Abstract
An 84 base pair sequence of the Streptococcus mutans virulence factor, known as dextranase, has been obtained from 10 individuals from the Bronze Age to the Modern Era in Europe and from before and after the colonization in America. Modern samples show four polymorphic sites that have not been found in the ancient samples studied so far. The nucleotide and haplotype diversity of this region have increased over time, which could be reflecting the footprint of a population expansion. While this segment has apparently evolved according to neutral evolution, we have been able to detect one site that is under positive selection pressure both in present and past populations. This study is a first step to study the evolution of this microorganism, analysed using direct evidence obtained from ancient remains.
Keywords: Streptococcus mutans, dextranase, neutral evolution, positive selection
1. Introduction
In the past few decades, genetic tools have made it possible to confirm the presence of the bacteria responsible for some of the diseases observed in ancient human remains. The aims of these studies have been numerous: to confirm the early diagnosis based on skeletal evidence, to identify the causative agent of a disease that has notably influenced some periods of human history, or to improve the general knowledge on the interaction between the human beings and the bacteria. Moreover, it has been used to reconstruct historical migrations by characterizing the diseases of the past, their morbidity, their expansion/diffusion and their evolution over time.
The diseases that have been molecularly studied so far are those believed to have caused the majority of the documented human epidemics, such as tuberculosis (Mycobacterium tuberculosis) [1,2], plague (Yersinia pestis) [3,4], leprosy (Mycobacterium leprae) [5,6] and syphilis (Treponema pallidum pallidum) [7,8], among others [9–11]. However, there are other microorganisms, such as bacteria from the dental plaque, that have accompanied humans since remote times for which their history is still not well established, and that could help us to understand prehistoric populations in depth. Thus, besides assessing issues such as the kind of diet and whether the meals were correctly handled and prepared [12], the general hygienic conditions [13], the gathering of people in populations [14], the routine contact of humans with animals, especially during the process of domestication [15], the recent discovery that some ignored illnesses can be traced back to far more ancient times than previously thought [16] implies that palaeogeneticists should be able to study human-bacterial interactions in dental plaque since their emergence.
In fact, oral infections have generally been overlooked when studying infections from the past even considering that caries has been the most persistent infection in the history of humanity. Palaeopathological studies have shown that their incidence go back to at least 1.5 Myr ago, being already found in a Paranthropus robustus specimen [17]. However, the most dramatic increase in caries frequency occurred in the transition towards agriculture owing to a change in diet. This change can be seen in both Europe and America [18–20], although there are places where such a relationship is not so clear, as in mainland southeast Asia, where the relative non-cariogenicity of rice, at that time an increasingly important subsistence mode in the region, and the retention of broad-spectrum livelihood strategies put the global application of this theory into question [21]. But this looks like the exception rather than the rule. Thus, study of Mesolithic–Neolithic transition has become a matter of great interest for learning about the infectivity of the bacteria involved in the formation of this lesion. Moreover, in recent times, the evidence that dental problems like the periodontal diseases and caries can cause other serious health problems have increased, so their development has been a matter of growing interest. New research suggests that periodontal diseases may contribute to the development of heart diseases [22], increase the risk of stroke [23] and may be a serious threat for people whose health is already harmed by diabetes, respiratory diseases or osteoporosis [24]. Thus, characterizing the microorganisms involved in these processes would be a step forward to gain a better knowledge of their interaction with human beings and the consequences that result from it. Following this reasoning, this study focuses on DNA characterization of the main agents responsible for the carious lesions from ancient remains.
Even though caries is the consequence of the loss of enamel as a result of the acidification of the oral plaque by a quite heterogeneous group of bacteria [25], Streptococcus mutans has been consistently associated with its presence [26]. The current advances in biomolecular technology offer the possibility of genetically characterizing these bacteria in human ancient remains, as well as determining the characteristics of the virulence factors that they need to carry out a successful infection. Preceding this study, in 2007 our group showed that it was possible to recover S. mutans DNA from human ancient remains [27]. On that basis, it has been possible to begin an evolutionary study of S. mutans in relation to the development of caries in humans [28], which has been extended in this work.
Caries lesions could be used to obtain information about the health of the individuals and their way of life in ancient times. Two combined factors increase the feasibility of carrying out the genetic analysis of this disease in ancient times. First, the resistance of teeth, which makes them particularly suitable for the study of the evolution of an organism that has one of its niches there. Second, the results of the activity of the cariogenic agents are evident in the teeth and make caries easy to detect.
Good markers of the S. mutans adaptation to its human host can be found in its virulence factors, thus their study may clarify some aspects of the evolution of this widely extended infection that has accompanied humankind during millennia, and therefore give us a more complete image of our evolutionary history. One of the most relevant points would be to know whether this microorganism has evolved according to the changes in the way of life of the host.
Moreover, the direction and strength with which natural selection has acted in different genomic regions of this bacterium could provide some clues over how good the adaptation to its host has been, and open the possibility to predict its evolution. Some things must be taken into account: (i) in many proteins, a high proportion of amino acids may be largely invariable owing to strong functional constraints [29], and (ii) adaptive evolution most probably occurs at a few time points and affects a few amino acids [30]. Thus, it is necessary to detect the ratio of non-synonymous to synonymous rates (ω), which measures the selective pressure at the protein level and can take values of ω < 1, =1 or >1 indicating negative, neutral or adaptive evolution, respectively. In addition, a method that can measure it in a whole nucleotidic fragment, on the one hand, and at each single codon on the other hand, will be especially important for this matter, because in cases of very local action of positive selection, the overall ω ratio will not be significantly greater than 1. The aim of this study is to recover genetic material from the caries of individuals from the Bronze Age up to the twentieth century in Europe and America, and to characterize a fragment of the gene that encodes the virulence factor known as dextranase. This will permit us to carry out comparisons with current strains in order to determine whether the genetic diversity in ancient times was as high as it is nowadays, and whether there was any other relevant difference in relation to geographical or chronological differentiation. Moreover, we want to assess whether any specific site has been submitted to positive selection pressure in past and present populations, and whether its intensity has changed over time.
