loss has frequently occurred during evolution. A presence/absence mapping on all the genomes from our initial JGI dataset is provided in SI Data File 3.
Aryl polyenes might function as protective agents against oxidative stress
Xanthomonadin has been proposed to play a role in protection from photodamage by visible light (Poplawsky et al., 2000; Rajagopal et al., 1997), an effect that is thought to be due to its ability to quench the reactive oxygen species (ROS) that are generated when the photosensitizer used in these studies, toluidine blue, is exposed to visible light (Poplawsky et al., 2000). Additionally, xanthomonadin has been shown to protect cellular lipids from peroxidation in vitro (Rajagopal et al., 1997) and xanthomonadin mutants show reduced epiphytic survival under conditions of natural light exposure (Poplawsky et al., 2000).
Similarly, we hypothesize that other APEs play a role in protecting bacterial cells from exogenous oxidative stress. Membrane-bound APEs could reduce the concentration of free radicals that would otherwise cause damage to other cellular lipids, proteins, or nucleic acids. Notably, many bacteria that harbor APE BGCs are either commensals or pathogens of a eukaryotic host; consequently, they are likely to encounter oxidative stress from immune cells during colonization or infection.
A role for APEs in protecting Gram-negative bacteria against oxidative stress would make them analogous to the chemically similar but biosynthetically distinct Gram-positive carotenoids, whose antioxidant activity is well established. An important example is staphyloxanthin, a membrane-bound carotenoid virulence factor that is responsible for the characteristic yellow pigmentation of S. aureus and proposed to protect S. aureus from immune-mediated oxidative stress. A S. aureus mutant defective in the first committed step of staphyloxantin biosynthesis exhibits higher susceptibility to various reactive oxygen species and in a neutrophil killing assay (Clauditz et al., 2006; Liu et al., 2005). This mutant was also attenuated in murine models for subcutaneous abscess (Liu et al., 2005) and systemic infection (Liu et al., 2008). Experiments to test whether APE-deficient mutants of Gram-negative bacteria harbor colonization or pathogenesis defects will be an important step in testing this model and gaining insight into why APE gene clusters are so widely distributed throughout the Gram-negative tree of life.
Using systematic searches to prioritize BGCs for experimental characterization
BGCs are commonly selected for characterization on the basis of chemical or enzymatic novelty. Following the example of the APE family, we anticipate that our global BGC map will enable gene clusters to be selected in a new way that is based on a criterion biologists have long used to prioritize genes: what are the most widely distributed gene clusters of unknown function? Various other prioritization criteria could be used to select BGCs of interest (Frasch et al., 2013). For example, one could select BGCs likely to encode new chemical scaffolds by searching for clusters that do not harbor conventional monomer-coupling enzymes.
Many gene cluster families still await characterization: even with conservative assumptions, we estimate the total number of bacterial BGC families (such as those encoding carotenoids or calcium-dependent lipopeptides) present in the biosphere to be ∼6,000 (SI Figure 1g), less than half of which are identified in our current set of genomes (∼2,400). Importantly, each of these 6,000 families will likely contain a range of molecules with distinct biological activities. As developments in single-cell genomics and metagenomics are opening up the exploration of a vast microbial dark matter, this number may grow even further: just in the 201 single-cell genomes of uncultivated organisms recently obtained by the JGI (Rinke et al., 2013), our method identified 947 candidate BGCs, of which 655 fall outside all known BGC classes (SI Figure 1h). Even among cultivated organisms, there are still many underexplored taxa (Letzel et al., 2012) (SI Text 2). For the foreseeable future, the number of gene clusters encoding molecules with distinct scaffolds will continue to rise as new genomes are sequenced, and computational approaches to systematically study their relationships will be of great value in prioritizing them for experimental characterization.
Experimental Procedures
Genome sequences
A set of 1154 complete genome sequences was obtained from JGI-IMG (Markowitz et al., 2012), version 3.2 (08/17/2010).
ClusterFinder algorithm and training data
The ClusterFinder prediction algorithm for BGC identification is a two-state Hidden Markov Model (HMM), with one hidden state corresponding to biosynthetic gene clusters (BGC state) and a second hidden state corresponding to the rest of the genome (non-BCG state). The training set for the BGC state was gathered using a comprehensive search of the scientific literature, which yielded 732 clusters. From these, 55 redundant BGCs were filtered out by selecting one random member from each biosynthetic gene cluster family, with a cluster family defined as a connected component in the >0.7 similarity network (see below). Thus, the final BGC state training set consisted of 677 experimentally characterized gene clusters. For the non-BGC state, non-BGC regions were collected from 100 randomly selected genomes, defined as those regions without significant sequence similarity to the BGC state training set sequences (Pfam domain similarities with E-value > 1e-10). ClusterFinder source code is available from the GitHub repository (https://github.com/petercim/ClusterFinder).
