Abstract
The biological processes that unfold during the G1-phase of the cell cycle are dependent on extracellular mitogenic factors which signal the cell to enter a state of quiescence, or commit to a cell cycle round by passing the restriction point (R-point) and enter the S-phase. Unlike normal cells, cancer cells evolved the ability to evade the R-point and continue through the cell cycle even in the presence of extensive DNA damage or absence of mitogenic signals. The purpose of this study was to perform a quantitative proteomic evaluation of the biological processes that are responsible for driving MCF-7 breast cancer cells into division even when molecular checkpoints such as the G1/S R-point are in place. Nuclear and cytoplasmic fractions of the G1 and S cell cycle phases were analyzed by LC-MS/MS to result in the confident identification of >2700 proteins. Statistical evaluation of the normalized data resulted in the selection of proteins that displayed ≥2-fold change in spectral counts in each cell state. Pathway mapping, functional annotation clustering and protein interaction network analysis revealed that the top-scoring clusters that could play a role in overriding the G1/S transition point included DNA damage response, chromatin remodeling, transcription/translation regulation and signaling proteins.
Keywords: proteomics, mass spectrometry, cell cycle, breast cancer
Introduction
Cancer is a complex disease characterized by a deregulation of the cell cycle clock which leads to uncontrolled proliferation of cells. In normal cells, the eukaryotic cell cycle is initiated by the binding of growth factors to transmembrane receptors, events that activate intracellular signaling pathways that promote cell growth and orderly entry into the four consecutive phases of cell division: G1, S, G2 and M [1-4]. The transition into the different cell cycle phases is controlled by Ser/Thr cyclin-dependent kinases (CDKs) that become active by forming complexes with cyclins (D1, E, A and B). Cyclins are proteins that oscillate in concentration through the cell cycle and thus periodically activate different CDKs [5-8]. In contrast to other cyclins, the level of the G1 cyclin D1 can be influenced by stimulation with growth factors, as well [3]. In fact, the G1 phase is susceptible to mitogenic growth and anti-growth signaling events that guide cellular entry into a quiescent state (G0), terminal post-mitotic differentiation, or commitment to cell cycle progression [5-8]. The R-point is a critical event in cell cycle control as it governs the G1-to-S transition process. Cells that pass the R-point are committed to a cell cycle round and are no longer dependent on mitogenic stimulation. Many cancer cells become self-sufficient of growth signals and insensitive to antigrowth signals, traits that allow them to evade the R-point and divide indefinitely. These capabilities, along with unlimited replicative potential, evasion of apoptosis, metastasis, sustained angiogenesis, deregulation of metabolism and evasion of immune destruction represent the major hallmarks of cancer [9].
Interestingly, over-expression of R-point cyclins D1 and E, and subsequent deregulation of the G1/S transition, has been observed in the majority of breast cancers [3, 8, 10]. Expression of cyclin D1 can be induced by several mitogens, including hormones such as estrogen. Cyclin D can bind to, and stimulate the activity of several transcription factors and nuclear receptors such as the estrogen receptor α. Approximately 70 % of breast cancers display an over-expression of the estrogen receptor (ER+), slower growth, enhanced differentiation, and better prognosis and response to hormone therapy [11,12]. The luminal epithelial MCF-7 cell line is the first, most representative and widely studied ER+ mammary carcinoma cell line, as demonstrated by the vast amount of information found in the literature [13]. The effects of mitogens, insulin, 17β-estradiol (E2) and Ca on cell cycle progression has been investigated by many authors [14-16]. In recent years, in addition to traditional molecular biology approaches, the power of advanced technologies such as mass spectrometry (MS), for assessing large-scale changes in the proteome and mapping post-translational modifications, has been recognized. One of the most advanced studies on MCF-7 cell cycle was performed by Emili and coworkers [17]. Their research included cell cycle synchronization, E2 stimulation, and comparison to HMEC184 normal human mammary epithelial cells. A total of 1481 MCF-7 proteins were identified, with the main protein categories being catalogued into cell cycle, signal transduction, apoptosis, transcription, translation, protein metabolism and catabolism. To advance the understanding of the molecular mechanisms that differentiate cancer from normal cells, and that control entry into the biological cell cycle, in the current study, the proteomic profiling of the G1 and S stages of the MCF-7 breast cancer cell cycle was pursued. Our aim was to investigate the mechanisms that drive aberrant cancer cells into division and control entry into the cell cycle even when molecular checkpoints that prevent the replication of damaged cells are in place.
Materials and Methods
Materials
MCF-7 breast cancer epithelial cells, EMEM, fetal bovine serum (FBS), 0.25 % trypsin/0.53 mM EDTA solution, and phosphate-buffered saline (PBS) were purchased from the American Tissue Culture Collection (ATCC, Manassas, VA). Insulin (bovine pancreas), 17-β estradiol, L-glutamine, Cell Lytic™ NuCLEAR™ extraction kit, phosphatase inhibitors (Na3VO4and NaF), urea, dithiothreitol (DTT), acetic acid, trifluoroacetic acid (TFA), NH4HCO3and all bovine protein standards (hemoglobin α/β, carbonic anhydrase, α-lactalbumin, fetuin, α-casein, β-casein and cytochrome c) were acquired from Sigma (St. Louis, MO). Sequencing-grade trypsin was obtained from Promega Corporation (Madison, WI), phenol-red free DMEM from Invitrogen (Carlsbad, CA) and charcoal/dextran treated FBS from Hyclone (Logan, UT). SPEC-PTC18 and SPEC-PTSCX solid-phase extraction pipette tips were from Varian (Lake Forest, CA). HPLC-grade methanol and acetonitrile were purchased from Fisher Scientific (Fair Lawn, NJ), and DI water was from a MilliQ Ultrapure water system (Millipore, Bedford, MA).
Cell processing
MCF-7 cells were cultured in an incubator (5 % CO2, 37 °C) in EMEM supplemented with 10 % FBS and 10 μg/mL insulin. The cells were arrested in G1 by serum-deprivation for 48 h in a medium consisting of DMEM and 4 mM L-glutamine. The cells were released into S by treatment for 24 h with DMEM containing physiological levels of E2 (1 nM), 10 % charcoal/dextran treated FBS, 4 mM L-glutamine and 1 μg/mL insulin. After harvesting, G1 and S-stage cells were stored at −80 °. Three biological replicates were processed. FACS analysis was performed on a Beckman Coulter EPICS XL-MCL analyzer (Brea, CA, USA). For analysis, the cells were thawed, lysed and separated into nuclear-enriched and cytoplasmic fractions by using the Cell Lytic™ NuCLEAR™ extraction kit and following the manufacturer’s protocol. Protein concentrations were measured using the Bradford assay (SmartSpec Plus spectrophotometer, Bio-Rad, Hercules, CA). Protein extracts (5 mg/mL) were denatured and reduced with 8 M urea and 4.5 mM DTT (1 hour, 60° C). Alkylation with iodoacetamide was not performed to avoid further increase in sample complexity due to incomplete reactions and/or the generation of side products. After a 10-fold dilution with 50 mM NH4HCO3, the extract was spiked with a solution of 8 standard bovine proteins (5 μM each), digested with trypsin (50:1 substrate:enzyme ratio, 24 hours, 37°C), quenched with glacial CH3COOH, and subjected to C18/SCX clean-up [18]. For MS analysis, each sample was re-suspended in CH3CN/H2O/TFA (5:95:0.1) to a final concentration of ~2 μg/μL MCF-7 proteins and 0.2 μM bovine standards.
LC-MS analysis and data processing
Each sample was analyzed five times (5 technical replicates) using a micro-LC system (Agilent Technologies, Palo Alto, CA) coupled to an LTQ-MS (Thermo Electron Corporation, San Jose, CA) using an on-column/no split injection set up described in detail elsewhere [18]. The LC separation columns were packed in-house using 100 μm i.d. × 12 cm fused silica capillaries and 5 μm Zorbax SB-C18 particles, and run at a flow rate of ~160-180 nL/min with a 3-hour long gradient (0-100 % B). Mobile phases A and B consisted of H2O:CH3CN:TFA in 95:5:0.01 and 20:80:0.01 v/v ratios. MS data were acquired via a data-dependent method by performing Zoom/MS2scans on the 5 most intense peaks in each MS scan [18]. The Bioworks 3.3 software (Thermo Electron) was used for searching the raw data files against a minimally redundant human protein database (SwissProt, 40,009 entries, 2008). Only fully tryptic fragments with up to two missed cleavages were allowed in the search, the peptide and fragment ion tolerances were 2 amu and 1 amu, respectively, % fragment ion coverage >30 % (from any combination of theoretical b, y and a ions) and all peptides were matched to unique proteins in the database. No posttranslational modifications were allowed. At the peptide level, MS filtering was performed with the Xcorr vs. charge state parameter set at 1.9, 2.2 and 3.8 for 1+, 2+ and 3+ peptides, respectively. At the protein level, only proteins with Bioworks p-score<0.001 were considered. We note that the Bioworks p-scores are not p-values, per se, but scores. As a result, false discovery rates (FDR) were assessed by searching the raw data against a forward-reversed sequence database.