2. Material and methods
(a). Study species and sampling
Samples of different antiquity and geographical origin, European and American, were chosen from different archaeological sites and from a skeletal collection housed at Universitat Autònoma de Barcelona (UAB) (see table 1 for Genbank accession numbers, the electronic supplementary material for technical details and electronic supplementary material, table S1 for further description). The selection was made visually, as caries is macroscopically detectable.
Table 1.
Alignment of the obtained sequences. (Sample references can be found in the electronic supplementary material, table S1. En dashes (–) indicate the same nucleotide as the reference sequence D4930.1 in that position, while polymorphisms are shown in bold. Samples U1 and LO1 were duplicated at the Laboratorio Nacional de Genómica para la Biodiversidad from Mexico.)
| strain |
accession no. |
site |
period |
364 |
367 |
368 |
380 |
385 |
390 |
400 |
410 |
420 |
429 |
430 |
432 |
437 |
440 |
447 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D4930.1 | D4930.1 | England | Modern | T | Aa | Aa | A | Ga | T | C | A | G | Ab | G | Gb | Ca | C | A | |
| modern samples | NN 2025 | APO 10655 | Japan | Modern | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – |
| LJ23 | APO12336.1 | Japan | Modern | – | – | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| U159 | AEO 14133 | North America | Modern | – | G | C | – | A | – | – | – | – | G | – | – | T | – | – | |
| GS5 | CP003686.1 | North America | Modern | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| ATCC 25175 | HQ711852.1 | England | Modern | – | – | – | – | – | – | – | – | G | – | – | – | – | – | ||
| 5DC8 | AOBX00000000 | England | Modern | – | – | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| KK21 | AOBY00000000 | Germany | Modern | – | G | C | – | A | – | – | – | – | G | – | – | T | – | – | |
| KK23 | AOBZ00000000 | Germany | Modern | – | G | C | – | A | – | – | – | – | G | – | A | – | – | – | |
| AC4446 | AOCA00000000 | Germany | Modern | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| NCTC11060 | AOCC00000000 | Denmark | Modern | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| ancient samples | U1 | KJ950640 | Granollers, Catalonia | twentieth century | – | – | C | – | A | – | – | – | – | G | – | – | – | – | – |
| M1 | KJ950631 | Montanissell, Lleida | Bronze Age | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| CR1 | KJ950636 | Can Reiners, Majorca | seventh century | – | – | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| V1 | KJ950637 | Vilomara, Catalonia | ninth to tenth centuries | – | – | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| SP1 | KJ950634 | Sant Pere Churches, Catalonia | fifth to twelfth centuries | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| SP2 | KJ950635 | Sant Pere Churches, Catalonia | fifth to twelfth centuries | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| T1 | KJ950638 | Tlatelolco, Mexico | precontact twenty-fifth to twenty-sixth centuries | – | – | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| T2 | KJ950639 | Tlatelolco, Mexico | precontact twenty-fifth to twenty-sixth centuries | – | – | T | – | A | – | – | – | – | G | – | – | – | – | – | |
| LO1 | KJ950632 | Los Olmos, Mexico | postcontact twenty-sixth century | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – | |
| LO2 | KJ950633 | Los Olmos, Mexico | postcontact twenty-fifth century | – | G | C | – | A | – | – | – | – | G | – | – | – | – | – |
aPositions where the polymorhism results in a non-synonymous change.
bPositions where the polymorphism results in a synonymous change.
The samples were stored and analysed in the Palaeogenetics Laboratory of the UAB. The conditions of sterility and the precautionary measures taken are previously described [11]: sample preparation (see the electronic supplementary material), DNA extraction and PCR reactions were performed in a laboratory dedicated specifically to work with ancient DNA (aDNA), positively pressurized and physically isolated from the laboratories used to carry out post-PCR processes. Laboratory overalls covering the whole body of the investigator, masks and protective lenses were also used. All the samples were amplified twice at the UAB, and the majority of them (7 out of 10) were cloned. In addition, samples T1, U1 and LO1 were analysed in the Laboratorio Nacional de Genómica para la Biodiversidad (Langebio, CINVESTAV-IPN, Mexico). In the Mexico laboratory, DNA extractions were performed essentially with the same protocol and procedures as in the UAB (see below) in an especially dedicated facility for aDNA analysis. Also, PCR amplifications of a dextranase gene fragment were conducted as described below, although in Langebio an endpoint PCR apparatus (Veriti, Applied Biosystems) was used. Two of the samples (U1 and LO1) yielded positive amplification and were cloned and sequenced, and from the U1 sample two independent extractions were obtained.
The genome region of S. mutans chosen to be amplified was an exonic fragment of 84 base pairs (bp) in length of the first variable region of the dextranase gene. This gene is one of the so-called virulence factors of the pathogen. It codes for an enzyme which cleaves α-1,6-linkages of glucans and is thought to be responsible both for the control of the amount and content of extracellular glucans and for the metabolic utilization of extracellular glucans ([31] and references therein). The primers used to carry out the PCR process were L-344 (forward primer) and R-467 (reverse primer) [27].
