Abstract
The innate immune system responds to infections and tissue damage by activating cytosolic sensory complexes called inflammasomes. Cytosolic DNA is sensed by AIM2-like receptors (ALRs) during bacterial and viral infections and in autoimmune diseases. Subsequently, recruitment of the adaptor protein ASC links ALRs to the activation of caspase-1. A controlled immune response is crucial for maintaining homeostasis, but ALR inflammasome regulation is poorly understood. Here, we identified the PYRIN domain (PYD)-only protein 3 (POP3), which competes with ASC for recruitment to ALRs, as an inhibitor of DNA virus-induced ALR inflammasome activation in vivo. Using a mouse model with macrophage-specific POP3 expression, the data emphasizes the importance of ALR inflammasome regulation in the monocytic/macrophage.
Germline-encoded pattern recognition receptors (PRRs) of the innate immune system are essential for host defense against pathogens through the production of pro-inflammatory mediators and antimicrobial factors. Cytosolic PRRs of the AIM2-like receptor (ALR, PYHIN or HIN-200 protein) and Nod-like receptor (NLR) families are necessary for the maturation and release of the pro-inflammatory cytokines interleukin (IL)-1β and IL-18 and the induction of pyroptotic cell death, which requires the activation of pro-inflammatory caspases-1, -4 and -5 (caspase-1 and -11 in mice) within inflammasomes1. The inflammasome adaptor ASC bridges PRRs to caspase-1, which facilitates caspase-1 activation by induced proximity2,3. In response to viral infections, inflammasomes sense pathogen-associated molecular patterns (PAMPs), including dsRNA, uncapped ssRNA and dsDNA. Thus, unique PRRs exist that sense cytosolic DNA and RNA4. In particular, ALRs sense viral DNA and assemble an inflammasome. Although absent in melanoma 2 (AIM2) senses bacterial and viral cytosolic DNA5–7, the interferon γ-inducible protein 16 (IFI16) detects modified nuclear viral DNA and assembles an initially nuclear inflammasome8. ALRs directly bind DNA with their HIN-200 domain9,10 and recruit ASC through their PYD11–14. ALRs are highly diverse between mice and humans. While only four proteins exist in humans, mice contain 13 predicted members, but their function is largely unknown15,16. The antiviral host response depends on the production of interferons. The production of the type II interferon, IFN-γ, by NK-cells, depends on IL-18 production in an AIM2-inflammasome dependent manner6, but ALRs also regulate the type I IFN response4. Although inflammasome activation is essential for host defense and clearance of intracellular bacteria, viruses, fungi and parasites, excessive and uncontrolled inflammasome activation causes auto-inflammatory diseases and impaired activation promotes metabolic disease. Recognition of self-nucleic acids by PRRs directly contributes to the pathology associated with several autoimmune diseases, emphasizing the importance of a balanced inflammasome regulation4.
The PYD-only proteins (POPs) represent a family of inflammasome regulators, which bind and occupy the PYD in ASC and PYD-containing PRRs and thereby abolish the PYD-PYD interactions necessary for inflammasome formation17–19. Although POPs are encoded in several species, including humans, they are lacking from mice, suggesting development of a more complex mechanism of inflammasome regulation in some organisms20. However, POP family members have not yet been studied in vivo. POP1 (also known as PYD-containing protein 1, PYDC1) is highly similar to the PYD of ASC and interacts with ASC, thereby affecting inflammasome activation and activation of NF-κB19. POP2 (also known as PYD-containing protein 2, PYDC2) interacts with the PYD of ASC, as well as with the PYD of several NLRPs18. POP2 also prevents NLR-mediated activation of NF-κB21. The significance of POPs in regulating host defense is emphasized by certain poxviruses encoding a viral POP in order to evade the host immune response by blocking inflammasome and NF-κB activation17,22.
Here we report the discovery of a previously undescribed type I IFN-inducible POP family member, POP3, which interacts with the ALRs AIM2 and IFI16 to inhibit ALR inflammasome formation. Thus we provide evidence that each major branch of inflammasome-activating PRRs evolved a POP regulator in humans. Silencing of POP3 in human macrophages enhances DNA and DNA virus-induced ALR inflammasome formation and hence the maturation and release of IL-1β and IL-18. Consistently, POP3 expression specifically in cells of the monocytic/macrophage lineage in transgenic mice, which is naturally not encoded in mice, impaired an ALR inflammasome response. Thus, POP3 functions as a key ALR inflammasome regulator in human and mouse macrophages and in an in vivo virus infection mouse model. The data also represents the first conditional inflammasome mouse model, emphasizing that inhibition of ALRs in macrophages is sufficient to impair systemic IL-18 and subsequent IFN-γ production to regulate the antiviral host response, which recapitulates global AIM2 deficiency.