For differential protein expression analysis, in-house developed Perl-scripts for the alignment of spectral count data were used [19]. Twelve samples were analyzed (G1 and S nuclear/cytoplasmic fractions, 5 LC-MS/MS technical and 3 biological replicates of each cell state). Proteins matched by a single peptide were allowed in the analysis, but only when the peptides were identifiable in multiple biological replicate analyses and had p-scores<0.001. The proteins that qualified for differential expression analysis were matched by a total of at least 4 spectral counts, representing almost invariably more than 2 unique peptides. To analyze the data in a biological context, a set of bioinformatics tools provided by the ExPASy Proteomics Server [20], GeneCards [21], GoMiner [22], DAVID Bioinformatics Resources 6.7 [23], and STRING functional protein association networks 8.3 [24] software packages, was used. The GoMiner parameters included all evidence codes; STRING parameters were set to medium/high confidence, ≤10 interactors, network depth 1, all active prediction methods; DAVID functional clustering/enrichment p-scores were calculated by comparing the lists of differentially expressed protein to the Homo sapiens background. The enrichment p-scores are shown as -log transformed values, and represent the geometric mean of all enrichment p-values for each annotation term in that group. The p-score threshold was 1.3, corresponding to a non-log scale of p=0.05 [23,25].
Results and Discussion
Sample and data processing outline
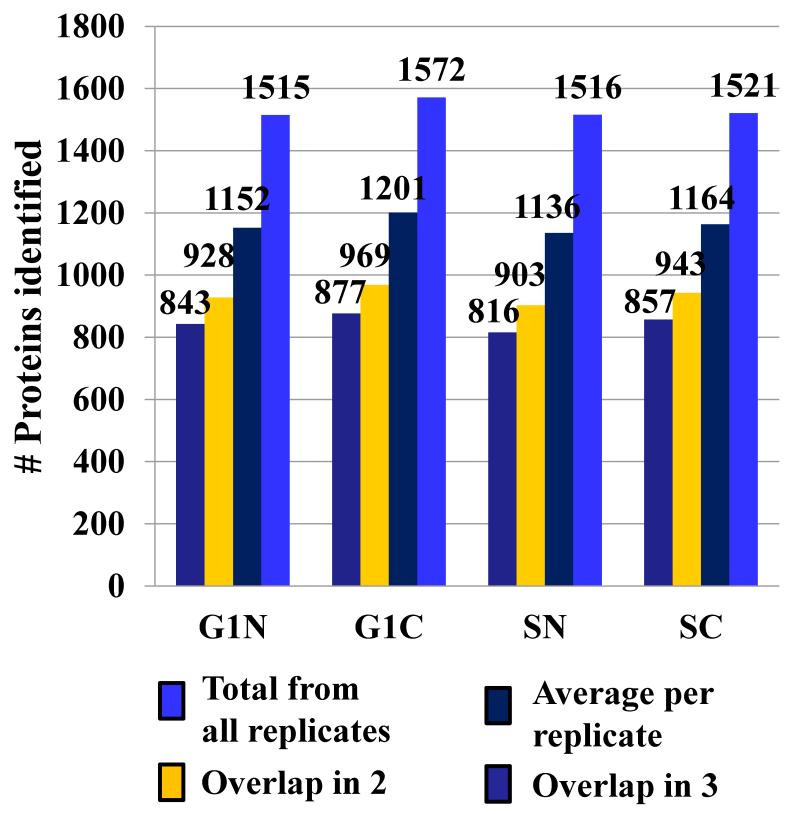
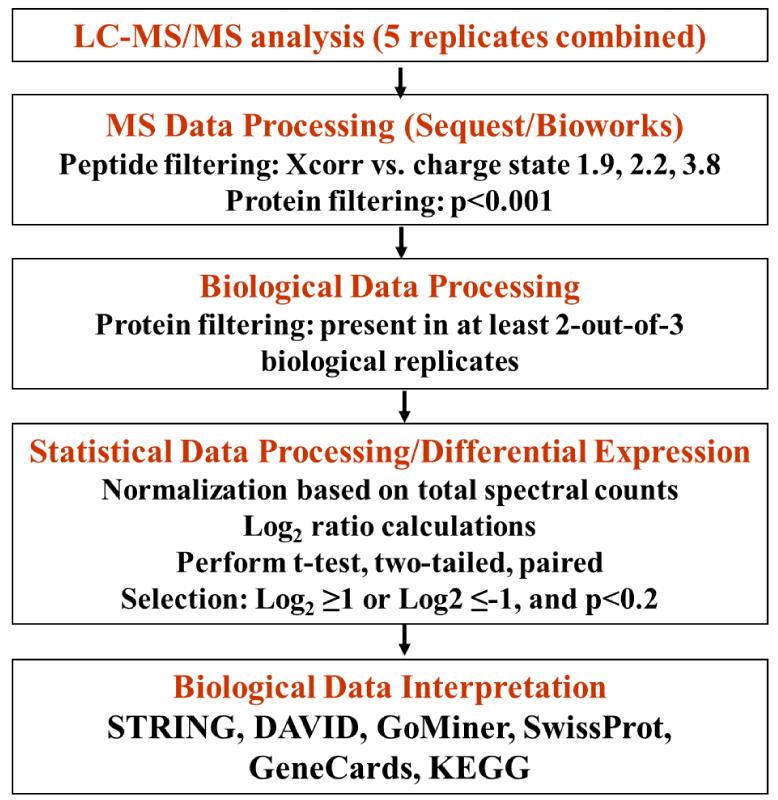
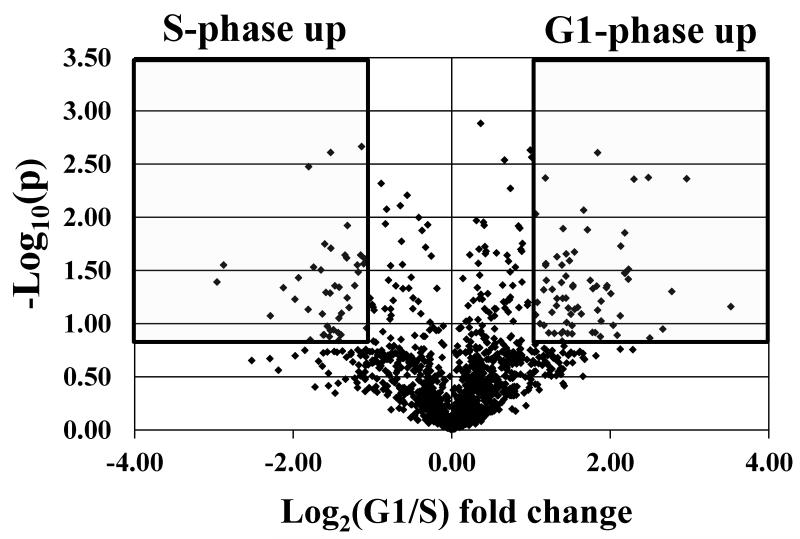
For quantitative proteomic experiments that involve multiple sample preparation and analysis steps, an advanced strategy for sample and data processing must be developed to ensure meaningful selection of differentially expressed proteins. To minimize the impact of experimental variability on protein identification and quantitation, in this work, the following measures were taken: (a) three biological replicates of the G1 and S cell cycle stages, with further separation into nuclear and cytoplasmic fractions, were analyzed (i.e., G1 nuclear-G1N, S nuclear-SN, G1 cytoplasmic-G1C and S cytoplasmic-SC; a biological replicate is defined as the analysis of a new batch of liquid N2frozen cells; a cell state is defined as a nuclear or cytoplasmic fraction of G1 or S cells, respectively); (b) five LC-MS/MS technical replicates were performed for each cell state to maximize the number of spectral counts per protein and improve reproducibility; (c) qualitative data filtering was performed at both protein and peptide levels with Xcorr vs. charge state set at 1.9, 2.2 and 3.8, and p-score<0.001, respectively; the protein FDR was <2.9 %, and the peptide FDR was <0.9 %; (d) proteins were qualified for quantitative analysis only if they were identified in two-out-of-three biological replicates; and (e) reproducibility was assessed for every step of the analysis. FACS profile data (Supplemental Figure 1) indicated that the G1/S/G2 percent-distribution of cells was roughly 80/10/7 and 28/60/10 in the G1 and S phases, respectively, with CV=2-12 % for the three G1 and S biological replicates. The G1-to-S ratio of cells changed by a factor of ~17 in going from G1 to S. After mass spectral filtering, a total of 2725 proteins were identified (Figure 1): the average number of proteins identified per biological replicate and cell state was 1163 (CV=2.4 %), with a combined value in all three biological replicates of 1531 (CV=1.8 %); the average number of proteins identified in at least two-out-of-three biological replicates was 936 (CV=2.9 %); and, the average number of proteins that overlapped in all three replicates was 848 (CV=3.0 %). The reproducibility of nuclear/cytoplasmic fractionation was assessed through GoMiner analysis: the nuclear cell fractions comprised 53-62 % nuclear and 59-66 % cytoplasmic protein designations, while the cytoplasmic fractions comprised 83-84 % cytoplasmic and 32-33 % nuclear protein designations (we note that some proteins had dual categorization). For quantitative comparisons, the raw MS data were subjected to three levels of data selection: raw MS data filtering, biological data filtering, and statistical data filtering. For the latter, in-house developed Perl scripts compiled the MS/MS database search results and produced an alignment of proteins with their respective spectral counts (12 samples, each having 5 technical replicates). The spectral counts for each protein in the 5 technical replicates were averaged to generate the final count for the protein in that sample. For data normalization purposes, all protein counts were summed up to generate the total counts for that particular sample. The total spectral counts for the 12 samples were averaged, and used for the normalization of individual protein counts in each sample. The average spectral count per cell state was 4082 (CV=7.5 %). After normalization, for handling missing values (proteins with zero counts), a count of 0.2 was added to each protein (equivalent to the addition of 1 count to any of the 5 technical replicates). Differential expression analysis was performed by: (a) calculating the G1-to-S spectral count ratios for each biological replicate in the nuclear and cytoplasmic fractions, (b) calculating the log2values of each spectral count ratio, (c) calculating the average of the three log2values for each protein, (d) and subjecting the data-sets to a two-tailed/paired student t-test. Proteins that displayed at least a 