(b). Experimental protocol
For DNA extraction, 0.5 g of powder were collected from the teeth cavities of each individual. Samples were divided into groups of three to five for the DNA extraction process, to keep a low sample-to-blank control ratio. A real-time PCR reaction using the Qiagen Rotor-Gene Q (QIAgen, Turnberry Lane, USA) and the Type-it HRM PCR kit(400) (QIAgen, Turnberry Lane, USA) was then carried out in a final volume of 25 μl. The PCR process consisted of the following steps: an initial denaturation step at 94°C for 5 min, followed by 45 cycles of PCR including 10 s at 94°C and 30 s at 55°C. The obtained product was purified using the PCRapace kit (Invitrogen, Carslbad, CA, USA) following supplier instructions.
In the cloned samples, after purification the amplified product was cloned into the TOPO TA cloning kit (Invitrogen). Cloned fragments were amplified by colony-PCR using pM13 forward and reverse primers with the following profile: an initial denaturation step at 94°C for 5 min, followed by 35 cycles of PCR including 1 min at 94°C, 1 min 30 s at 55°C, 1 min at 72°C and a final extension step of 7 min at 72°C. The amplified product was purified again as described above and then sequenced.
Sequence reactions were carried out using the sequencing kit BigDye Terminator v. 3.1 (Applied Biosystems, Carslbad, USA) according to the manufacturer's specifications, and run in an ABI 3130XL sequencer (Applied Biosystems, Foster City, USA). The BLAST program [32] was used to search for similar sequences in the GenBank database (NCBI). The consensus sequence for each gene fragment was determined by alignment of the forward and reverse sequences using BioEdit v. 7.0.5.3 (Ibis Biosciences, Carslbad, USA).
Finally, two teeth were purposely chosen to be of different geographical origins for parallel DNA extraction from dentine. MtDNA haplogroup identification was carried out in order to have a general overview of the degree of conservation of the samples, and to check whether the results were consistent with the population of origin. The samples were amplified in the second half of the mitochondrial Hypervariable Region I sequence and the obtained haplogroup was corroborated by means of restriction fragment length polymorphisms.
The data sequence assembly is available at the electronic supplementary material.
(c). Statistical analyses
The nucleotide diversity per site (π) and haplotype diversity (H) were calculated using SPSS 15.0.1 software (IBM, New York, NY, USA). The best-fit model of nucleotide evolution was selected using the Bayesian Information Criterion implemented in the best-fit model test included in MEGA 5.05 software [33,34]. The FST distance between the two groups was calculated with Arlequin v. 3.11 [35] and finally, a median-joining network was constructed using the Network 4.1.5.6 software (Fluxus Technology Ltd, Suffolk, UK).
Maximum-likelihood estimations (MLEs) of the dN/dS ratio (ω) were obtained by using the Codeml program from the PAML package [36], and normalized values of dN–dS on a codon-by-codon analysis were obtained using the HyPhy software [37]. The ML phylogenetic tree was reconstructed with the PHYML package [38] (see the electronic supplementary material for technical details).
A branch-site test of positive selection was applied to both groups of samples to check whether they were evolving according to neutral evolution. Moreover, three of the tests supported by PAML package, M0 versus M3 to test for variable ω among sites, and M1a versus M2a and M7 versus M8 to test for possible positive selection at specific sites, were carried out [39]. To calculate the posterior probabilities that each site belongs to a particular site class, a Bayes Empirical Bayes approach [29] was applied, and sites coming from the class with ω > 1 with a high posterior probability (p > 0.95) were inferred to be under positive selection. Branch lengths were fixed at their MLEs under M0 (one-ratio). In addition, the ω rate ratios, estimated using the method of Nei & Gojobori [40] for pairwise sequence comparison, were compared between the total amount of sequences and those from current populations to check whether any change in their values could be detected over time (see the electronic supplementary material for technical details).
3. Results
A 84 bp DNA fragment of the dextranase gene of S. mutans was obtained from caries samples from 10 individuals (see table 1 for Genbank accession numbers). Six samples belonged to ancient European populations and four belonged to ancient American ones. The results are summarized in table 1, with the sequences obtained in this study aligned with the 11 modern sequences of this segment of the dextranase gene currently available in the Genbank database (see the electronic supplementary material, table S1). Seven of the samples (M1, CR1, LO1, LO2, SP1, U1 and T2) were cloned (see the electronic supplementary material, table S2) and the consensus sequences obtained from each one always matched the one obtained by direct PCR. The sequences from the samples that belong to the caries of the ancient individuals were identical to those observed in some modern populations, with the exception of the American sample T2, a sample prior to European contact. In addition, two of the ancient samples (U1 and LO1) were independently replicated in Langebio, Mexico, giving coincident results.
The results showed that five of the nine ancient samples (M1, SP1, SP2, LO1 and LO2) matched up exactly with one of the two modern Japanese strains (NN2025) and also with modern North American (GS5), Danish (NCTC11060) and German (AC4446) ones. The remaining five showed a G to A transition in the nucleotide position 367 (U1, CR1, V1, T1 and T2), which is also currently extended worldwide as observed in current Japanese (LJ23) and English (5DC8) strains. One sample (T2) also harboured a C to T transition in the position 368 that was not seen in any other sample, as the other two samples that showed a change in that position were two modern English strains harbouring a C to A transversion (ATCC25175 and D49430.1).
Amplifications of human mtDNA, as quality controls, were successful and yielded the expected results. One sample originally coming from Mexico (LO1, accession no. KJ950642) harboured an haplogroup of American origin (A), and one coming from Catalonia (M1, accession no. KJ950641) harboured one of European origin (K) (see the electronic supplementary material, table S3).
The nucleotidic (π) and haplotypic (H) diversity indexes were higher in the modern than in the ancient sequences (0.019 ± 0.004 versus 0.009 ± 0.002 for the π values, and 0.848 ± 0.074 versus 0.639 ± 0.126 for the H values). Both the increase in nucleotide and haplotype diversity was statistically significant (one-tailed Mann–Whitney test [41], p < 0.01 and p < 0.05, respectively) (see the electronic supplementary material, table S4).