ClusterFinder validation
The algorithm was validated in three ways. First, its output was compared to 10 bacterial genomes manually annotated for BGCs (leading to an area under the ROC curve of 0.84). Second, its performance was assessed on 74 experimentally characterized BGCs outside the training set. Out of these, 70 (95%) were detected successfully. When tested alongside antiSMASH (Medema et al., 2011) on the genomes of Pseudomonas fluorescens Pf-5, Streptomyces griseus IFO13350 and Salinispora tropica CNB-440 (SI Table IV), antiSMASH detected 62 out of 65 (95%) manually annotated secondary metabolite gene clusters, while ClusterFinder detected 59 of these (91%). However, ClusterFinder identified 43 (66%) unannotated gene clusters that appeared likely to synthesize small molecule metabolites on manual inspection, whereas antiSMASH detected only five (8%). This highlights the strength of ClusterFinder in detecting gene clusters irrespective of whether they belong to known or a priori specified classes. Among the additional gene clusters detected by ClusterFinder are known gene clusters encoding the biosynthesis of, e.g., alginate and lipopolysaccharides, as well as an uncharacterized cluster that was previously predicted to encode a novel type of secondary metabolite (Hassan et al., 2010).
Type classification of BGCs
ClusterFinder-detected biosynthetic gene clusters were classified by antiSMASH (Medema et al., 2011) to determine their subtypes (e.g., type I polyketide, nonribosomal peptide, terpenoid). The native antiSMASH types were supplemented by a list of profile HMMs for protein domains characteristic of saccharide gene clusters (SI Table V), as well as by fatty acid gene clusters, which could be assigned based on the HMMs that antiSMASH uses in polyketide synthase annotation. Gene clusters lacking protein domains characteristic of gene cluster classes included in antiSMASH were binned in a separate class.
BGC distance metric and similarity network
BGC similarity networks were calculated using a modified version of the distance metric from Lin and coworkers (Lin et al., 2006) for multi-domain proteins. The modified version consists of two different indices: the Jaccard index (which measures the similarity in Pfam domain sets from two BGCs) and the domain duplication index, with weights of 0.36, and 0.64, respectively. The Goodman-Kruskal γ index, which was included in the original similarity metric with a low weight of 0.01, was omitted, since the conservation of the order between two sets of domains does not appear to have an important effect on the structure of the small molecule product, except in the case of NRPS and PKS gene clusters (Fischbach et al., 2008). BGC families were calculated with a Lin similarity threshold of 0.5 and MCL clustering with I = 2.0. The similarity network was obtained using the same Lin similarity threshold and visualized using Cytoscape (Smoot et al., 2011).
Bioinformatic analysis of APE gene clusters
Expansion of the APE BGC family was performed using manual parsing of MultiGeneBlast (Medema et al., 2013) architecture search results (with the E. coli, V. fischeri, X. campestris and F. johnsonii APE gene clusters as query) against GenBank version 197 (08/2013), with a 20% sequence identity cut-off and 2000 blastp hits mapped per query sequence. APE Clusters of Orthologous Groups (COGs) were obtained using OrthoMCL (Li et al., 2003) (MCL I = 1.5, sequence identity cutoff 20%), and were used to construct a cladogram with hierarchical clustering using the Lin modified distance metric. Structure-guided multiple sequence alignments of APE A and KS domains were performed using PROMALS3D (Pei et al., 2008), and phylogenetic trees were inferred with MEGA5 (Tamura et al., 2011) using the Maximum Likelihood method.
Construction of the V. fischeri ES114 APE-cluster deletion mutant
Oligonucleotide primers, plasmids and bacterial strains used and generated in this study are summarized in SI Tables VI-VIII. A deletion construct was generated by fusing the ∼1 kb up- and downstream regions of the V. fischeri cluster into a counterselectable suicide plasmid backbone using circular polymerase extension cloning (CPEC; (Quan and Tian, 2011)). This construct was introduced into V. fischeri ES114 by tri-parental mating and integrants were identified by selection for kanamycin resistance. Second homologous recombination events were enriched by non-selective growth, followed by induction of the counterselectable marker to identify cells that had lost the integrated plasmid backbone. Successful deletion mutants were separated from revertants and verified by colony PCR and sequencing.
Heterologous expression of APE gene clusters
The E. coli CFT073 and V. fischeri ES114 APE clusters were amplified by PCR in three parts from genomic DNA and assembled into the SuperCos I vector backbone using either the CPEC (Quan and Tian, 2011) or Gibson (Gibson et al., 2009) method. The V. fischeri APE cluster was further modified by introducing an apramycin-resistant cassette containing the ermE* promoter upstream of the operon starting with VF0844 using PCR targeting (Gust et al., 2004). Correct insertion of ermE*p was verified by sequencing. The heterologous expression constructs for the E. coli CFT073 and V. fischeri APE clusters were introduced into chemically competent E. coli Top10 yielding strains JC087 and JC090, respectively.