2-fold change in spectral counts (i.e., average log2values ≥1 or ≤-1) and p-values≤0.2 were taken into consideration for quantitative analysis. Figure 2 summarizes the protocol developed for this study. We note that in the context of large-scale exploratory studies, a p-value<0.2 is acceptable for datasets with a limited number of replicates, as the ultimate biological significance needs to be evaluated at the system level, not the isolated protein level [25]. Moreover, statistical significance of differential expression for single proteins may not reflect correctly biological significance. Selecting only proteins with p<0.1 or p<0.05 from our dataset enabled essentially the identification of the same differentially expressed clusters, but with a smaller number of protein members in the cluster. From a total of 2725 identified proteins, 1944 were identified in at least two biological replicates, of which 1234 were present in the nuclear-enriched and 1273 in the cytoplasmic fractions, respectively. After statistical filtering, only 395 nuclear and 312 cytoplasmic proteins qualified for quantitative comparisons. A total of 95, 63, 76 and 48 differentially expressed proteins were identified in the G1N, G1C, SN and SC fractions, respectively. The distribution of differential expression p-values for these proteins was p≤0.05 (42%), 0.05<p≤0.1 (20%), 0.1<p≤0.15 (15%), 0.15<p≤0.2 (23%). A volcano plot provided in Figure 3 illustrates the outcome of the statistical data evaluation process for the G1 and S nuclear fractions. As a multi-level experimental control, each cell extract was spiked with 8 standard bovine proteins. Validation of the differential expression data was performed by: (a) assessing the results for the standard protein spikes (in contrast to the MCF-7 proteins, none of the standards passed the combined 2-fold/p<0.2 threshold that was selected as an indicator of protein differential expression; Supplemental Table 1); (b) assessing the “change” in spectral counts for proteins that are known to change upon G1-to-S transition; three proteins, indicators of cell proliferation, that all passed the 2-fold/p<0.2 differential expression threshold were selected: CDK1, PCNA (a cyclin) and KI67 in the nuclear fraction (see discussion on SN up-regulation vs. G1N); (c) assessing the “no-change” in spectral counts for housekeeping proteins that are not expected to change upon G1-to-S transition; three proteins, that are often used as internal standards due to relatively constant expression level in the cell, were selected: GAPDH in the nuclear fraction, and TBA1A and TBB2A in the cytoplasmic fraction; none of them passed the 2-fold/p<0.2 double-threshold (Supplemental Table 2); and (d) FACS experiments that confirmed progression through the cell cycle (Supplemental Figure 1).
Figure 1.
Reproducibility of protein identifications in three biological replicates. The bar graphs represent overlapping proteins between biological replicates, and the average and total number of protein IDs per cell state (G1N, G1C, SN and SC).
Figure 2.
Sample and data processing protocol. Three biological replicates of G1 and S, nuclear and cytoplasmic cell fractions, were analyzed by LC-MS/MS. Data filtering was performed at three levels: mass spectrometric, biological and statistical.
Figure 3.
Volcano plot illustrating protein differential expression in the G1 (95 proteins) and S (76 proteins) nuclear fractions.
Protein differential expression
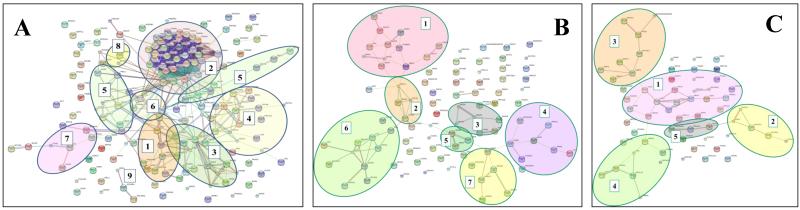
The list of 1944 proteins comprised a considerable number of cell cycle and cancer hallmark proteins (Figure 4A). The proteins that displayed differential expression between G1 and S were queried with STRING to identify the network of interacting proteins within each group. Kegg pathway charts [26] and protein descriptions provided by SwissProt, STRING, DAVID and GoMiner facilitated the interpretation of the biological processes associated with the highlighted networks. The significance of the major protein clusters is discussed, with focus on the changes in the nuclear fractions, in particular G1, which has the highest relevance to cancer (Supplemental Table 3). Enrichment p-scores for the most relevant members of the group are provided. Up-regulated proteins are highlighted in bold, while proteins that were identified in the dataset, but did not pass the differential expression thresholds, are underlined. Gene abbreviations are provided in Supplemental Table 4.
Figure 4.
STRING diagrams. (A) Cell cycle proteins identified in all MCF-7 fractions: (1) cell cycle regulation; (2) proteasome degradation; (3) chromatin maintenance; (4) DNA repair; (5) mitosis/cytokinesis; (6) nuclear import/export; (7) integrin signaling; (8) G1 arrest proteins; (9) ribosome biogenesis. (B) Up-regulated proteins in G1N: (1) DNA damage repair/transcription repression; (2) transcription activation/repression; (3) transcription initiation (Mediator proteins); (4) transcription activation/cell motily/adhesion; (5) nuclear lamina organization; (6) ribosome biogenesis/translation initiation; (7) translation/amino acid metabolism; (C) Up-regulated proteins in SN: (1) cell proliferation/cell cycle regulation; (2) transcription/rRNA processing, (3) mitochondrial protein metabolism; (4) mitochondrial metabolism; (5) protein folding.
G1N up vs. SN(Figure 4B)
The G1N up-regulated functional clusters included DNA damage repair, transcription regulation/chromatin remodeling and ribosome biogenesis/translation proteins. Smaller sets included cell cycle, signaling, differentiation, endocytosis and cell motility.