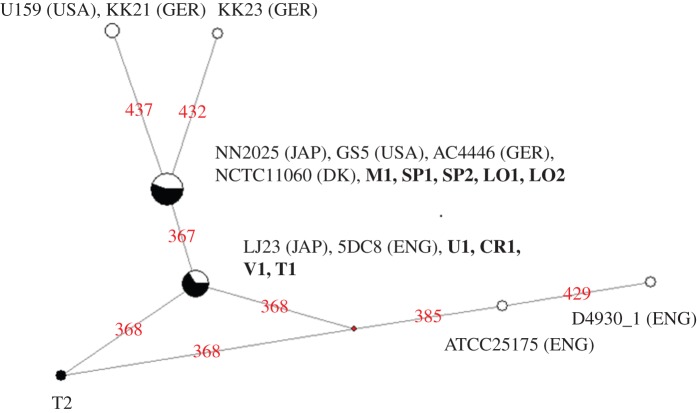
The best-fit model of nucleotide evolution was the Kimura two-parameter model [42], with a discrete Gamma distribution of rate variation among sites (+G), with five gamma rate categories and alpha shape parameter of 0.06 and a transition to transversion ratio of 6.31. Pairwise FST distances between the ancient and the current population were calculated, showing that there were no significant differences (p > 0.05; see the electronic supplementary material, table S5). A phylogenetic network was constructed under the assumptions of this model (figure 1).
Figure 1.
Median-joining network of the sequences presented in this study. Modern sequences (11 modern strains from Genbank obtained in this study) are: U159 and GS5 (USA), NN2025 and LJ23 (Japan), D4930.1, ATCC 25175 and 5DC8 (England), KK21, KK23 and AC4446 (Germany) and NCTC11060 (Denmark). Ancient samples frequency is in black inside the circles and their names are highlighted in bold, and modern samples frequency is represented in white. (Online version in colour.)
The maximum-likelihood phylogenetic tree was calculated with PHYML [38] using the best-fit model of nucleotide evolution obtained, and used as the basis to estimate the mean number of changes per codon per branch. The test statistics M0 versus M3, M1a versus M2a and M7 versus M8 gave significant results in both the current and the ancient populations, and also whether all the samples were considered as a single population (p < 0.01 in all cases) (see the electronic supplementary material, table S6). We considered the signal of positive selection to be strong when both M1a versus M2a and M7 versus M8 were significant at the 5% level. Only codon site 2 showed positive selection with a high posterior probability (p > 0.99) in both sets of sequences, once the Bayes Empirical Bayes approach was applied [15]. Site 25 fell near the threshold value to reject neutral evolution in the M7 versus M8 test (see the electronic supplementary material, table S7).
At site 2, the selective strength seemed stronger in ancient populations than in modern ones, using all the models of amino acid evolution available at the HyPhy package (see the electronic supplementary material, table S8).
Among the ancient sequences, only non-synonymous substitutions were observed. As no orthologous sequences from a closely related species could be found performing a BLAST search (see the electronic supplementary material, table S9), two samples from this study, M1 and NN2025, were used as background branches to carry out the branch-site test for positive selection. These tests showed that ω was not significantly greater than 1, neither in the ancient nor in the modern population (see the electronic supplementary material, table S10).
4. Discussion
The physical presence of S. mutans in ancient samples was first detected by our team in 2007 [27], and some of the preliminary sequences of this study were published in 2011 [28]. In addition, this bacterium was recently detected by other groups using the gold-labelled antibody transmission electron microscopy [43], and in 2012 an aDNA segment from S. mutans DNA was amplified from an ancient dental calculus of an individual dating from approximately 500 years BP [44]. Ten sequences from the Bronze Age to the beginning of the twentieth century from ancient dental carious lesions are presented in this study, allowing our team to carry out a phylogenetic analysis of the ancient strains of this bacterium.
This sample size is related to the difficulties inherent to the analysis of aDNA. The work on aDNA is subjected to important difficulties: (i) the biochemical damage it can suffer [45,46], (ii) the risk of amplifying exogenous (contaminant) DNA that may outcompete aDNA in downstream analyses [47], and (iii) the possible inhibitors that the samples may carry [48]. Owing to this, all the samples suspicious of bearing postmortem damage, as an excess of type II transitions [45,46], and those that did not amplify after three amplifications were discarded to avoid obtaining sequences affected by miscoding lesions or products resulting from carryover contamination.
Regarding other authenticity criteria, no positive controls had been used, and no DNA from S. mutans had been previously amplified in our laboratories. In addition, some of the obtained sequences were cloned in order to obtain the consensus sequences, which always matched up with the one obtained by direct sequencing of PCR products. Moreover, two samples from different origins were amplified with primers for human mtDNA, and the results coincided with the expectations, as each sample harboured a haplogroup that made phylogenetic sense, considering their different geographical sources. The diversity of the results further guaranteed that they were not the product of a general process of contamination. Finally, two of the samples were successfully replicated in the aDNA laboratory of Langebio, CINVESTAV-IPN, in Mexico, showing that the products obtained were not affected by intra-laboratory contamination. Thus, the authenticity of these results can be verified [47], and it can be stated that it is possible to isolate DNA from this bacterium in archaeological remains from periods as ancient as the Bronze Age (Montanissell sample, M1).
Focusing on the sequences, six polymorphic positions can be seen (367, 368, 385, 429, 432 and 437) in present-day populations, while in ancient samples just the first two of the cited polymorphisms have been observed. In the near future, more ancient samples sharing a similar origin with the samples used in this study, such as Asia, shall be analysed to warrant a minimum representation of geographical variability. The African continent, not represented here, must also be a focus of interest in future studies regarding past and present populations. Therefore, it will be necessary to increase the sample size of both modern and ancient groups. Nevertheless, and although the small sample size recommends taking these results with caution, it seems that both the nucleotide and haplotype diversities of this region of the dextranase gene are increasing over time (both values are significantly higher in the modern than in the ancient populations).