RESULTS
POP3 is expressed in response to type-I interferons
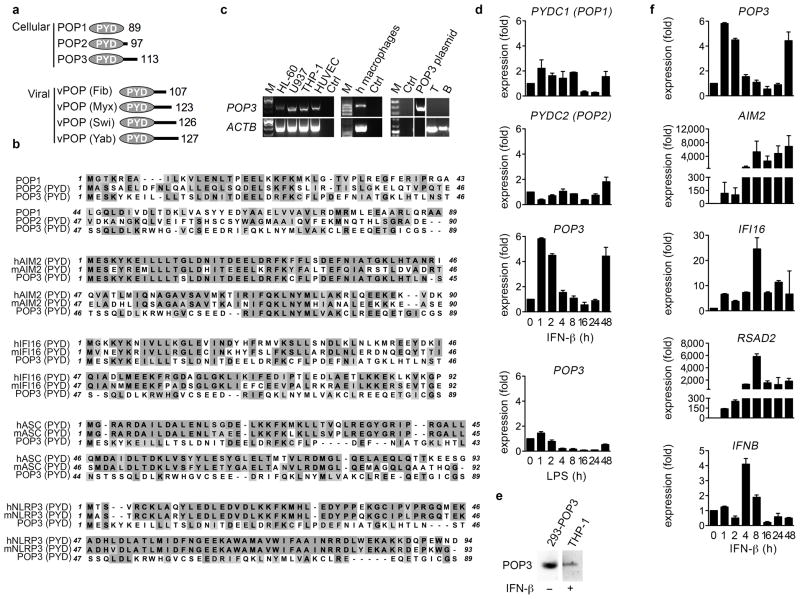
We discovered a previously undescribed human POP family member, which we termed POP3 (Genbank accession number: KF562078) (Fig. 1a). The POP3 cDNA revealed an open reading frame of 342 bp (Supplementary Fig. 1a) encoded from a single exon located within the IFN-inducible gene cluster between IFI16 and pyrin and HIN domain family member 1 (PYHIN1) on chromosome 1q23, which also contains AIM2 and myeloid cell nuclear differentiation antigen (MNDA) (Supplementary Fig. 1b). In comparison, the syntenic mouse chromosomal region 1H3 is amplified and contains 13 predicted genes, but not a POP3 ortholog15,16. POP3 encodes a single PYD of 113 aa with 5 α-helices, whereas the PYD of AIM2 consists of 6 α-helices. Hence, the 3rd AIM2-PYD α-helix appears to be unstructured in POP3 (Supplementary Fig. 2a), reminiscent to the structure observed for the PYD of NLRP1, which forms a flexible loop instead of α-helix 3 and is predicted to become stabilized upon PYD-PYD interaction23. All 3 POPs exhibited low sequence homology to each other, implying that they might have unique functions (Fig. 1b). In contrast to POP1 and POP2, POP3 showed high sequence similarity to the PYD of AIM2 (Fig. 1b, Table 1), and showed overall high similarity to the PYDs of HIN-200 family members (Supplementary Fig. 2b)24. POP3 shared several of the characteristic sequence motifs within α-helices 1 and 2 of HIN-200 PYDs, but not those present within α-helices 5 and 6 (Supplementary Fig. 2c). Phylogenetic tree analysis of all PYD proteins also placed POP3 within the HIN-200 family (Supplementary Fig. 2d). Thus, POP3 most likely originated from exon duplication of the AIM2-PYD, reminiscent to POP1, which is derived from the PYD of ASC19. Consistently, POP3 revealed low sequence homology with the PYDs of mouse and human ASC and NLRP3 (Fig. 1b, Table 1). POP3 mRNA was expressed in monocytic cell lines and human primary macrophages (hMΦ), but not in B and T cells (Fig. 1c). Similar to ALRs, POP3 expression was upregulated in response to IFN-β in hMΦ, but the TLR4 agonist LPS did not induce POP3 expression (Fig. 1d). Accordingly, POP3 was also detected by immunoblot in IFN-β-treated THP-1 macrophages (Fig. 1e). The POP3 expression pattern was unique, since neither POP1 nor POP2 were regulated by IFN-β, emphasizing a selective role of POP3 within the type I-IFN-mediated host response. POP3 expression was upregulated as an early response gene within the first two hours, as well as a late response gene after 48 hours of IFN-β treatment, which was distinctive from AIM2, IFI16, IFNB and the IFN-stimulated gene RSAD2 (also known as VIPERIN) (Fig. 1f). Thus, the IFN-β-inducible POP3 is a member of the POP family and shows similarity to the PYDs of HIN-200 proteins.
Figure 1. POP3 is a novel type-I interferon-inducible member of the POP family.
(a) Schematic diagram of cellular and viral POP family members and their size in amino acids. (b) Clustal W alignment of human POP members and POP3 and the PYDs of human and mouse AIM2, IFI16, ASC and NLRP3. (c) BLOSSUM identity scores (%) for POP3 and the proteins aligned in b. (d) RT-PCR of POP3 expression. (e, f, g) Real-time PCR analysis in response to LPS and/or IFN-β in hMΦ at the indicated times (n=3 ± s.e.m.). POP3 was also detected by immunoblot in THP-1 cells treated for 48 hrs with IFN-β, using HEK293 cells transfected with a POP3 cDNA as a control. Data are representative of two experiments (c–f).
Table 1. BLOSSUM identity scores.
Identity in % is shown for POP3 and the proteins aligned in Fig. 1b.
| hAIM2 | mAIM2 | hIFI16 | mIFI16 | hASC | mASC | hNLRP3 | mNLRP3 | POP1 | POP2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| POP3 identity (%) | 60.9 | 43.5 | 17.4 | 15.2 | 18.9 | 16.8 | 20.2 | 12.5 | 21.7 | 11.1 |
POP3 binds to ALRs and inhibits inflammasome formation
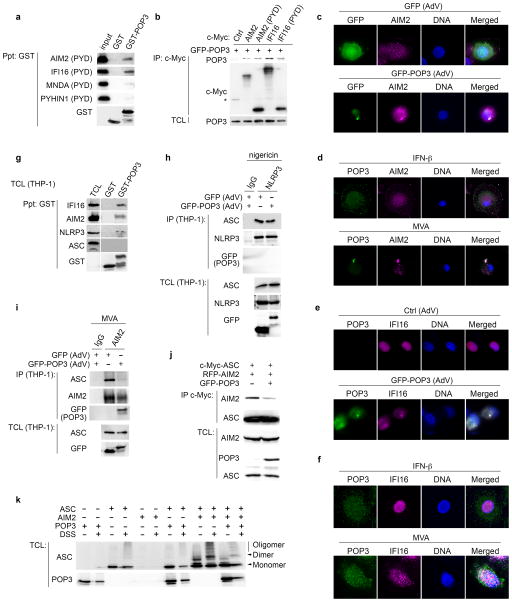
Since PYDs usually exhibit homotypic interactions and POP3 contained several HIN-200 PYD -specific sequence motifs and displayed high homology to the PYD of AIM2, we investigated if POP3 was able to bind to the PYD of HIN-200 proteins. GST-POP3, but not GST control, bound to the PYD of AIM2 and IFI16. However, GST-POP3 showed no significant interaction with the PYDs of MNDA and PYHIN1 (Fig. 2a). Upon transient co-transfection of POP3 with AIM2 and IFI16 in HEK293 cells, their interaction was confirmed by co-immunoprecipitation (Fig. 2b).
Figure 2. POP3 interacts with ALRs.