DNA damage repair (4-16-fold enrichment/p-score 2.2)
Genomic stress induced by various factors results in the activation of the DNA cell cycle check point and transcriptional programs at the G1/S and G2/M boundaries, and of DNA repair, mRNA decay and apoptosis pathways. The DNA damage repair cluster identified in this study encompassed members of the entire plethora of mechanisms used by cells to respond to mutagenic events: nucleotide excision repair (XPC and PNKP), base excision and single strand repair (XRCC1), double-strand break repair (RAD50 and TP53BP), DNA mismatch repair (MSH3), and other possible DNA repair proteins (MEN1). Cells have developed various DNA repair mechanisms to correct for genomic damage: mismatch repair (MMR) corrects miscopied DNA bases after DNA synthesis, base excision repair (BER) targets minor chemical nucleotide alterations by removal of the affected base, nucleotide excision repair (NER) corrects bulky DNA lesions that cause helical distortions by removal of a short strand containing the damaged bases, homologous recombination (HR) uses sister chromatids as a template to restore genomic sequences in case of double strand breaks (DSB), while the less accurate non-homologous endjoining (NHEJ) process targets DSBs by rejoining the broken ends using DNA ligases [9,27,28]. The signal transduction pathways that sense DNA damage and activate the repair mechanisms are known as the DNA damage response (DDR) pathways. The DDR mechanism is initiated primarily by a set of PIKK (phosphatidylinositol 3-kinase-like protein kinase) proteins such as ATM, ATR and DNAPK (DNA protein kinase), and PARP family proteins that activate the DNA damage checkpoint (Chk1/Chk2) machinery. The DDR is further mediated by essential components of this pathway such as p53 (tumor protein 53), TP53BP1 (tumor protein 53 binding protein 1), BRCA1, MDC1 (mediator of DNA damage), H2AX (histone 2A variants), CDK inhibitor p21, and the pro-apoptotic BAX and PUMA proteins, just to name a few. Ciccia described four independent sensors of single and double-strand breaks: PARP, Ku70/Ku80, MRN (MRE11, RAD50 and NBS1) and RPA (replication protein A) [28]. DNAPK is a DSB repair enzyme composed of a catalytic subunit, PRKDC, and a KU protein heterodimer (KU70 and KU80 or KU86). PRKDC, KU70 and KU80 were identified in all fractions of the dataset. In NHEJ, DSBs rapidly bind the KU heterodimer, which subsequently loads and activates PRKDC, causing stabilization of the DSB ends and recruitment of the X-ray repair cross-complementing protein 4 (XRCC4)/DNA ligase 4 (LIG4) complex which promotes religation of the ends [28]. NHEJ is active throughout all stages of the cell cycle, but is the preferred mechanism in G1, as there is no homologous template present for recombination. Alternatively, in HR, PARP1 recruits the MRN complex to the DSB. The MRN complex is the primary sensor of DSBs and is composed of the meiotic recombination 11 homolog 1 (MRE11), the DNA repair protein RAD50 and the Nijmegen breakage syndrome protein 1 (NBS1) [28]. MRE11 and RAD50 stabilize the DSB ends and participate in their initial resection. DNA end resection is essential for HR and is primarily promoted in S and G2, where sister chromatids are available for HR. As HR depends on the availability of intact DNA strand templates for repair, it is the preferred mechanism in late S/G2. Limited resection can be carried out during G1, however, through an alternate NHEJ mechanism which utilizes PARP1 as a sensor, the MRN complex, and a complex formed by the X-ray repair cross-complementing protein 1 (XRCC1) and the DNA ligase 3 (LIG3) to religate DSB ends [28]. TP53BP1 was shown to inhibit DSB resection and promote NHEJ, while the deletion of TP53BP1 was shown to induce DSB resection via the alternate NHEJ in G1.
In single strand BER, protein complexes formed by PARP/XRCC1/APEX/Lig3/polβ or PARP/PCNA/FEN1/APEX/Lig1/polβ/polδκ are assembled at the sites of DNA damage to assist repair by short-patch or long-patch BER, respectively [26,29]. PNKP (bifunctional polynucleotide phosphatase/kinase) has been shown to interact with XRCC1, and has a role in processing single and double strand break termini in a variety of DNA repair mechanisms [30]. The XPC (xeroderma pigmentosum, complementation group C) repair complex (XPC/hHR23B/CETN2) and the Cul4/DDB1 (damaged DNA binding 1)/DDB2 complex are involved in the first step of damage recognition in global genome repair (GGR)-NER and transcriptional coupled repair mechanisms, recognizing a broad spectrum of DNA helix distortions such as single-stranded loops, mismatched bubbles or single stranded overhangs [21,26,27]. Damage recognition by XPC is followed by the recruitment of the TFIIH (basal transcription factor II) complex for additional scanning for lesions in the opposite strand, and as part of this complex, ERCC2, another NER protein, was identified. Components of the MMR machinery, that correct DNA mismatches generated during DNA replication to prevent genome-wide instabilities, have been also identified in the nuclear fractions. MutS protein homolog 2 (MSH2) forms heterodimers with MSH3 and MSH6 to recognize DNA damage sites (mismatches and insertion/deletion loops-IDLs). By forming higher order complexes, it promotes the excision of the lesion and strand correction by DNA re-synthesis and ligation [26,29]. Ultimately, the tumor suppressor MEN1, with a transcriptional role in blocking the G1/S transition by promoting the expression of CDK inhibitors, was also observed in G1 [31].
Altogether, the global activation of the DNA damage repair machinery is in accord with greater genomic surveillance prior to entry into S. Inherently, some of the differentially expressed repair proteins are implicated in a variety of other biological processes, e.g., TP53BP1 plays a role in checkpoint signaling and also enhances TP53-mediated transcriptional activation, RAD50 is involved in maintenance of telomere integrity along with TERF2 (telomeric repeat binding factor 2), and XPC is involved in chromatin remodeling and ubiquitin mediated proteolytic pathways. Moreover, a number of damage repair proteins that did not pass the 2-fold differential expression threshold displayed an increased level of spectral counts in G1: PARP1, PRKDC, KU70, KU80, MDC1. The complex interactions between the DNA damage repair proteins and the biological processes implicated in cell cycle regulation are highlighted in the STRING diagram of all 119 repair proteins identified in MCF-7 (Figure 5). The interactions between DNA repair and essential cell cycle/proliferation and chromosome/chromatin maintenance proteins represent the hubs of the most feverish interactions, re-enforcing the key role that these proteins play not only in the maintenance of genome integrity, but also in controlling the entire cell cycle proliferative apparatus.
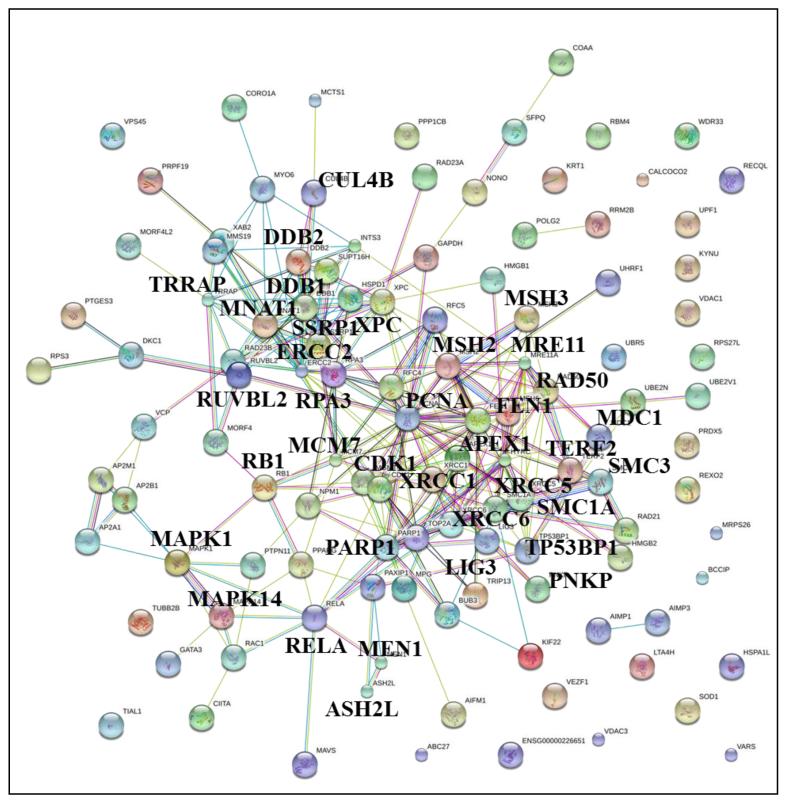
Figure 5.
STRING diagram of 119 proteins with roles in DNA damage repair. DNA damage repair proteins: TP53BP, XPC, XRCC, MSH, RAD, DDB1/2, RPA3, PARP1, ERCC2, HYRC, APEX1, FEN1, MDC1, MRE11, LIG3, CUL4; Cell cycle/proliferation proteins: PCNA, CDK1, RB1, RELA, MNAT; and, Chromosome/chromatin maintenance protein: TERF2, ASH2, TRRAP, RUVBL2, SMC1A, SMC3, SSRP1 and MCM.