Alternatively, the increase in the genetic diversity we found might be attributed to a bias in the choice of the modern strains of S. mutans. However, this seems unlikely because in any of the original papers reporting the modern sequences it is specified that the strains were chosen for any particular reason other than characterizing partially or totally the genome from different strains of this organism [49–54], or distinguishing the different functional regions of the dextranase gene sequence in relation to its enzymatic activity [55]. Therefore, no sampling bias is evident.
This increase in diversity is not translated into an increase of the ω value, as the new observed substitutions bring a negative value in the overall dN – dS difference (see the electronic supplementary material, table S8), pointing to a slight decrease of ω over time, and suggesting a constraint of the selective pressure in this dextranase segment over S. mutans recent history. In fact, empirical data have demonstrated that in closely related microbial sequences, a relative preponderance of non-synonymous changes is seen, leading to a high value of ω [56]. The interpretations of this fact have been numerous, from statistical artefact [57], to relaxed [58] or positive [59] selection, recent ancestry [60,61] or a lag in the removal of slightly deleterious non-synonymous mutations that have survived via hitch-hiking to a nearby strong adaptive mutation [56]. Whatever the reason, the advantage of our study is that obtaining samples from archaeological remains allows comparison of the ω of this population at different points in time, separated by hundreds and thousands of years not by inferred phylogenetic reconstruction, but by directly observed data. As a fall in the ω value is detected, we can rule out the possibilities that neither relaxed nor positive selection have been the driving forces in the full segment over time. The fact that no positive selection is observed when considering the segment as a whole, in spite of being a known factor of virulence, agrees with the fact that dextranase has not been included among the genes that were under Darwinian positive selection in previous studies [62,63]. Nevertheless, using likelihood-ratio tests of codon evolution one specific site (site 2) appears to have been subjected to positive selection throughout the evolutionary history of the segment and continue to do so, although to a lesser extent, and a second one (site 25) could be departing from neutral evolution.
The majority of the samples, both modern and ancient, belong to the two central nodes, reflecting that the full segment is still evolving neutrally while, with the exception of sample T2, the extremes of the phylogenetic network are represented by modern-day sequences, thus reflecting the increase in genetic variability over time. This is supported by the previously mentioned rise in the genetic diversity of the segment and a recent study showing that the S. mutans population started expanding exponentially around 10 000 years ago, approximately coinciding with the onset of human agriculture [63].
Finally, as observed in previous studies focusing in S. mutans comparative genomics of current strains [50,63,64], no characteristic differences in relation to geographical distribution were seen either in the modern or ancient population.
Nevertheless, more samples of different periods and longer sequences will be needed in future studies in order to fully certify the constriction of selective pressure in this segment inferred from our results. Tracing back S. mutans history demands the review of the history of caries. As stated previously, adaptive evolution most probably occurs at a few time points [17], where the frequency of caries has significantly changed, so those periods in time are the ones most likely to show a process of non-neutral selection, marking interesting moments to check the evolution of the bacteria involved in the process.
Six such moments in the history of this illness stand out and it will be worth studying them in depth in future works: the first stage of farming in the American continent (8000–5000 BP) when caries has been recorded with relatively high indices as a consequence of the consumption of endemic fruits rich in maltodextrines and sugar [65–67]; the introduction and spread of cereals in the Old World, reaching a 75% in caries frequency around 4500 BC [68]; its increment in America since 2300 BC, clearly related to maize consumption (Zea mays) [69,70] and more specifically to a gradual replacement of popcorn for a more cariogenic and amylaceous maize [66,71]; the contact between the people of the Old and the New World, especially from before and after AD 1550 when sugar and sugar cane started being imported in large scales from America to Europe [72,73]; the advent of Industrial Revolution that gave rise to cooking technologies that broadened the dietary breath and implied a major dietary shift to increased animal fat, sugar and processed foods [74]; and finally, the decrease since the 1970s throughout industrialized countries related to dental treatment, the introduction of fluoride water and toothpaste [75,76] on the one hand, and a range of changing social factors linked to improvements in general health indicators [77–79] on the other. Caries has such a well-documented story that once this technology can be routinely applied the palaeogeneticists will know which populations warrant this kind of study.
The analysis of S. mutans in individuals from these historical periods could start an interesting field that aims to understand more issues about the way of life of ancient people. The information obtained could also be used to reconstruct past population movements. In this sense, this work reports molecular genetic evidence obtained from ancient bacteria associated with archaeological caries lesions that will help in this aim. Analysing such an ancient genetic material also brings about the possibility of determining whether its evolution can be somehow correlated with changes in the way of life of its hosts. Owing to S. mutans being, to some extent, maternally transmitted [80], it could consequently reflect the migration pattern of women, which may be inferred by mtDNA analysis. A first paper reporting this kind of study was published by Moodley et al. [81] by using another human pathogen, Helicobacter pylori, demonstrating that two different strains of this bacterium accompanied the two human prehistoric migrations along the Pacific.
Also, it would be important to check the adaptation of this bacterium at molecular level to the different niches that it can find inside a human host.
The answer to some of these questions may be related to the changes that the virulence factors of the bacterium have experienced over time; so this field needs to continue expanding in order to be able to respond to them.
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
Funding statement
This work was supported by Generalitat de Catalunya (grant no. 2014 SGR 1420), and Conacyt (Mexico) (grant no. CB-2008-01-105481).