(a) GST-precipitation (Ppt) between GST-POP3 and in vitro transcribed/translated PYDs of AIM2, IFI16, MNDA and PYHIN1, using GST as a control. (b) Co-immunoprecipitation of co-transfected POP3 and ALRs. HEK293 cells were transfected with GFP-POP3 and either ctrl, full length AIM2 and IFI16 or the PYD of AIM2 and IFI16, as indicated and myc-tagged proteins were immunoprecipitated (IP) and analyzed for co-purified POP3, alongside total cell lysates (TCL). *denotes a cross-reactive protein. (c, e) hMΦs were either infected with c, GFP or e, Ctrl and GFP-POP3-expressing AdV and 36 h post infection immunostained for c, AIM2 and e, IFI16 and POP3 and analyzed by immunofluorescence microscopy. (d, f) hMΦs were either treated with IFN-β or infected with d, MVA (2 h) or f, KSHV (8 h) and immunostained as above and endogenous POP3. (g) Interaction of GST-POP3 with endogenous proteins from IFN-β-treated THP-1 TCL using GST as negative control. (h, i) THP-1 cells were infected with GFP or GFP-POP3 AdV, LPS primed and h, nigericin-treated or i, infected with MVA, followed by IP of h, NLRP3 or i, AIM2 and control IgG and analyzed alongside TCL by immunoblot. (j) HEK293 cells were transfected with ASC, AIM2 and POP3 as indicated and myc-tagged ASC was IP and analyzed for bound AIM2 in the presence or absence of POP3 by immunoblot. (k) HEK293 cells were transfected as above and lysates were cross-linked with DSS where indicated and analyzed by immunoblot for ASC oligomerization. Data are representative of two experiments (a–k).
This observation was further supported by co-localization studies. AIM2 and IFI16 have been shown to co-localize with ASC, albeit at different sites and in response to different stimuli8,12,13. While DNA from Modified Vaccinia virus Ankara (MVA) and murine cytomegalovirus (MCMV) is sensed by AIM2 in the cytosol11–14, modified DNA originating from latent Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) infection is recognized by IFI16 within the nucleus in vitro8. AIM2 was predominantly localized to cytosolic punctate structures, and this pattern was not altered in response to GFP adenovirus (AdV) infection (Fig. 2c), while adenovirus-mediated GFP-POP3 expression resulted in co-localization in cytoplasmic punctate structures (Fig. 2c). We also observed very limited co-localization of endogenous AIM2 with endogenous POP3 in a few cytosolic punctate structures in hMΦ treated with IFN-β to up-regulate AIM2 and POP3 expression (Fig. 2d), but co-localization was greatly enhanced 2 hours after MVA infection (Fig. 2d). In contrast to the predominantly cytosolic AIM2 localization, IFI16 localizes within the nucleus in endothelial cells, where it interacts with ASC8. Similarly, we observed solely nuclear IFI16 localization in hMΦ, which was not altered in response to control adenovirus infection (Fig. 2e). However, in response to adenovirus-mediated expression of GFP-POP3, IFI16 was re-distributed to the cytosol, where it partially co-localized with GFP-POP3 (Fig. 2e). In agreement, we did not observe co-localization of endogenous IFI16 and POP3 in IFN-β-treated hMΦ (Fig. 2f), nor did MVA infection alter IFI16 distribution or promote co-localization with POP3 at the tested times (data not shown). Yet, KSHV infection caused partial cytosolic redistribution of IFI16 as quickly as 2 hours p.i. (data not shown), which was more prominent 8 hours p.i.. At that time we observed partial co-localization of IFI16 with POP3 (Fig. 2f), although KSHV did not cause aggregation of IFI16 in hMΦ at the titer used in our experiments. These results further supported an interaction of POP3 with ALRs.
Accordingly, GST-POP3 also purified endogenous AIM2 and IFI16 (Fig. 2g). However, in contrast to POP1 and POP218,19, POP3 did not bind to the inflammasome adaptor ASC. Unexpectedly, we also observed weak binding of recombinant POP3 to NLRP3 in vitro (Fig. 2g), in spite of the rather low degree of homology and the presence of the HIN-200 PYD-specific sequence motifs within POP3. Assembly of the inflammasome through PYD-PYD interactions is a key step for its activation and subsequent cytokine release. The PYD of ALRs and NLRP3 interact with the PYD of ASC and we hypothesized that the PYD-containing POP3 could interfere with this interaction. Contrary to the interaction of recombinant POP3 with NLRP3 in vitro, POP3 was not recruited to and did not disrupt the NLRP3-ASC complex in LPS-primed and nigericin-treated THP-1 cells (Fig. 2h). However, POP3 was recruited to, and disrupted the endogenous AIM2-ASC complex in response to MVA infection in THP-1 cells (Fig. 2i). Moreover, POP3 also caused a reduced interaction of ectopically expressed ASC and AIM2 in HEK293 cells by co-immunoprecipitation, indicating that POP3 is able to disrupt ALR inflammasome complex assembly by competing with ASC for the PYD binding site in AIM2 (Fig. 2j). These data suggest that POP3 functions selectively as an ALR inflammasome inhibitor.
AIM2 inflammasome assembly causes the formation of ASC oligomers12. Only co-transfection of AIM2 and ASC in HEK293 cells caused the formation of ASC dimers and oligomers, but not transfection of ASC and POP3 or POP3, ASC or AIM2 alone. However, in the presence of POP3, AIM2-mediated ASC dimers and oligomers were significantly reduced (Fig. 2k), thus supporting our hypothesis that POP3 can inhibit the PYD-dependent recruitment of ASC to AIM2. These data indicate that POP3 is a previously undescribed IFN-β-inducible protein, which directly interacts with the ALRs AIM2 and IFI16 through PYD-PYD interaction to prevent inflammasome formation.