APE compound purification
For large-scale isolation and purification of APEEC and APEVF, all steps were performed in a way that avoided exposure to light. Cells were harvested from 32 L of E. coli JC087 and 80 L of V. fischeri ES114 liquid cultures, respectively. Following lyophilization, the cell material was extracted four times with 1:2 methanol/dichloromethane and the extracts were concentrated, resuspended in 1:2 methanol/dichloromethane and subjected to mild saponification with 0.5 M potassium hydroxide for 1 hour. The mixture was neutralized and the organic layer was collected, washed, dried, and resuspended in acetone for further purification by a two-step RPHPLC method. For both extracts, the peaks with absorbance at 441 nm were collected, dried under vacuum and stored at −20 °C in an amber vial prior to structural analysis (SI Figure 7e-f).
APE structural characterization
Purified APE methyl esters were analyzed by a combination of high-resolution uPLCESI-TOF mass spectrometry and 1D and 2D-NMR experiments, enabling the determination of their molecular formula: C21H22O3 for APEEC ([M-H]− adduct at 321.1496 m/z (Δppm = −0.310)) and C22H24O3 for APEVF ([M-H]− adduct at 335.1652 m/z (Δppm = 0.0)). Analysis of the 1H-NMR, COSY, HSQC, HMBC, ROESY and TOCSY spectra of APEVF in D6 DMSO and APEEC in D6 acetone enabled the determination of their solution structure (Figure 3b). This procedure is described in detail in the Extended Experimental Procedures section and shown in SI Figure 7c-d and SI Figure 9.
For further details regarding the materials and methods used in this work, see the Extended Experimental Procedures.
Supplementary Material
Highlights.
The ClusterFinder algorithm detects BGCs of both known and unknown classes
There exist large and widely distributed BGC families with no characterized members
We show that the most prominent family encodes the biosynthesis of aryl polyenes
The aryl polyene clusters constitute the largest known family of BGCs
Acknowledgments
We thank members of the Fischbach lab for helpful discussions, and an anonymous reviewer for constructive feedback on the manuscript. We thank Edward Ruby (University of Wisconsin) for providing us with V. fischeri ES114, Didier Mazel (Institut Pasteur) for plasmid pSW8197, and Mervyn Bibb (John Innes Centre) for plasmids pIJ773, pIJ790 and pIJ10257. This work was supported by an HHMI Predoctoral Fellowship (PC), a Boehringer Ingelheim Fonds travel grant (MHM), Grant 10463 from the GenBiotics programme of the Dutch Technology Foundation STW to ET (MHM), an NWO-Vidi fellowship (RB), NIH grant TW006634 (RGL), the James and Eleanor Delfino Charitable Trust (KK), a Medical Research Program Grant from the W.M. Keck Foundation (MAF), a Fellowship for Science and Engineering from the David and Lucile Packard Foundation (MAF), DARPA award HR0011-12-C-0067 (MAF), and NIH grants OD007290, AI101018, AI101722 and GM081879 (MAF). This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No.: HHSN272200900018C. M.A.F. is on the scientific advisory board of Warp Drive Bio.
Footnotes
Author Contributions: P.C., M.H.M., J. Claesen, K.K., R.G.L. and M.A.F. designed the research, analyzed the data and wrote the paper, with substantial input from E.T., J. Clardy, and A.S. P.C. and M.H.M. performed the computational research. J. Claesen and K.K. performed the experimental research. K.M. and A.P. provided input data and data integration into the JGI-IMG database. L.C.W.B., P.A.G., M.K., B.W.B., and M.A.F. designed an earlier version of the gene cluster identification algorithm that served as a model for the current version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- Challis GL. Genome mining for novel natural product discovery. J Med Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- Clauditz A, Resch A, Wieland KP, Peschel A, Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T, Schellhorn HE. Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics. 2009;10:349. doi: 10.1186/1471-2164-10-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fautz E, Reichenbach H. Biosynthesis of flexirubin: Incorporation of precursors by the bacterium Flexibacter elegans. Phytochemistry. 1979;18:957–959. [Google Scholar]
- Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci USA. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt PM, Mahmud T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat Prod Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- Franke J, Ishida K, Hertweck C. Genomics-driven discovery of burkholderic acid, a noncanonical, cryptic polyketide from human pathogenic Burkholderia species. Angew Chem Int Ed Engl. 2012;51:11611–11615. doi: 10.1002/anie.201205566. [DOI] [PubMed] [Google Scholar]