References
- 1.Donoghue HD. 2011. Insights gained from palaeomicrobiology into ancient and modern tuberculosis. Clin. Microbiol. Infect. 17, 821–829. ( 10.1111/j.1469-0691.2011.03554.x) [DOI] [PubMed] [Google Scholar]
- 2.Müller R, Roberts CA, Brown TA. 2014. Biomolecular identification of ancient Mycobacterium tuberculosis complex DNA in human remains from Britain and continental Europe. Am. J. Phys. Anthropol. 153, 178–189. ( 10.1002/ajpa.22417) [DOI] [PubMed] [Google Scholar]
- 3.Bos KI, et al. 2011. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478, 506–510. ( 10.1038/nature10675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner DM, et al. 2014. Yersinia pestis and the Plague of Justinian 541—543 AD: a genomic analysis. Lancet Infect Dis. 14, 319–326. ( 10.1016/S1473-3099(13)70323-2) [DOI] [PubMed] [Google Scholar]
- 5.Montiel R, García C, Cañadas MP, Isidro A, Guijo JM, Malgosa A. 2003. DNA sequences of Mycobacterium leprae recovered from ancient bones. FEMS. Microbiol. Lett. 226, 413–414. ( 10.1016/S0378-1097(03)00617-7) [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, et al. 2010. Detection of Mycobacterium leprae DNA from archaeological skeletal remains in Japan using whole genome amplification and polymerase chain reaction. PLoS ONE 5, e12422 ( 10.1371/journal.pone.0012422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolman CJ, Centurion-Lara A, Lukehart SA, Owsley DW, Tuross N. 1999. Identification of Treponema pallidum subspecies pallidum in a 200-year-old skeletal specimen. J. Infect. Dis. 180, 2060–2063. ( 10.1086/315151) [DOI] [PubMed] [Google Scholar]
- 8.Montiel R, Solórzano E, Díaz N, Álvarez-Sandoval BA, González-Ruiz M, Cañadas MP, Simoes N, Isidro A, Malgosa A. 2012. Neonate human remains: a window of opportunity to the molecular study of ancient syphilis. PLoS ONE 7, e36371 ( 10.1371/journal.pone.0036371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donoghue HD. 2007. Molecular palaeopathology of human infectious disease. In Advances in human palaeopathology (eds Pinhasi R, Mays S.), pp. 147–176. Chichester, UK: Wiley & Sons Ltd. [Google Scholar]
- 10.Wilbur AK, Stone AC. Using ancient DNA techniques to study human disease. In The global history of paleopathology: pioneers and prospects (eds Buikstra JE, Roberts CA.), pp. 703–717. New York, NY: Oxford University Press. [Google Scholar]
- 11.Malgosa A, Montiel R, Díaz N, Solórzano E, Smerling A, Isidro A, García C, Simon M. 2005. Ancient DNA: a modern look at the infections of the past. In Recent research developments in microbiology (ed. Pandalai SG.), pp. 213–236. Trivandrum, India: Research Signpost. [Google Scholar]
- 12.Bäumler AJ, Hargis BM, Tsolis RM. 2000. Tracing the origins of Salmonella outbreaks. Science 287, 50–52. ( 10.1126/science.287.5450.50) [DOI] [PubMed] [Google Scholar]
- 13.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. 2014. Evolution and diversity in human herpes simplex virus genomes. J. Virol. 88, 1209–1227. ( 10.1128/JVI.01987-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W, et al. 2010. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467, 420–425. ( 10.1038/nature09442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourhy H, et al. 2008. The origin and phylogeography of dog rabies virus. J. Gen. Virol. 89, 2673–2681. ( 10.1099/vir.0.2008/003913-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trueba G, Dunthorn M. 2012. Many neglected diseases may have originated in the Paleolithic or before: new insights from genetics. PLoS Negl. Trop. Dis. 6, e1393 ( 10.1371/journal.pntd.0001393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grine FE, Gwinnett AJ, Oaks JH. 1990. Early hominid dental pathology: interproximal caries in 1.5 million-year-old Paranthropus robustus from Swartkrans. Arch. Oral. Biol. 35, 381–386. ( 10.1016/0003-9969(90)90185-D) [DOI] [PubMed] [Google Scholar]
- 18.Pezo-Lanfranco L, Eggers S. 2010. The usefulness of caries frequency, depth, and location in determining cariogenicity and past subsistence: a test on early and later agriculturalists from the Peruvian coast. Am. J. Phys. Anthropol. 143, 75–91. ( 10.1002/ajpa.21296) [DOI] [PubMed] [Google Scholar]
- 19.Da-Gloria P, Larsen CS. 2014. Oral health of the Paleoamericans of Lagoa Santa, central Brazil. Am. J. Phys. Anthropol. 154, 11–26. ( 10.1002/ajpa.22467) [DOI] [PubMed] [Google Scholar]
- 20.Gómez S, Romero A, de Juan J. 2009. ‘Patrón de subsistencia o modo de producción: caries y desgaste dental en poblaciones del Valle Alto del río Vinalopó (Alicante)’. In Investigaciones histórico-médicas sobre salud y enfermedad en el pasado’ (eds Cerdà P, Prósper G.), pp. 659–664. Valencia, Spain: Grupo Paleolab y Sociedad Española de Paleopatologia. [Google Scholar]
- 21.Halcrow SE, Harris NJ, Tayles N, Ikehara-Quebral R, Pietrusewsky M. 2013. From the mouths of babes: dental caries in infants and children and the intensification of agriculture in mainland Southeast Asia. Am. J. Phys. Anthropol. 150, 409–420. ( 10.1002/ajpa.22215) [DOI] [PubMed] [Google Scholar]
- 22.Janket S-J, Baird AE, Chuang S-K, Jones JA. 2003. Meta-analysis of periodontal disease and risk of coronary heart disease and stroke. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 95, 559–569. ( 10.1067/moe.2003.107) [DOI] [PubMed] [Google Scholar]