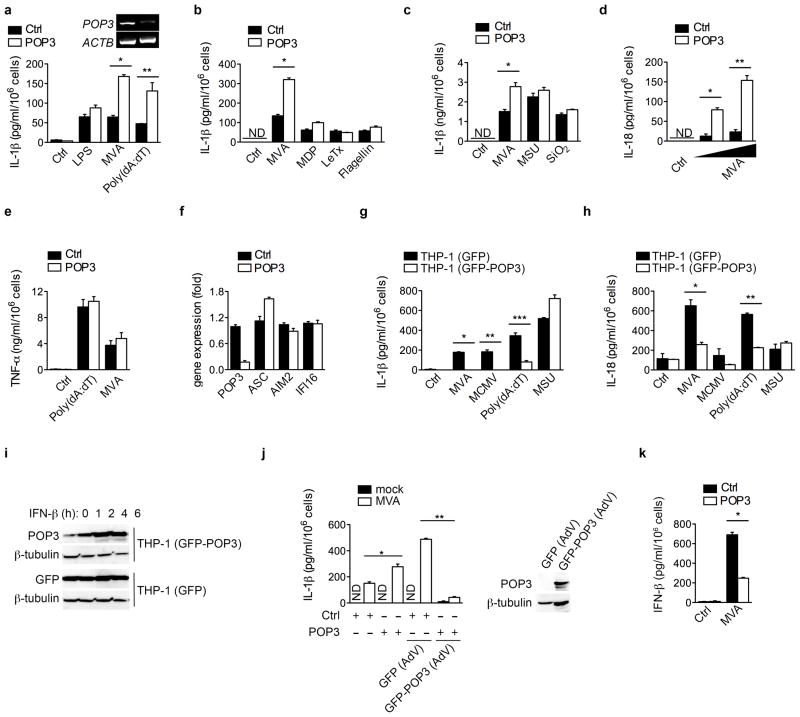
POP3 inhibits ALR-mediated IL-1β and IL-18 release
In response to DNA virus infection, AIM2 and IFI16 function as cytosolic and nuclear inflammasome-activating DNA-sensors, respectively5–8. In the absence of POP3, following siRNA-mediated silencing in hMΦ, as determined by RT-PCR (Fig. 3a), IL-1β release in response to cytosolic double strand (ds) DNA, such as transfection of poly(dA:dT) or infection with MVA, was significantly enhanced (Fig. 3a). Comparable results were also obtained with a second POP3-targeting siRNA (Supplementary Fig. 3a). However, POP3 silencing did not affect IL-1β release triggered upon activation of non-ALR inflammasomes, including inflammasome responses to LPS, the NLRP1 inflammasome in response to B. anthracis lethal toxin (LeTx)25 and muramyldipeptide (MDP)26, the NLRP3 inflammasome in response to monosodium urate (MSU) crystals27 or silica (SiO2)28–30 and the NLRC4 inflammasome in response to S. typhimurium flagellin31,32 (Fig. 3a, b, c). THP-1 cells are widely used to study inflammasome responses and we also observed elevated AIM2-dependent release of IL-1β in POP3 silenced THP-1 cells in response to AIM2-, but not NLRP3-dependent stimuli (Supplementary Fig. 3b). We also observed increased MVA-induced IL-18 release upon silencing of POP3 (Fig. 3d). This effect of POP3 was specific for inflammasome-dependent cytokines, since the release of the inflammasome-independent cytokines TNF (Fig. 3e) and IL-6 (Supplementary Fig. 3c) was not affected by POP3 silencing. POP3 silencing also did not affect mRNA expression of ASC, AIM2 and IFI16, as determined by real-time PCR (Fig. 3f) or protein expression (Supplementary Fig. 3d). Conversely, THP-1 cells stably expressing GFP-POP3, but not GFP control, showed significantly reduced release of IL-1β (Fig. 3g) and IL-18 (Fig. 3h), but not TNF (Supplementary Fig. 3e), in response to MVA and MCMV infection and transfection of poly(dA:dT), but not in response to MSU crystals, further supporting our observations obtained in hMΦ upon POP3 silencing. Moreover, this cell system also recapitulated the IFN-β-inducible expression of POP3 (Fig. 3i). We next restored POP3 expression in POP3 silenced hMΦ by adenoviral delivery of GFP-POP3, as determined by immunoblot (Fig. 3j). While transduction with a GFP-expressing adenovirus into control siRNA transfected cells slightly increased MVA-induced IL-1β secretion, transduction with a GFP-POP3 expressing adenovirus strongly suppressed this response (Fig. 3j). Overall our data suggest that POP3 functions as an inhibitor of DNA-induced inflammasome activation, while showing no impact on NLRP1, NLRP3 and NLRC4 inflammasomes. In addition to being defective in inflammasome activation, Aim2−/− macrophages show elevated IFN-β production in response to dsDNA, MVA or bacterial infection through a yet unknown mechanism6,12,13, and IFI16 functions as a sensor promoting IFN-β production in response to DNA virus infection33. In agreement with the elevated IFN-β secretion in Aim2−/− macrophages, which indicates that it may negatively regulate IFN-β production, we observed that silencing of POP3 decreases IFN-β production in response to MVA infection of hMΦ (Fig. 3k), and poly(dA:dT) transfection in THP-1 cells (Supplementary Fig. 3f). Conversely, stable GFP-POP3, but not GFP expressing THP-1 cells displayed elevated IFN-β production in response AIM2-specific stimuli (Supplementary Fig. 3g). Further studies will be necessary to determine, whether also dsVACV70-induced and IFI16-dependent IFN-β production is affected by POP3 and the mechanism by which it regulates IFN-β production. Type I IFNs block IL-1α and IL-1β synthesis through an IL-10-STAT3-dependent autocrine mechanism34, while simultaneously upregulating expression of IL-1RA and IL-18BP to compete with IL-1β and IL-18 for receptor binding, respectively35,36. Consistently, we observed elevated Il1ra and Il18bp transcripts in POP3 expressing and MVA-infected BMDM (Supplementary Figure 3h). Collectively, these results indicate that POP3 functions as an inhibitor of ALR inflammasome-mediated release of IL-1β and IL-18 in human macrophages and promotes a type I IFN response.
Figure 3. Silencing of POP3 in hMΦ enhances ALR-mediated IL-1β and IL-18 release.