(b) Transcription regulation/chromatin remodeling and organization
(2-17-fold enrichment/p-scores 1.3-2.1). The transcriptional regulators that were up-regulated in G1 had repression, activation or mixed functions, and were part of interacting clusters that involved DNA damage response and chromatin remodeling proteins. Five components of the Mediator complex (MED8, MED22, MED24, MED13 and MED25), all transcriptional coactivators thought to be required for the transcription of almost all RNA polymerase II-dependent genes, were identified, three being 2-3 fold up-regulated. RNA polymerase II controls DNA transcription to generate mRNA, snRNA (small nuclear RNA) and miRNA (MicroRNA). snRNAs, by association with proteins in complexes, are involved in the regulation of transcription factors, telomere maintenance and RNA splicing. miRNAs are posttranslational regulators that control gene expression by transcript degradation or translational repression to ultimately result in gene silencing. Recent work has shown that aberrant expression of miRNA is correlated with cancer [32], that coregulatory complexes that involve histone acetyl transpherases (HAT) and deacetylases (HDAC) can modulate the transcriptional activity of ligand-bound ERs, and that overexpression of MED1, a component of the Mediator complex, renders ER+ breast cancer cells resistant to tamoxifen [33].
The regulators that displayed interactions with the DNA damage repair proteins in the STRING diagram included mainly transcriptional repressors (ZNF217, ZNF512B, MBD2, CTBP2, ASH2L, DR1 and PELP1). The data suggest that the DNA damage repair machinery is intertwined with cellular processes that result in epigenetic modifications of target genes, to facilitate the assembly of transcription repressor complexes that mediate chromatin remodeling [34,35], a sought outcome in a cell presenting multiple mutations. One mechanism that controls the accessibility of the DNA template for transcription involves posttranslational modifications of the core histone tail Lys residues. In our dataset, ZNF217 (zinc finger protein 217) which is a transcriptional repressor oncoprotein, promotes cell proliferation by silencing the expression of CDK inhibitor p15(ink4b) (CDKN2B), a tumor suppressor that controls cell cycle progression in G1 [36]. Amplification of ZNF217 was associated with loss of responsiveness to growth-inhibitory signals via the TGF-beta pathway, and with roles in evasion of apoptosis [36-38]. ZNF217 is also a component of the histone deacetylase complexes that contain HDAC1, HDAC2, CTBP1 and 2, which regulate cell cycle via epigenetic repression. ZNF512B was shown to have roles in decreasing the transcriptional activity of SMADSs in the TGF-β pathway, the expression of E2F1 and cell proliferation, while increasing apoptosis and the expression of RAS proteins [39]. MBD2, a transcriptional repressor that recruits histone deacetylases and DNA methyltransferases, has roles in gene silencing, including the tumor suppressor CDK inhibitors p14 and p16 [40]. CTBP2 (C terminal binding protein 2) acts as a transcriptional co-repressor of transcription regulators and of target genes of intracellular signaling pathways such as Wnt and Notch [41-44], and it associates with chromatin modifiers such as HDACs. DR1, another repressor of activated and basal transcription of class II genes, can interact with acetyltransferases to alter the activity of Histones H3 and H4. ASH2 functions as a transcription regulator component of a histone methyltransferase complex involved in the methylation of Lys-4 histone H3 tails, an epigenetic event associated with active gene transcription and activation [45]. Interestingly, MEN1 was shown to interact with complexes that include ASH2 to possibly regulate the transcription of CDK inhibitors p18 and p27 and inhibit cell proliferation [45]. Finally, PELP1 (proline, glutamate and leucine rich protein 1) which facilitates estrogen receptor (ER) genomic and non-genomic signaling, is a corepressor of some nuclear hormone receptors and a coactivator of estrogen receptor-mediated transcription. It was found to be implicated in the phosphorylation and activation of ERK1/ERK2, expression of cyclin D1, hyperphosphorylation of retinoblastoma protein (RB) and cell cycle progression [20,24,46,47]. PELP1 also displayed interactions with TAF2E, a component of the transcription factor IID (TFIID) and PCAF (P300/CBP-associated factor) histone acetylase complexes. The TFIID complex is coordinating the initiation of transcription by RNA polymerase II, and plays an essential role in mediating promoter responses to transcriptional activators and repressors [21]. Targets of PCAF’s acetyltransferase activity include p53, and of its ubiquitin-ligase activity include the oncoprotein HDM2-a ubiquitin ligase itself that targets p53 for degradation [48,49]. DNA accessibility can be controlled through an alternate mechanism, as well, via ATP-dependent complexes that use the energy of ATP hydrolysis to alter or disrupt the association of DNA with the histone proteins. G1N up-regulated proteins, part of such complexes, included transcriptional regulators with repressor/activator roles such as SMARCA5, SMARCD2 and HLTF. SMARCA5 facilitates transcription by RNA polymerase II, and HLTF has helicase and ligase activities and is also involved in error-free postreplication repair of damaged DNA [21].
Two additional up-regulated proteins, significant to maintaining telomere and chromosome integrity, were TERF2 and RAD50. Telomeric DNA shortens after every replication event, and after reaching a critical length, cells become senescent [50,51]. Unlimited proliferative capacity is a hallmark of tumorigenesis, and cancer cells acquire it by maintaining or lengthening their telomeres using two independent mechanisms: telomerase-mediated telomere synthesis or alternative telomere-lengthening (ALT) [9,50,51]. TERF2 mediates T-loop formation, stabilizes telomere structure, participates in negative regulation of telomere length, and binds DNA repair proteins that also play a role in telomere homeostasis [50,52]. Some of the DNA repair proteins that bind TERF2 have been discussed above in a DSB repair context: DNAPK, RAD50, MRE11 and PARP1 [50]. A complex consisting of MRE11, RAD50 and TERF2 localizes to the telomeres during interphase, and possibly plays an important role in stabilizing T-loop formation. The association of PARP1 with TERF2 seems to localize the former to sites of telomere damage. Upon poly(ADP-rybosyl)ation of TERF2, it dissociates from telomeres which gives the DNA repair machinery access to repair damaged telomeres. The STRING networks also revealed that TERF2 interacts with TP53BP1 and ZNF217. As part of the DNA damage response, TP53BP and other proteins accumulate at the chromosome ends upon inhibition of TERF2 or critical telomere shortening [24]. Gene amplification of ZNF217, which has been observed in MCF-7 cells, is associated with increased telomerase activity and infinite lifespan of cells [37].
Additional contributions to chromatin organization were provided by the lamin proteins. The nuclear envelope (NE) separates the nucleus from the cytoplasm, and functions in chromatin regulation, gene expression, signaling, cell proliferation and DNA repair have been described [53]. Isoform beta/gamma of the NE transmembrane protein lamina-associated polypeptide 2 (LAP2B or TMPO) is transcriptionally up-regulated in a variety of tumors and may have a regulatory role in nuclear growth. LAP2B binds to barrier-to-autointegration factor (BAF), identified in all fractions, to participate in chromatin folding, and its overexpression has been linked to inhibition of E2F-dependent transcription [53]. STRING network predictions revealed that LAP2B interacts with both lamins A/C (LMNA) and B1 (LMNB1), both of which displayed up-regulation in G1N. Lamins, the only known filament system in the nucleus, participate in signaling by binding and sequestering transcription factors [53]. Lamin A, interacts with lamina-associated polypeptide 2 isoform alpha (LAP2A) and the retinoblastoma-associated (RB1) proteins, both of which were identified in the nuclear fractions, to sequester the RB1/E2F complex and arrest the cell cycle in the G1 phase [53]. Thus, up-regulation of LMNA in G1N is in accordance with the arrested state of the MCF-7 cells.
In addition to transcriptional repressors, transcriptional activators with roles in cell growth, proliferation and differentiation were also identified. These included a general transcription factor (GTF2E1) and activators of target genes of signaling pathways such as Wnt (CTNND1), estrogen signaling (PELP), or of pathways associated with development and differentiation (YY1 or Yin Yang 1). YY1 was also found to negatively regulate p53 by ubiquitination and subsequent degradation [54], prevent cyclin D1 accumulation by blocking the gene promoter, and bind RB to promote progression to the S phase [55]. CTNND1 and YY1 displayed STRING interactions with a group of proteins with roles in endocytosis, actin polymerization and cell motility (DNM1, DNM2, WASL, ARPC5, ARFGAP2), proteins that have been associated in turn with cytokinesis, cell adhesion and metastasis. Along with YY1, other proteins such as CTBP2, LIMS1 and ROD1, had all additional roles in regulating cell differentiation, a program that is often distorted in cancer cells. LIMS, that has shown STRING interactions with an integrin linked kinase (ILK), is an effector of integrin and growth factor signaling pathways [21], and overexpression of ROD1 has been shown to block differentiation of human leukemia cells without affecting their proliferative ability [56].