- 23.Grau AJ, et al. 2004. Periodontal disease as a risk Factor for ischemic stroke. Stroke 35, 496–501. ( 10.1161/01.STR.0000110789.20526.9D) [DOI] [PubMed] [Google Scholar]
- 24.Jagelaviciene E, Kubilius R. 2006. The relationship between general osteoporosis of the organism and periodontal diseases. Medicina (Kaunas) 42, 613–618. [PubMed] [Google Scholar]
- 25.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, Mira A. 2012. The oral metagenome in health and disease. ISME J. 6, 46–56. ( 10.1038/ismej.2011.85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forssten SD, Björklund M, Ouwehand AC. 2010. Streptococcus mutans, caries and simulation models. Nutrients 2, 290–298. ( 10.3390/nu2030290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smerling A, Solórzano E, Díaz N, Montiel R, Chimenos E, Malgosa A. 2007. Análisis de ADN de Streptococcus mutans recuperado en restos antiguos. In Enfermedad, muerte y cultura en las sociedades del pasado. Importancia de la contextualización en los estudios paleopatológicos (eds Barca Durán J, Jiménez Ávila J.), pp. 554–566. Fundación Academia Europea de Yuste. [Google Scholar]
- 28.Solórzano E, Díaz N, Smerling A, Montiel R, Malgosa A. 2011. Rastreo genético del Streptococcus mutans. Acta Odontológica Venezolana 49, 2. [Google Scholar]
- 29.Yang Z, Wong WSW, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. ( 10.1093/molbev/msi097) [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Nielsen R, Yang Z. 2004. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. ( 10.1093/molbev/msi237) [DOI] [PubMed] [Google Scholar]
- 31.Morisaki H, Igarashi T, Yamamoto A, Goto N. 2002. Analysis of dextran-binding domain of the dextranase of Streptococcus mutans. Lett. Appl. Microbiol. 35, 223–227. ( 10.1046/j.1472-765X.2002.01160.x) [DOI] [PubMed] [Google Scholar]
- 32.Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. ( 10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York, NY: Oxford University Press. [Google Scholar]
- 34.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. ( 10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Excoffier L, Laval G, Schneider S. 2005. ARLEQUIN v. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z. 2007. PAML 4: a program package for phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 37.Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. ( 10.1093/bioinformatics/bti079) [DOI] [PubMed] [Google Scholar]
- 38.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 39.Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozime evolution. Mol. Biol. Evol. 15, 568–573. ( 10.1093/oxfordjournals.molbev.a025957) [DOI] [PubMed] [Google Scholar]
- 40.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- 41.Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60. ( 10.1214/aoms/1177730491) [DOI] [Google Scholar]
- 42.Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. ( 10.1007/BF01731581) [DOI] [PubMed] [Google Scholar]
- 43.Preus HR, Marvik OJ, Selvig KA, Bennike P. 2011. Ancient bacterial DNA (aDNA) in dental calculus from archaeological human remains. J. Archaeol. Sci. 8, 1827–1831. ( 10.1016/j.jas.2011.03.020) [DOI] [Google Scholar]
- 44.De la Fuente C, Flores S, Moraga M. 2013. DNA from human ancient bacteria: a novel source of genetic evidence from archaeological dental calculus. Archaeometry 55, 767–778. ( 10.1111/j.1475-4754.2012.00707.x) [DOI] [Google Scholar]
- 45.Binladen J, et al. 2006. Assessing the fidelity of ancient DNA sequences amplified from nuclear genes. Genetics 172, 733–741. ( 10.1534/genetics.105.049718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert MTP, Binladen J, Miller W, Wiuf C, Willerslev E, Poinar H, Carlson JE, Leebens-Mack JH, Schuster SC. 2007. Recharacterization of ancient DNA miscoding lesions: insights in the era of sequencing-by-synthesis. Nucleic Acids Res. 35, 1–10. ( 10.1093/nar/gkl483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montiel R, Francalacci P, Malgosa A. 2007. Ancient DNA and biological anthropology: believers vs. skeptics. In Recent advances in molecular biology and evolution: applications to biological anthropology (eds Santos C, Lima M.), pp. 209–250. Kerala, India: Research Signpost. [Google Scholar]
- 48.King CE, Debruyne R, Kuch M, Schwarz C, Poinar HN. 2009. A quantitative approach to detect and overcome PCR inhibition in ancient DNA extracts. BioTechniques 47, 941–949. ( 10.2144/000113244) [DOI] [PubMed] [Google Scholar]
- 49.Igarashi T, Yamamoto A, Goto N. 1995. Sequence analysis of the Streptococcus mutans Ingbritt dexA gene encoding extracellular dextranase. Microbiol. Immunol. 39, 853–860. ( 10.1111/j.1348-0421.1995.tb03282.x) [DOI] [PubMed] [Google Scholar]
- 50.Maruyama F, et al. 2009. Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content. Genomics 10, 358 ( 10.1186/1471-2164-10-358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aikawa C, et al. 2012. Complete genome sequence of the serotype k Streptococcus mutans strain LJ23. J. Bacteriol. 194, 2754–2755. ( 10.1128/JB.00350-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ajdic D, et al. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl Acad. Sci. USA 99, 14 434–14 439. ( 10.1073/pnas.172501299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biswas S, Biswas I. 2012. Complete genome sequence of the Streptococcus mutans GS-5, a serotype c strain. J. Bacteriol. 194, 4787–4788. ( 10.1128/JB.01106-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song L, Sudhakar P, Wang W, Conrads G, Brock A, Sun J, Wagner-Döbler IW, Zeng AP. 2012. A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains. BMC Genomics 13, 128 ( 10.1186/1471-2164-13-128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YM, et al. 2011. Truncation of N- and C-terminal regions of Streptococcus mutans dextranase enhances catalytic activity. Appl. Microbiol. Biotechnol. 91, 329–339. ( 10.1007/s00253-011-3201-y) [DOI] [PubMed] [Google Scholar]