- Frasch HJ, Medema MH, Takano E, Breitling R. Design-based re-engineering of biosynthetic gene clusters: plug-and-play in practice. Curr Opin Biotechnol. 2013;24:1144–1150. doi: 10.1016/j.copbio.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338:387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- Fuchs SW, Bozhuyuk KA, Kresovic D, Grundmann F, Dill V, Brachmann AO, Waterfield NR, Bode HB. Formation of 1,3-cyclohexanediones and resorcinols catalyzed by a widely occurring ketosynthase. Angew Chem Int Ed Engl. 2013;52:4108–4112. doi: 10.1002/anie.201210116. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Goel AK, Rajagopal L, Nagesh N, Sonti RV. Genetic locus encoding functions involved in biosynthesis and outer membrane localization of xanthomonadin in Xanthomonas oryzae pv. oryzae. J Bacteriol. 2002;184:3539–3548. doi: 10.1128/JB.184.13.3539-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol. 2004;54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LD, Hartney S, Duboy R, Goebel NC, Zabriskie TM, et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ Microbiol. 2010;12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- Jenkins CL, Starr MP. The pigment of Xanthomonas populi is a nonbrominated aryl-heptaene belonging to xanthomonadin pigment group 11. Curr Microbiol. 1982;7:195–198. [Google Scholar]
- Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, Ziemert N, Gonzalez DJ, Duggan BM, Nizet V, Dorrestein PC, Moore BS. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc Natl Acad Sci USA. 2013;110:E4407–E4416. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- Letzel AC, Pidot SJ, Hertweck C. A genomic approach to the cryptic secondary metabolome of the anaerobic world. Nat Prod Rep. 2012;30:392–428. doi: 10.1039/c2np20103h. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MH, Ung PM, Zajkowski J, Garneau-Tsodikova S, Sherman DH. Automated genome mining for natural products. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Zhu L, Zhang DY. An initial strategy for comparing proteins at the domain architecture level. Bioinformatics. 2006;22:2081–2086. doi: 10.1093/bioinformatics/btl366. [DOI] [PubMed] [Google Scholar]
- Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, Nizet V, Wang AH, Oldfield E. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319:1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry B, Robbins T, Weng CH, O'Brien RV, Cane DE, Khosla C. In vitro reconstitution and analysis of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2013;135:16809–16812. doi: 10.1021/ja409048k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol. 2009;75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Takano E, Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol. 2013;30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of transacyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol. 2008;26:225–233. doi: 10.1038/nbt1379. [DOI] [PubMed] [Google Scholar]
- Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:W30–34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer S, Sussmuth R, Heckmann D, Recktenwald J, Huber P, Jung G, Wohlleben W. Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908. Antimicrob Agents Chemother. 1999;43:1565–1573. doi: 10.1128/aac.43.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- Poplawsky AR, Urban SC, Chun W. Biological role of xanthomonadin pigments in Xanthomonas campestris pv. campestris. Appl Environ Microbiol. 2000;66:5123–5127. doi: 10.1128/aem.66.12.5123-5127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc. 2011;6:242–251. doi: 10.1038/nprot.2010.181. [DOI] [PubMed] [Google Scholar]
- Rajagopal L, Sundari CS, Balasubramanian D, Sonti RV. The bacterial pigment xanthomonadin offers protection against photodamage. FEBS Lett. 1997;415:125–128. doi: 10.1016/s0014-5793(97)01109-5. [DOI] [PubMed] [Google Scholar]
- Rehm BH. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol. 2010;8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- Reichenbach H, Kohl W, Bottger-Vetter A, Achenbach H. Flexirubin-type pigments in Flavobacterium. Archives of Microbiology. 1980;126:291–293. [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Samuel G, Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydrate research. 2003;338:2503–2519. doi: 10.1016/j.carres.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- Segata N, Bornigen D, Morgan XC, Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 2013;4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic A, Zucko J, Simunkovic J, Long PF, Cullum J, Hranueli D. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008;36:6882–6892. doi: 10.1093/nar/gkn685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr MP, Jenkins CL, Bussey LB, Andrewes AG. Chemotaxonomic significance of the xanthomonadins, novel brominated aryl-polyene pigments produced by bacteria of the genus Xanthomonas. Arch Microbiol. 1977;113:1–9. doi: 10.1007/BF00428572. [DOI] [PubMed] [Google Scholar]
- Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MH, Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep. 2011;28:663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qian G, Li Y, Wang Y, Wang Y, Wright S, Li Y, Shen Y, Liu F, Du L. Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS One. 2013;8:e66633. doi: 10.1371/journal.pone.0066633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Rausch C, Lopez P, Hoof I, Gaykova V, Huson DH, Wohlleben W. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J Biotechnol. 2009;140:13–17. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tu57 and production of new antibiotics. Chem Biol. 2001;8:569–581. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.