(c) Translation initiation/Ribosome biogenesis
(3-10-fold enrichment/p-scores 1.6-2.14). Production of ribosomes and translation are critical, high energy-consuming processes in cell growth and proliferation [57,58]. Cancer cells are known to display elevated ribosomal biogenesis and protein synthesis processes [58], and tumorigenesis has been reported to be promoted by the over-expression of individual subunits of translation initiation factors [59]. The subunit alpha of eukaryotic translation initiation factor 2 (EIF2A or EIF2S1) that catalyzes the first step of protein synthesis by forming a complex with tRNA and GTP and binding to a 40S ribosomal subunit, along with the eukaryotic release factor 1 (ERF1 or ETF1), which recognizes termination codons and terminates the nascent peptide synthesis, were both up-regulated in G1N. The phosphorylation of EIF2A protein product halts protein synthesis and activates the NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells) signaling pathway [60]. NFκB is a transcription factor that is activated in response to various stimuli (stress, radiation, free radicals), and its inappropriate regulation was linked to a number of biological processes such as immune response, cell growth and development, differentiation, tumorigenesis and apoptosis. Both EIF2A and ETF1 displayed interactions with RAN, a member of the RAS oncogene family, which is required for protein import into the nucleus and RNA export into the cytoplasm, as well as for chromatin condensation and cell cycle control. In addition, EIF2A and ETF1 showed multiple interactions with a set of up-regulated proteins involved in the assembly of the ribosomal subunits 40s and 60s (RPL18A, NOP56, BRIX1, RSL1D1, PDCD11, EXOSC4 and NVL). Two tyrosyl-/aspartyl-tRNA synthase proteins (YARS, DARS2), with key role in linking amino acids with nucleotide triplets contained in tRNA, displayed STRING interactions with transferases and hydrolases involved in amino acid metabolism (SHMT1, SHMT2, NDRG1).
(d) Other functional clusters
Additional up-regulated protein clusters included mitochondrial respiration, amino acid metabolism and RNA processing. Most importantly, several smaller sets of proteins that were discussed in the previous sections, but had additional functions in cell cycle regulation and proliferation, were identified. Positive regulators of cell cycle included proteins with role in DNA damage repair, proteasome degradation, transcriptional activation and signaling (TERF2, CUL1, PELP1, RAN), while negative regulators included proteins with role in transcriptional repression, signaling, apoptosis and stress response (TP53BP1, CCAR1, CDK11A, NDRG1, PPP2R1A, PPP2CB). CUL1, for example, as part of the Skp/Cullin-F-box complex (SCF), regulates the degradation of CDK inhibitors p21 and p27. Its up-regulation, may have, therefore, an important role in G1/S cell cycle progression in MCF-7 cells [61]. CCAR1 is involved in transcriptional regulation and induction of apoptosis by altering the expression of some genes that control cell cycle regulation (MYC, CCNB1 and CDKN1A), while CDK11A, with roles in apoptosis, is believed to also act as a negative regulator of cell cycle progression [21]. NDRG1 is implicated in growth inhibition through its participation in biological processes related to stress response, differentiation and p53-mediated caspase activation and apoptosis [62]. PPP2R1A and PPP2CB are major Ser/Thr phosphatases that have been implicated in signaling processes with negative role in cell division [21]. Additional proteins with roles in cancer progression included GLYR1 (glyoxylate reductase 1 homolog), ILK (integrin linked kinase), CDC42BPB (Serine/threonine-protein kinase MRCK beta), MTA2 (metastasis associated protein) and CROP (cysplatin resistance associated overexpressed protein). GLYR1 regulates p38/MAPK14 signaling by phosphorylating MAPK14 and activating ATF2 (Activating transcription factor 2) [63]. In turn, ATF2 which is a HAT, also stimulates CRE (cAMP-responsive element)-dependent transcription, and its abnormal activation was linked to the development of aggressive epithelial tumors [21]. ILK and CDC42BPB, through their roles in mediating cell architecture and cytoskeleton organization, respectively, have potential roles in anchorage-dependent cell growth and invasion [21]. MTA2 and CROP, both widely expressed proteins, did not pass the 2-fold up-regulation threshold, but their relevance and low p-value warrants their discussion (log2=0.88/p=0.02 and log2=0.74/p=0.01). MTA2 has roles in transcription regulation and chromatin remodeling by covalent modifications of histone proteins. Through its implication in p53 deacetylation, MTA2 is believed to be involved in loss of growth inhibition and metastasis [64]. CROP is a nuclear protein that has been isolated from cysplatin resistant cell lines, and is believed to be involved in the formation of the spliceosome [65].
SN up vs. G1N (Figure 4C)
As expected, the up-regulated clusters in the SN fraction are consistent with progression through the cell cycle. These clusters included mainly proteins involved in cell proliferation/cell cycle regulation, metabolism (e.g., energy production, oxidative phosphorylation, redox reactions, mitochondrial respiration), nuclear import/export and transcription/translation (2-16-fold enrichment/p-scores 1.4-4.8). After release in S, the predominant cell proliferation network carried at its center three important proliferation markers: cyclin-dependent kinase 1 (CDK1 or CDC2), required for entry into S-phase and mitosis in higher cells [66], antigen Ki-67 (KI67), expressed throughout the cell cycle of proliferating cells but not in the early G1/G0 resting phase [67], and proliferating cell nuclear antigen (PCNA), a cofactor of DNA polymerase δ, involved in the control of eukaryotic DNA replication [68]. An entire set of 14-3-3 proteins (α/β, ε, γ, θ, ζ), with roles in growth, signal transduction and cell adhesion, showed some level of up-regulation even if not passing the two-fold threshold. One of these, 14-3-3 ε (YWHAE) displayed interactions with CDC2. YWHAE is an adapter protein that participates in cell division and signaling regulation (including insulin sensitivity), and modulates the activity of a large number of binding partners [69]. A few additional proteins that had been associated with breast cancer cell proliferation included GRB2, CUTL1 and G3BP2. Growth-factor receptor binding protein 2 (GRB2) has been reported to be over-expressed in breast cancers, interestingly, with predominant localization in the nucleus [70]. GRB2, which was found up-regulated in both the nuclear and cytoplasmic fraction of our S-stage MCF7 cells, is an essential signal transduction adapter protein that links cell surface receptors to the RAS signaling pathway. The Ras-Raf-Mek-Erk branch of the receptor tyrosine kinase (RTK) pathways lies at the center of signaling networks that govern proliferation, differentiation and cell survival. Homeobox protein cut-like-1 (CUTL1) is a transcriptional regulator associated with TGF-β-promoted cell invasion and motility, and its expression is inversely correlated with breast cancer survival [71]. Ras GTPase-activating protein-binding protein 2 (G3BP2) displays over-expression in a majority of breast tumors. The protein has nuclear localization in the active cell cycle, and a possible role in cell cycle control as an RNA transporter has been suggested [72]. As part of the mitochondrial metabolism cluster, heat shock protein 70 kDa 9 (HSP9) and prohibitin (PHB) were two other up-regulated proteins that were ascribed roles in cellular proliferation. HSPA9 has been also associated with stress response and aging, and PHB serves as a foldase/unfoldase molecular chaperon in the mitochondria, and as a transcription modulator in the nucleus [73,74]. PHB was also shown to be estrogen regulated in MCF cells [75]. Both have been suggested to be potential predictive markers of breast cancer [73].
G1C vs. SC
Protein up-regulation in the G1C and SC fractions represented a broad spectrum of biological processes that supported the events in the nucleus, and detailed discussions go beyond the purpose of this paper. Briefly, in G1C, these clusters included protein transport/turnover/ubiquitination, degradation of misfolded proteins, glycoprotein and lipid metabolism, mitochondrial respiration/electron transport/protein processing, cytoskeleton organization and control of cell shape and motion, and signal transduction (2-24-fold enrichment/p-scores 1.3-2.5). In SC, the up-regulated clusters encompassed DNA replication (mcm complex proteins)/histone acetylation/transcription initiation, RNA processing/splicing, translation initiation/protein folding, protein transport/nuclear import (displaying interactions with mcm proteins), vesicle trafficking/endocytosis, and various RNA processing and metabolic functions (4-180-fold enrichment/p-scores 1.3-2.1). Proteins that were selected as differentially expressed changed expression ratio in either the nuclear or the cytoplasmic fractions. Only two proteins were found to change expression in both, by translocation: glia maturation factor beta (GMFB) and dynamin 2 (DYN2). The functional role of these proteins is commensurate with their translocation from one cellular subfraction to the other [20,21]. GMFB was up-regulated in G1C, and upon cell stimulation in SN. GMFB is an enhancer of the p38 MAPK signaling pathway, with roles in regulating protein turnover in the cytoplasm. In the nucleus, in response to stimuli, p38 MAPKs are responsible for the activation of transcription factors (ATF 1, 2 and 6, ELK1, PTPRH, DDIT3, TP53/p53 and MEF2C and 2A), the regulation of chromatin accessibility, NF-kappa-B recruitment, and G2 delay. DYN2 which was up-regulated in G1N, and upon stimulation in SC, is a microtubule associated proteins with roles in endocytosis.