(a–f, j, k) hMΦ were transfected with either control or POP3 siRNAs, confirmed for POP3 silencing by RT-PCR (a, insert), and treated with LPS, MDP, MSU or SiO2, transfected with poly(dA:dT), LeTx, or flagellin, or infected with MVA as indicated for 16 h and analyzed a–c, for mature IL-1β; d, mature IL-18; e, TNF and k, IFN-β by ELISA (n = 3 ± s.e.m.). (f) POP3 silencing and ASC, AIM2 and IFI16 expression were determined by Real-Time PCR. (g–i) Stable THP-1 (GFP) or THP-1 (GFP-POP3) cells were analyzed for secretion of g, IL-1β and h, IL-18 in response to MVA and MCMV infection, transfection of poly(dA:dT) and treatment with MSU by ELISA (n = 3 ± s.e.m.). i, THP-1 (GFP) or THP-1 (GFP-POP3) cells were treated with IFN-β for the indicated times and expression of GFP and POP3 was analyzed by immunoblot and compared to β-tubulin. (j, k) hMΦ were transfected with siRNAs as above, j, transduced with either control GFP or GFP-POP3 expressing AdV, and either mock or MVA infected and analyzed for mature IL-1β by ELISA (n = 3 ± s.e.m.) or k, analyzed for secretion of IFN-β by ELISA (n = 3 ± s.e.m.). Data are representative of 3 experiments (a–f) and 2 experiments (g, h, j, k) or one experiment (i). (a) *P<0.0001, **P=0.0139; (b) *P<0.0001; (c) *P=0.0039; (d) *P=0.0008, **P=0.0006; (g) *P<0.0001, **P=0.0015, ***P=0.0011; (h) *P=0.0054, ** P<0.0001; (j) *P=0.0197, **P<0.0001; (k) *P<0.0001;
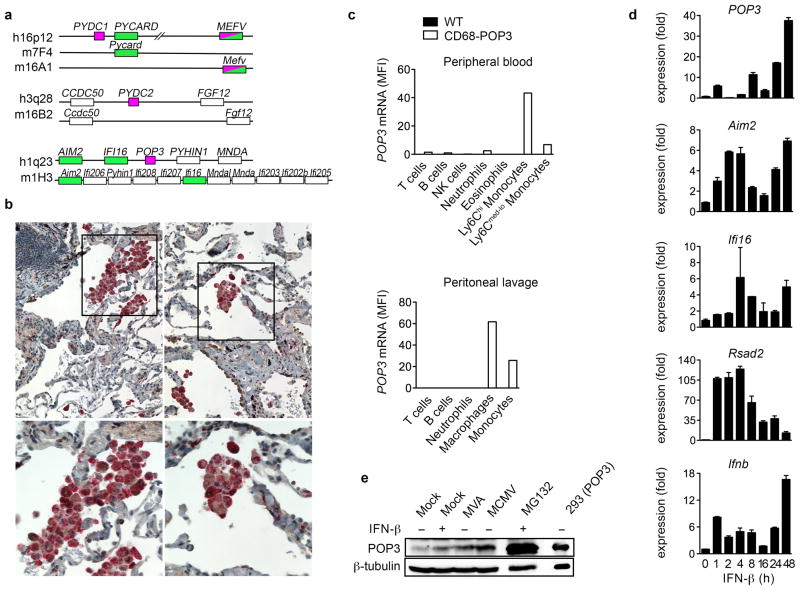
Macrophage-specific POP3-expressing transgenic mice
POP1 and POP2 are lacking from mice20. Similar to the close chromosomal location of POP1 and ASC, POP3 is found next to AIM2 and IFI16, and POP3 is also absent in mice, despite significant amplification of this gene cluster (Fig. 4a). To study the POP3 function in vivo, and in particular its role in inflammasome regulation in macrophages, we generated transgenic (TG) mice expressing POP3 from the human CD68 (hCD68) promoter in combination with the IVS-1 intron containing a macrophage-specific enhancer37,38, which seemed the most promising after testing several commonly used macrophage-specific promoters (data not shown). Our promoter choice was further based on our observation that POP3 was specifically expressed in human CD68+ macrophages in inflamed lung lesions (Fig. 4b), using a custom-raised antibody that did not cross react with other POPs, nor with the related IFI16 and AIM2 PYDs (Supplementary Fig. 3i, j). Analysis of POP3 mRNA expression in CD68-POP3 TG mice by flow cytometry using SmartFlares, verified expression specifically in the monocyte/macrophage lineage, and particularly in the CD11b+Ly6Chi classical monocytes in peripheral blood and in CD11b+F4/80+ peritoneal macrophages (Fig. 4c, and Supplementary Fig. 4), thus making this the first macrophage-specific mouse model to study inflammasomes and the first mouse model to study POPs. The low abundance POP3 expression in BMDM generated from CD68-POP3 TG mice was induced by IFN-β as an early and late response gene at the transcriptional level (Fig. 4d), thus closely resembling its regulation observed in hMΦ. Aim2, Ifi16 and Rsad2 show higher inducibility in hMΦ than in BMDM, despite the similar Ifnb transcription. Similarly, POP3 protein was inducibly expressed in response to IFN-β or MVA and MCMV infection (Fig. 4e). We also observed that POP3 protein expression could be stabilized in the presence of the proteasome inhibitor MG132 (Fig. 4e), indicating that POP3 expression is not only tightly regulated on the transcriptional-, but also on the post-translational level. Post-translational regulation is further supported by the observation that THP-1 cells expressing GFP-POP3 from the constitutive CMV promoter express elevated levels of POP3 protein after treatment with IFN-β (Fig. 3i). Overall these results support the rationale for using this particular POP3 mouse model.
Figure 4. Monocyte/macrophage-lineage-specific expression of POP3 in CD68-POP3 TG mice.
(a) Genomic analysis of POP-encoding human chromosomal regions and synthenic mouse chromosomes. Inflammasome-activating proteins are shaded in green and POPs in magenta. (b) Immunohistochemical staining of human inflamed lung tissue for CD68 (red) and POP3 (brown). The boxed area of the top panels (10x) is magnified in the bottom panels (40x). (c) POP3 mRNA expression analysis of WT and CD68-POP3 TG mice by flow cytometry (mean fluorescent intensity, MFI) with SmartFlares in cell populations identified with specific lineage markers in peripheral blood cells (top panel) and peritoneal lavage cells (bottom panel) isolated 6 h after MCMV infection. (d) Real-time PCR expression analysis in CD68-POP3 TG BMDM in response to IFN-β treatment or infection with MVA or MCMV for 16 h (n=3 ± s.e.m.). (e) Analysis of POP3 protein levels in CD68-POP3 TG BMDM after 16 h of IFN-β treatment or infection with MVA or MCMV. As a control, BMDM were also treated with the proteasome inhibitor MG132 and run alongside a control lysate of HEK293 cells that were transiently transfected with POP3. Membranes were stripped and re-probed for β-tubulin as loading control. Data are representative of two (b, e, d) and three (c) experiments.