- 56.Rocha EPC, Smith JM, Hurst LD, Holden MTG, Cooper JE, Smith NH, Feil EJ. 2006. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 239, 226–235. ( 10.1016/j.jtbi.2005.08.037) [DOI] [PubMed] [Google Scholar]
- 57.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. 2002. Microevolutionary genomics of bacteria. Theor. Popul. Biol. 61, 435–447. ( 10.1006/tpbi.2002.1588) [DOI] [PubMed] [Google Scholar]
- 58.Read TD, et al. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296, 2028–2033. ( 10.1126/science.1071837) [DOI] [PubMed] [Google Scholar]
- 59.Baker L, Brown T, Maiden MC, Drobniewski F. 2004. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg. Infect. Dis. 10, 1568–1577. ( 10.3201/eid1009.040046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gutacker MM, et al. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feil EJ, et al. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185, 3307–3316. ( 10.1128/JB.185.11.3307-3316.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anisimova M, Bielawski J, Dunn K, Yang Z. 2007. Phylogenomic analysis of natural selection pressure in Streptococcus genomes. BMC Evol. Biol. 7, 154 ( 10.1186/1471-2148-7-154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cornejo OE, et al. 2012. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol. Biol. Evol. 30, 881–893. ( 10.1093/molbev/mss278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Do T, et al. 2010. Generation of diversity in Streptococcus mutans genes demonstrated by MLST. PLoS ONE 5, e9073 ( 10.1371/journal.pone.0009073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernal V, Novellino P, González P, Perez SI. 2007. Role of wild plant foods among Late Holocene hunter-gatherers from Central and North Patagonia (South America): an approach from dental evidence. Am. J. Phys Anthropol. 133, 1047–1059. ( 10.1002/ajpa.20638) [DOI] [PubMed] [Google Scholar]
- 66.Pezo-Lanfranco L. 2010. Reconstrução de padres paleopatológicos dentais em agricultores incipientes e desenvolvidos do litoral dos Andes Centrais. MSc. dissertation Instituto de Biociências. Universidade de São Paulo, São Paulo. [Google Scholar]
- 67.Piperno D. 2011. Northern Peruvian early and middle preceramic agriculture in Central and South American contexts. In From foraging to farming in the Andes. New perspectives on food production and social organization (ed. Dillehay T.), pp. 274–284. New York, NY: Cambridge University Press. [Google Scholar]
- 68.Caselitz P. 1998. Caries ancient plague of humankind. In Dental anthropology. fundamentals, limits, and prospects (eds Alt KW, Rösing FW, Teschler-Nicola M.), pp. 203–226. Wien, NY: Springer. [Google Scholar]
- 69.Larsen CS, Shavit R, Griffin MC. 1991. Dental caries evidence for dietary change: an archaeological context. In Advances in dental anthropology (eds Kelley MA, Larsen CS.), pp. 179–203. New York, NY: Wiley-Liss. [Google Scholar]
- 70.White CD. 1994. Dietary dental pathology and cultural change in the Maya. In Strength in diversity (eds Herring DA, Chan L.), pp. 279–302. Toronto, Canada: Canadian Scholar's Press. [Google Scholar]
- 71.Rodríguez JV. 2003. Dientes y diversidad humana, Avances de la antropología dental. Bogotá: Editorial Guadalupe Ltda. [Google Scholar]
- 72.Saunders S, De Vito C, Katzenberg A. 1997. Dental caries in nineteenth century Upper Canada. Am. J. Phys. Anthropol. 104, 71–87. () [DOI] [PubMed] [Google Scholar]
- 73.Parker M. 2011. The sugar barons: family, corruption, empire and war. London, UK: Hutchinson. [Google Scholar]
- 74.Lieberman LS. 2003. Dietary, evolutionary and modernizing influences on the prevalence of type 2 diabetes. Annu. Rev. Nutr. 23, 345–377. ( 10.1146/annurev.nutr.23.011702.073212) [DOI] [PubMed] [Google Scholar]
- 75.Shafer WG, Hine MK, Levy BW. 1983. A textbook of oral pathology. Philadelphia, PA: Saunders. [Google Scholar]
- 76.Brunelle JA, Carlos JP. 1990. Recent trends in dental caries in US children and the effect of water fluoridation. J. Dent. Res. 69, 723–727. [DOI] [PubMed] [Google Scholar]
- 77.Nikiforouk G. 1985. Understanding dental caries. New York, NY: Basel Karger. [Google Scholar]
- 78.Shaw JH. 1985. Diet and dental health. Am. J. Clin. Nutr. 41, 1117–1131. [DOI] [PubMed] [Google Scholar]
- 79.Haugejorden O. 1996. Using the DMF gender difference to assess the ‘major’ role of fluoride toothpastes in the caries decline in industrialized countries: a meta-analysis. Community Dent. Oral Epidemiol. 24, 369–375. ( 10.1111/j.1600-0528.1996.tb00881.x) [DOI] [PubMed] [Google Scholar]
- 80.Caufield PW. 2009. Tracking human migration patterns through the oral bacterial flora. Clin. Microbiol. Infect. 1(Suppl), 37–39. ( 10.1111/j.1469-0691.2008.02680.x) [DOI] [PubMed] [Google Scholar]
- 81.Moodley Y, et al. 2009. The peopling of the Pacific from a bacterial perspective. Science 323, 527–530. ( 10.1126/science.1166083) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.