Concluding remarks
Differential protein expression analysis of the G1 and S cell cycle stages of MCF-7 breast cancer cells has revealed functional protein clusters indicative of all hallmarks of cancer. At the proteomic level, the data uncovers for the first time new relationships between co-regulated protein networks with essential roles in transcription activation and repression, signaling, and cell cycle control. Up-regulated proteins in G1 included two main categories. On one hand, as expected for starved cancer cells, DNA damage response and cell cycle inhibitory proteins were identified. These clusters included DNA damage repair proteins (XPC, PNKP, XRCC1, RAD50, TP53BP, MSH3, MEN1), transcriptional repressors and chromatin remodeling (HLTF, TP53BP1, DR1, MBD2), apoptosis and negative regulators of cell cycle and growth (CDK11A, CCAR1, NDRG1), and inhibitory signaling (PPP2R1A, PPP2CB) and arrest proteins (LMNA). On the other hand, originators of proliferation, as possible drivers through the G1/S transition point in cancerous cell states, were found to be decisively abundant. These networks encompassed positive regulators of cell cycle/growth and transcriptional/translational activators (TERF2, EIF2A, EIF2S1), repressors of CDK inhibitors (CUL1, CTBP2, MBD2, ZNF217), enhancers of telomerase activity and limitless replicative potential (TERF2, ZNF217, ZNF512B, PP2A), mediators of cell survival (LIMS1) and evasion of apoptosis (ZNF217), activators of cell signaling (Wnt/CTNND1, estrogen/PELP, MAPK14/GLYR1, integrin/LIMS1/ILK), promoters of insensitivity to antigrowth signals (ZNF217, ZNF512B, CTBP1/2, HDAC1/2), blockers of differentiation (ROD1), and cell adhesion, spreading and metastasis proteins (LIMS1, MTA2), just to summarize a few. The relevant protein clusters up-regulated in S, while included mainly indicators of enhanced metabolic activity and cell proliferative signaling, pinpointed the presence of additional proteins with roles in angiogenesis, cell invasion and metastasis. The presence of oncoproteins was prevalent in all cell states. The enhanced proliferative capacity of these cancer cells spans the entire range of tumorigenesis hallmarks. The interdependence of biological mechanisms that encompass DNA repair and transcriptional activities associated with the repression of CDK inhibitors and acquisition of insensitivity to growth inhibition, results ultimately in capabilities that promote evasion of apoptosis, enhanced cell proliferation, limitless replicative potential and metastasis. By providing novel insights into the functional categories that drive cancer cells into division, the data points to a broad range of potential therapeutic targets that concurrently affect the cell cycle signaling and transcriptional/translational machinery.
Supplementary Material
Acknowledgments
This work was supported by funds from Virginia Tech and NCI (R21 CA126669-01A1) to I.M. Lazar. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI.
Abbreviations
- CDK
(cyclin dependent kinase)
- E2
(estradiol)
- ER+
(estrogen receptor positive)
Footnotes
The authors declare no conflict of interest.
References
- [1].Ford HL, Pardee AB. Cancer and the cell cycle. J. Cell Biochem. 1999;(Suppl. 32-33):166–172. doi: 10.1002/(sici)1097-4644(1999)75:32+<166::aid-jcb20>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- [2].Pardee AB, Dubrow R, Hamlin JL, Kletzien RF. Animal cell cycle. Annu. Rev. Biochem. 1978;47:715–750. doi: 10.1146/annurev.bi.47.070178.003435. [DOI] [PubMed] [Google Scholar]
- [3].Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Park M, Lee S. Cell cycle and cancer. J. Biochem. Mol. Biol. 2003;36:60–65. doi: 10.5483/bmbrep.2003.36.1.060. [DOI] [PubMed] [Google Scholar]
- [5].Pardee AB. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. U S A. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- [7].Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- [8].Fernandez PL, Jares P, Rey MJ, Campo E, Cardesa A. Cell cycle regulators and their abnormalities in breast cancer. Mol. Pathol. 1998;51:305–309. doi: 10.1136/mp.51.6.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hanahan D, Weinberg RA. The hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [10].Nielsen NH, Loden M, Cajander J, Emdin SO, Ladberg G. G1-S transition defects occur in most breast cancers and predict outcome. Breast Cancer Res. Treat. 1999;56:105–112. doi: 10.1023/a:1006208419350. [DOI] [PubMed] [Google Scholar]
- [11].Keen JC, Davidson NE. The biology of breast carcinoma. Cancer. 2003;97:825–833. doi: 10.1002/cncr.11126. [DOI] [PubMed] [Google Scholar]
- [12].Zwart W, Theodorou V, Carroll JS. Estrogen receptor-positive breast cancer: a multidisciplinary challenge. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:216, 230. doi: 10.1002/wsbm.109. [DOI] [PubMed] [Google Scholar]
- [13].Levenson AS, Jordan VC. MCF-7: The first hormone-responsive breast cancer cell line. Cancer Res. 1997;57:3071–3078. [PubMed] [Google Scholar]
- [14].Lai A, Sarcevic B, Prall OWJ, Sutherland RL. Insulin/insulin-like growth factor-I and estrogen cooperate to stimulate cyclin E-Cdk2 activation and cell cycle progression in MCF-7 breast cancer cells through differential regulation of cyclin E and p21WAF1/Cip1. J. Biol. Chem. 2001;276:25823–25833. doi: 10.1074/jbc.M100925200. [DOI] [PubMed] [Google Scholar]
- [15].Rodriguez-Mora OG, Lahair MM, McCubrey JA, Franklin RA. Calcium/calmodulin-dependent kinase I and calcium/calmodulin-dependent kinase kinase participate in the control of cell cycle progression in MCF-7 human breast cancer cells. Cancer Res. 2005;65:5408–5416. doi: 10.1158/0008-5472.CAN-05-0271. [DOI] [PubMed] [Google Scholar]
- [16].Caldon CE, Sergio CM, Schutte J, Boersma MN, et al. Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol. Cell. Biol. 2009;29:4623–4639. doi: 10.1128/MCB.00269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sandhu C, Connor M, Kislinger T, Slingerland J, Emili A. Global protein shotgun expression profiling of proliferating MCF-7 breast cancer cells. J. Proteome Res. 2005;4:674–689. doi: 10.1021/pr0498842. [DOI] [PubMed] [Google Scholar]
- [18].Sarvaiya HA, Yoon JH, Lazar IM. Proteome profile of the MCF7 cancer cell line: a mass spectrometric evaluation. Rapid Commun. Mass Spectrom. 2006;20:3039–3055. doi: 10.1002/rcm.2677. [DOI] [PubMed] [Google Scholar]
- [19].Yang X, Lazar IM. MRM screening/biomarker discovery: a library of human cancer-specific peptides. BMC Cancer. 2009;9:96. doi: 10.1186/1471-2407-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Safran M, Dalah I, Alexander J, Rosen N, et al. GeneCards Version 3: the human gene integrator. Database. 2010 doi: 10.1093/database/baq020. 2010, baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zeeberg BR, Feng W, Wang G, Wang MD, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dennis G, Jr., Sherman BT, Hosack DA, Yang J, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- [24].Jensen LJ, Kuhn M, Stark M, Chaffron S, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- [27].Morgan DO. The Cell Cycle: Principles of Control. New Science Press Ltd; London: 2007. [Google Scholar]
- [28].Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bronchud MH, Foote MA, Giaccone G, Olopade O, Workman P, editors. Principles of Molecular Oncology. Humana Press; New Jersey: 2008. pp. 269–279. [Google Scholar]
- [30].Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Balogh K, Patocs A, Huyady L, Racz K. Menin dynamics and functional insight: take your partners. Mol. Cell. Endocrinol. 2010;326:80–84. doi: 10.1016/j.mce.2010.04.011. [DOI] [PubMed] [Google Scholar]