POP3 inhibits ALR-mediated cytokine release in BMDM
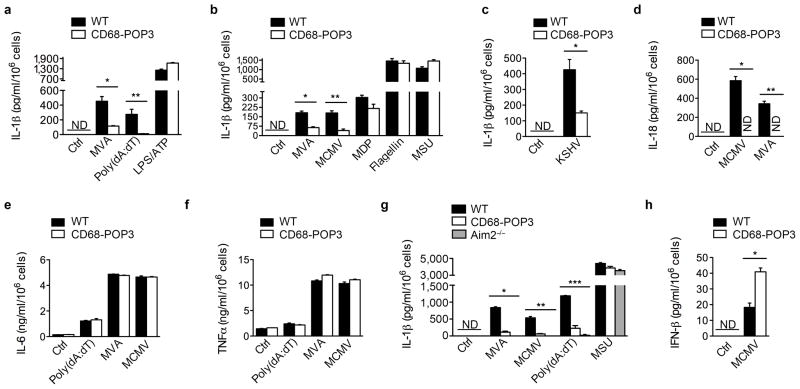
Next we analyzed the effect of POP3 expression on AIM2 and other inflammasomes. Consistent with the increased inflammasome response we observed in the absence of POP3 in hMΦ, POP3 expression resulted in a significant decrease of IL-1β release in response to poly(dA:dT) transfection and MVA or MCMV infections, but not in response to MDP, flagellin, LPS + ATP or monosodium urate crystals (MSU) in BMDM (Fig. 5a, b) and in peritoneal macrophages (Supplementary Fig. 5a). These results underscore the same selectivity of POP3 for AIM2 inflammasomes in mice and humans without affecting NLRP1b, NLRC4 and NLRP3 inflammasomes25,27,31,32,39. Whereas DNA from MVA and MCMV is sensed by AIM2 in the cytosol, modified DNA originating from latent KSHV infection is recognized by IFI16 within the nucleus in vitro8. Based on our observation that POP3 and IFI16 are able to interact, we investigated the release of IL-1β in response to KSHV infection, which was significantly decreased in the presence of POP3, establishing that POP3 impairs AIM2 and likely also IFI16 inflammasomes (Fig. 5c). However, IFI16-dependent KSHV sensing still needs to be validated in vivo by utilizing Ifi16−/− mice in the future. As for hMΦ, POP3 also inhibited DNA virus-induced IL-18 release (Fig. 5d), but did not alter the release of the caspase-1-independent inflammatory cytokines IL-6 and TNF (Fig. 5e, f). To ensure that random TG POP3 integration was not responsible for the observed phenotype, we validated our finding in an independent, second TG line with identical outcomes (Supplementary Fig. 5b). We further generated TG mice ubiquitously expressing the human coxsackie and adenovirus receptor with deleted cytoplasmic domain (hCARΔcyt)40 from the ubiquitin C promoter41 (Supplementary Fig. 5c), which allows efficient infection with recombinant adenovirus at low MOI (Supplementary Fig. 5d). Infection of UbiC-hCARΔcyt BMDM with GFP-POP3 or GFP control expressing adenovirus further confirmed the inhibitory function of POP3 on the AIM2-induced IL-1β release independently of POP3 integration (Supplementary Fig. 5e). As expected from low level ectopic expression of an inhibitor such as POP3, the AIM2-mediated IL-1β release by POP3 was not completely abolished, but nevertheless reached levels close to Aim2−/− macrophages in response to AIM2-dependent stimuli. Neither CD68-POP3 TG- nor Aim2−/− BMDM showed any diminished IL-1β release in response to MSU (Fig. 5g). In agreement with our observation that silencing of POP3 in hMΦ and THP-1 cells partially inhibited IFN-β production (Fig. 3k, Supplementary Fig. 3f), POP3 expressing BMDM showed elevated levels of IFN-β in response to MCMV infection (Fig. 5h), reminiscent of Aim2−/− macrophages6. Thus, TG expression of POP3 in mouse macrophages further confirmed a role of POP3 in inhibiting ALR-mediated cytokine release as initially observed by POP3 silencing in hMΦ, and confirmed that human POP3 is functional in mice.
Figure 5. POP3 expression in BMDM inhibits AIM2 and IFI16 inflammasome-mediated cytokine release.
(a–h) BMDM were infected with MVA, MCMV or KSHV, treated with LPS/ATP, MDP, MSU or transfected with poly(dA:dT) or flagellin for 16 h and analyzed for a–c, g, mature IL-1β, d, mature IL-18, e, IL-6 and f, TNF, h, IFN-β by ELISA, as indicated (n = 3 [a, b, d–h], n = 6 [c] ± s.e.m.). Data are representative of three (a–g), two (c, h) experiments. (a) *P=0.0064, **P=0.0188; (b) *P=0.0015, **P=0.0026; (c) *P=0.0020; (d) *P<0.0001, **P<0.0001; (g) *P<0.0001; **P=0.0004 (POP3), **P=0.0009 (AIM2−/−); ***P=0.0003 (POP3), ***P<0.0001 (AIM2−/−); (h) *P=0.0037.
POP3 inhibits ALR-mediated caspase-1 activation in BMDM
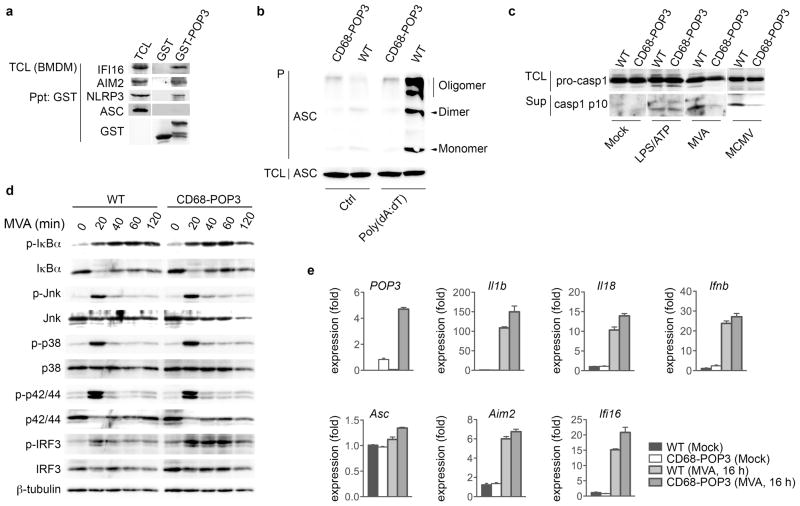
The PYDs of human and mouse ALRs are well conserved, and therefore it was not surprising that POP3 also co-purified IFI16 and AIM2, but not ASC from BMDM (Fig. 6a), similar to what we observed in THP-1 cells (Fig. 2g). As observed in human THP-1 cells, recombinant POP3 also weakly co-purified NLRP3 in BMDM in vitro (Fig. 6a). This result further assured that the function of human POP3 would be translated in our mouse model. To delineate the mechanism by which POP3 inhibits ALR inflammasomes in mouse macrophages, we analyzed ASC oligomerization in response to AIM2 inflammasome stimulation with poly(dA:dT) in WT and CD68-POP3 TG BMDM as a readout for AIM2 inflammasome formation12. Insoluble ASC monomers, dimers and oligomers were drastically decreased in the presence of POP3 (Fig. 6b), supporting impaired AIM2 inflammasome formation in the presence of POP3. Inflammasome formation is essential for caspase-1 activation, and although the protein amount of pro-caspase-1 was not altered in POP3 expressing BMDM, active caspase-1 p10 was significantly reduced in response to MVA and MCMV, but not in response to LPS + ATP in culture supernatants (Fig. 6c), further emphasizing the functional specificity of POP3 for AIM2, but not NLRP3 inflammasome formation. The POP3 effect was specific for caspase-1 and was not caused by modulating NF-κB activation, since the NF-κB-inducible cytokines TNF and IL-6 were equally secreted (Fig. 5e, f). In addition, we observed similar NF-κB activation and MAPK signaling responses upon MVA infection in the absence and presence of POP3 in BMDM (Fig. 6d). Furthermore, transcription of Il1b, Il18, Ifnb, Asc, Aim2 and Ifi16 was not reduced in POP3 expressing mock and MVA-infected BMDMs (Fig. 6e), further supporting a role of POP3 in regulating AIM2-mediated inflammasome activation and caspase maturation, but not in modulating the expression of inflammasome components. Enhanced IFN-β production in POP3 expressing BMDM was further supported by the increased and sustained phosphorylation of IRF3 in CD68-POP3 TG BMDM in response to MVA infection (Fig. 6d). POP3 expression was significantly elevated in response to MVA infection (Fig. 6e), similar to its binding partners IFI16 and AIM2, and reminiscent to what we observed by immunoblot in response to MVA infection and IFN-β treatment (Fig. 4d, e). These results demonstrate that POP3 affects cytokine release by inhibiting ALR-mediated caspase-1 activation.