- [32].Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nagalingam A, Tighiouart M, Ryden L, Joseph L, et al. Med1 plays a critical role in the development of tamoxifen resistance. Carcinogenesis. 2012;33:918–930. doi: 10.1093/carcin/bgs105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang GG, Allis D, Chi P. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol. Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- [35].Wang GG, Allis D, Chi P. Chromatin remodeling and cancer, part II: ATP-dependent chromatin remodeling. Trends Mol. Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Thillainadesan G, Isovic M, Loney E, Andrews J, et al. Genome analysis identifies the p15ink4b tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol. Cell. Biol. 2008;28:6066–6077. doi: 10.1128/MCB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Quinlan KG, Verger A, Yaswen P, Crossley M. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim. et Biophys. Acta. 2007;1775:333–340. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
- [38].Huang G, Krig S, Kowbel D, Xu H, et al. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum. Mol. Genet. 2005;14:3219–3225. doi: 10.1093/hmg/ddi352. [DOI] [PubMed] [Google Scholar]
- [39].Tili E, Michaille JJ, Liu CG, Alder H, et al. GAM/ZFp/ZN512B is central to a gene sensor circuitry involving cell-cycle regulators, TGFβ effectors, Drosha and microRNAs with opposite oncogenic potentials. Nucleic Acids Res. 2010;38:7673–7688. doi: 10.1093/nar/gkq637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Berger J, Bird A. Role of MBD2 in gene regulation and tumorigenesis. Biochem. Soc. Trans. 2005;33:1537–1540. doi: 10.1042/BST0331537. [DOI] [PubMed] [Google Scholar]
- [41].Bergman LM, Blaydes JP. C-terminal binding proteins: Emerging roles in cell survival and tumorigenesis. Apoptosis. 2006;11:879–888. doi: 10.1007/s10495-006-6651-4. [DOI] [PubMed] [Google Scholar]
- [42].Huelsken J, Behrens J. The Wnt signalling pathway. J. Cell Sci. 2002;115:3977–3978. doi: 10.1242/jcs.00089. [DOI] [PubMed] [Google Scholar]
- [43].Kopan R. Notch: a membrane-bound transcription factor. J. Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- [44].Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to suppressor of Hairless. Genes Dev. 2002;16:1964–1976. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Scacheri PC, Davis S, Odom DT, Crawford GE, et al. Genome-wide analysis of Menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2006;2:e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vadlamudi RK, Kumar R. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl. Recept. Signal. 2007;5:e004. doi: 10.1621/nrs.05004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Brann DW, Zhang QG, Wang RM, Mahesh VB, Vadlamudi RK. PELP1--a novel estrogen receptor-interacting protein. Mol. Cell Endocrinol. 2008;290:2–7. doi: 10.1016/j.mce.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Linares LK, Kiernan R, Triboulet R, Chable-Bessia C, et al. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat. Cell Biol. 2007;3:331–338. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- [49].Liu L, Scolnick DM, Trievel RC, Zhang HB, et al. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].De Boeck G, Forsyth RG, Praet M, Hogendoorn PC. Telomere-associated proteins: cross-talk between telomere maintenance and telomere-lengthening mechanisms. J. Pathol. 2009;217:327–344. doi: 10.1002/path.2500. [DOI] [PubMed] [Google Scholar]
- [51].Weinberg RA, Garland Science, Taylor & Francis Group . The Biology of Cancer. LLC; New York: 2007. [Google Scholar]
- [52].de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- [53].de las Heras JI, Batrakou DG, Schirmer EC. Cancer biology and the nuclear envelope: a convoluted relationship. Semin. Cancer Biol. 2012 doi: 10.1016/j.semcancer.2012.01.008. in press, doi: 10.1016/j.semcancer.2012.01.008. [DOI] [PubMed] [Google Scholar]
- [54].Sui G, Affar EB, Shi Y, Brignone C, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- [55].Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- [56].Yamamoto H, Tsukahara K, Kanaoka Y, Jinno S, Okayama H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 1999;19:3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J. Cell. Biochem. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol. BioSyst. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Grzmil M, Rzymski T, Milani M, Harris AL, et al. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene. 2010;29:4080–4089. doi: 10.1038/onc.2010.152. [DOI] [PubMed] [Google Scholar]
- [60].Jiang HY, Wek SA, McGrath BC, Scheuner D, et al. Phosphorylation of the α subunit of eukaryotic initiation factor 2 is required for activation of NF-κB in response to diverse cellular stresses. Mol. Cell. Biol. 2003;23:5651–5663. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sabile A, Meyer AM, Wirbelauer C, Hess D, et al. Regulation of p27 degradation and S-phase progression by Ro52 RING finger protein. Mol. Cell. Biol. 2006;26:5994–6004. doi: 10.1128/MCB.01630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang J, Chen S, Zhang W, Zhang J, et al. Human differentiation-related gene NDRG1 is a Myc downstream-regulated gene that is repressed by Myc on the core promoter region. Gene. 2008;417:5–12. doi: 10.1016/j.gene.2008.03.002. [DOI] [PubMed] [Google Scholar]
- [63].Fu J, Yang Z, Wei J, Han J, Gu J. Nuclear protein NP60 regulates p38 MAPK activity. J. Cell Sci. 2006;119:115–123. doi: 10.1242/jcs.02699. [DOI] [PubMed] [Google Scholar]
- [64].Futamura M, Nishimori H, Shiratsuchi T, Saji S, et al. Molecular cloning, mapping and characterization of a novel human gene, MTA1-L1, showing homology to a metastasis-associated gene. MTA1. J. Hum. Genet. 1999;44:52–56. doi: 10.1007/s100380050107. [DOI] [PubMed] [Google Scholar]
- [65].Nishii Y, Morishima M, Kakehi Y, Umehara K, et al. CROP/Luc7A, a novel serine/arginine-rich nuclear protein, isolated from cisplatin-resistant cell line. FEBS Lett. 2000;465:153–156. doi: 10.1016/s0014-5793(99)01744-5. [DOI] [PubMed] [Google Scholar]
- [66].Westbrook L, Manuvakhova M, Kern FG, Estes NR, II, et al. Cks1 regulates cdk1 expression: a novel role during mitotic entry in breast cancer cells. Cancer Res. 2007;67:11393–11401. doi: 10.1158/0008-5472.CAN-06-4173. [DOI] [PubMed] [Google Scholar]
- [67].Scholzen T, Gerdes J. The Ki-67 protein: From the known and the unknown. J. Cell Physiology. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- [68].Leonardi E, Girlando S, Serio G, Mauri FA, et al. PCNA and Ki67 expression in breast carcinoma: correlations with clinical and biological variables. J. Clin. Pathol. 1992;45:416–419. doi: 10.1136/jcp.45.5.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Craparo A, Freund R, Gustafson TA. 14-3-3 (epsilon) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem. 1997;272:11663–11669. doi: 10.1074/jbc.272.17.11663. [DOI] [PubMed] [Google Scholar]
- [70].Verbeek BS, Adriaansen-Slot SS, Rijksen G, Vroom TM. GRB2 overexpression in nuclei and cytoplasm of human breast cells: a histochemical and biochemical study of normal and neoplastic mammary tissue specimens. J. Pathol. 1997;183:195–203. doi: 10.1002/(SICI)1096-9896(199710)183:2<195::AID-PATH901>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [71].Michl P, Ramjaun AR, Pardo OE, Warne PH, et al. CUTL1 is a target of TGFβ signaling that enhances cancer cell motility and invasiveness. Cancer Cell. 2005;7:521–532. doi: 10.1016/j.ccr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- [72].French J, Stirling R, Walsh M, Kennedy HD. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem. J. 2002;34:223–231. doi: 10.1023/a:1021737413055. [DOI] [PubMed] [Google Scholar]
- [73].Czarnecka AM, Campanella C, Zummo G, Cappello F. Mitochondrial chaperones in cancer: from molecular biology to clinical diagnostics. Cancer Biol. Ther. 2006;5:714–720. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- [74].Mishra S, Murphy LC, Murphy LJ. The Prohibitins: emerging roles in diverse functions. J. Cell. Mol. Med. 2006;10:353–363. doi: 10.1111/j.1582-4934.2006.tb00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].He B, Kim TH, Kommagani R, Feng Q, et al. Estrogen-regulated prohibitin is required for mouse uterine development and adult function. Endocrinology. 2011;152:1047–1056. doi: 10.1210/en.2010-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.