Figure 6. POP3 interacts with AIM2 and IFI16 in BMDM.
(a) Co-purification of GST-POP3 with endogenous proteins from IFN-β-treated BMDM TCL using GST as negative control. (b) Inflammasome assembly was determined by analyzing ASC oligomerization by immunoblot in response to poly(dA:dT) transfection in BMDM after cross-linking of pellets (P). (c) Immunoblot of BMDM to detect activation of caspase-1 in response to MVA and MCMV infection and treatment with LPS/ATP in culture SN and of the caspase-1 pro-form in TCL. (d) WT and CD68-POP3 TG BMDM were infected with MVA for the indicated times (in min.), lysed and TCL were analyzed for activation of IκBα, JNK, p38, p42/44 and IRF3 signaling pathways using pan- and phospho-specific antibodies and β-tubulin as a loading control. (e) Real-time PCR analysis of transcript levels in BMDM before and after MVA infection (n=3 ± s.e.m.). Data are representative of two experiments (a–e).
POP3 blunts ALR-mediated anti-viral host defense in vivo
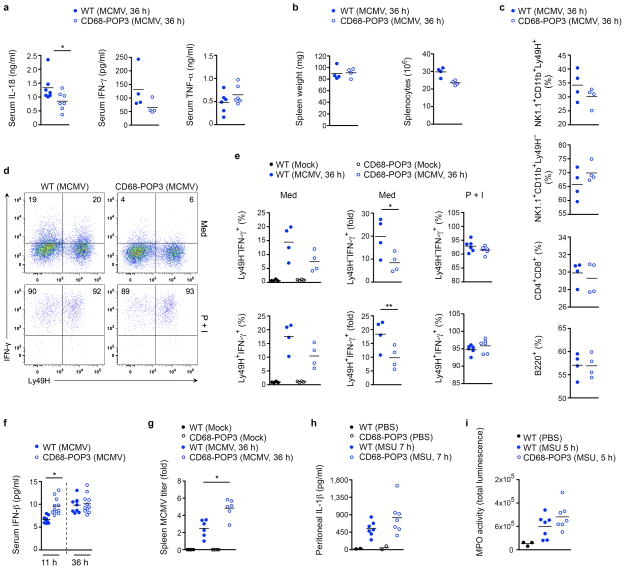
Although, there is support for an inflammasome inhibitory role of POPs upon overexpression, none have been studied in macrophages or in vivo. Aim2−/− mice are severely impaired in mounting an efficient host response to MCMV infection, due to a deficiency in the inflammasome-dependent systemic IL-18 release6. IL-18 acts in synergy with IL-12 to stimulate IFN-γ production by splenic NK cells, which is crucial for the early anti-viral response against DNA viruses, including MCMV42,43. Therefore, we challenged WT and CD68-POP3 TG mice with MCMV and found that, similar to Aim2 deficiency6, serum IL-18 and IFN-γ concentrations were also strongly decreased in CD68-POP3 expressing mice at 36 hours post i.p. infection. However, TNF serum concentrations were not affected (Fig. 7a). CD68-POP3 TG mice displayed a similar spleen weight as WT mice after MCMV infection, but showed slightly reduced splenocyte numbers (Fig. 7b). Since IL-18 is required for Ly49H+ NK cell expansion44 and we had already observed reduced IL-18 concentration in CD68-POP3 TG mice (Fig. 7a), it was not surprising that CD68-POP3 TG mice displayed a decreased number of NK1.1+Ly49H+ cells, whereas NK1.1+Ly49H− cells were increased. However, we found comparable numbers of T and B cells (Fig. 7c and Supplementary Fig. 6). Accordingly, CD68-POP3 TG mice had significantly less IFN-γ producing splenic NK cells ex vivo at 36 hours post infection (Fig. 7d, e), reminiscent of Aim2−/− mice6. This response was specific to MCMV and not due to an intrinsic defect of splenic NK cells from CD68-POP3 TG mice to produce IFN-γ, since activation of WT and POP3 splenic NK cells with the phorbol ester PMA and ionomycin ex vivo, produced comparable numbers of IFN-γ+ NK cells (~95%) (Fig. 7e). In addition to impaired IL-18 and IFN-γ production, CD68-POP3 TG mice displayed elevated serum IFN-β concentration at early (11 hours), but not at later time points (36 hours) post MCMV infection (Fig. 7f). These results suggest that the deficient IFN-γ response is due to impaired systemic IL-18 observed in CD68-POP3 TG mice upon MCMV infection. Since IFN-γ is essential for restricting in vivo MCMV amplification, we were also able to observe a significant increase in the splenic MCMV titer in CD68-POP3 TG mice (Fig. 7g). The 2-fold increase was comparable to the 2-fold increase observed in Asc−/− mice in a previously published experiment6.
Figure 7. CD68-POP3 TG mice are impaired in AIM2-dependent and viral DNA-induced host defense in vivo.
(a) WT and CD68-POP3 TG mice were infected with MCMV and serum levels of IL-18, IFN-γ and TNF were determined by ELISA 36 h after infection. (b) Spleens were isolated from above mice and analyzed for weight and number of splenocytes by flow cytometry. (c) Immunophenotyping of splenocytes after 36 h of MCMV infection. (d, e) Intracellular expression of IFN-γ by splenic NK cells 36 h after MCMV infection, cultured for 4 h ex vivo in medium (Med) alone or in the presence of PMA and ionomycin (P+I) and gated for CD11b+NK1.1+ cells, d, showing the result from a representative mouse with the percentage of IFN-γ+ Ly49H+ and Ly49H− NK cells indicated in the quadrants and e, results from 4–6 mice/group. (f) Mice were infected with MCMV as above and serum was analyzed for IFN-β by ELISA 11 and 36 h after infection. (g) Splenic viral titers 36 h after MCMV infection (n=6 per group). (h, i) WT and CD68-POP3 TG mice were i.p. injected with MSU crystals or PBS and h, IL-1β levels were determined by peritoneal lavage 7 h after injection by ELISA and i, mice were imaged for myeloperoxidase (MPO) activity in vivo and quantified after 5 h. Data are representative of two (a–h) and one (i) experiments. All data show the mean value. (a) *P=0.0451; (e) *P=0.0457, **P=0.0447; (f) *P=0.0003; (g) *P=0.0028.
A functional specificity of POP3 for ALRs is further supported by the observation that WT and CD68-POP3 TG mice did not show significant differences in their response to MSU crystal-challenge in vivo. Severity of MSU-induced peritonitis was comparable in both genotypes, showing similar peritoneal IL-1β concentrations 7 hours after MSU challenge (Fig. 7h). Since IL-1β produced by macrophages is essential for neutrophil infiltration into the peritoneal cavity45, it was not surprising that we observed no differences in neutrophil infiltration in vivo by utilizing a luminescent myeloperoxidase (MPO) probe (Fig. 7i, Supplementary Fig. 7a). These results clearly demonstrate that POP3 has a critical role in the host response to MCMV through regulating the AIM2 inflammasome in vivo, without functionally affecting the NLRP3 inflammasome. Collectively, our data support a model where POP3 functions in the anti-viral host response within a type I IFN-mediated inflammasome regulatory feedback loop (Supplementary Figure 7b).
DISCUSSION
Although inflammasome-produced cytokines are necessary for host defense and metabolic health, excessive and uncontrolled cytokine production contributes to pathological inflammation and autoinflammatory diseases. Hence, factors that promote a balanced inflammasome response are essential for maintaining homeostasis. However, the regulation of inflammasomes is still poorly understood. In particular, mechanisms of self- and foreign DNA discrimination have not been completely elucidated, but a better understanding likely supports decoding the underlying causes that contribute to the onset of autoinflammatory and autoimmune diseases, including Systemic lupus erythematosus (SLE), Aicardi-Goutières syndrome or inflammatory myocarditis46. Therefore, a strict control system that properly regulates DNA-induced immune responses is of utmost importance. Here we show that the type I IFN-inducible POP3 is one of the proteins that might function to maintain a balanced inflammasome response in humans by specifically inhibiting ALR inflammasome assembly in response to immunogenic DNA. While other POPs directly interact with the inflammasome adaptor ASC18,19, POP3 interacts with the PYD of ALRs and thereby prevents recruitment of ASC. Although, we find that recombinant POP3 interacted with NLRP3 in vitro, we did not observe a functional impairment of NLRP3-dependent inflammasome formation and activation in vitro and in vivo. Importantly, POP3 was not recruited to the endogenous ligand-induced NLRP3-ASC complex, but was recruited to MVA-induced endogenous AIM2, where it prevented ASC recruitment. Thus, POP3 evolved as a specific ALR inflammasome regulator.
Our study further revealed that the human HIN-200 cluster is more complex than previously described and differs from mice. However, mice may employ an alternative mechanism for ALR inflammasome regulation through the DNA-binding HIN-200 family member p202, which lacks the PYD and is not encoded in humans, but may function as an antagonist for AIM2 in mice14. However, p202 is barely detectable in C57BL/6 mice, which were used in our study, but is highly expressed in BALB/c and NZB mouse strains14. This may have been an important influence as to why POP3 expression in C57BL/6 mice was able to drastically alter the immune response upon MCMV infection in our study using ‘humanized’ C57BL/6 mice. Two proteins encoding only a PYD are predicted in mice within the HIN-200 cluster, which both lack a human ortholog. Pydc3 (Ifi208) is predicted to encode only a PYD, but is significantly larger than POPs and might encode a HIN-200 domain, according to expressed sequence tags16,47. Two predicted Pydc4 alternative transcripts encode only a PYD47, but the longest transcript encodes a 586 aa protein, which is 95.2% identical to PYDC3. However, neither gene shows similarity with POP3 and no expression data or functional data are available47. Thus, Aim2b, a predicted Aim2 splice form in mice, might most closely resemble the POP3 function in mice. Similar to the human HIN-200 locus, the rat HIN-200 chromosomal region is also predicted to encode four HIN-200 proteins (Rhin2, Rhin3, Rhin4, Aim2) and the putative POP Rhin5, which is twice the size of POP3 and shares less than 14% sequence identity and also lacks any expression data47. Contrary to mice, we speculate that humans evolved POP3 to interfere with ALR inflammasome assembly.
The type I IFN-responsiveness further distinguishes POP3 from other POP family members. Thus, POP3 represents one of the type I IFN-inducible proteins that antagonizes IFN-γ in macrophages and inflammasome activation34, and might contribute to the anti-inflammatory and immunosuppressive functions of type I IFNs48. Aim2−/− BMDM show elevated IFN-β production, although the mechanism by which AIM2 negatively regulates IFN-β production is unknown5,6. We however, observed that POP3 silencing, which increases AIM2 signaling, also reduces IFN-β production, and accordingly, POP3 expression in hMΦ, THP-1 cells and BMDM promotes IFN-β production. Thus, besides promoting its own IFN-β-dependent production, POP3 might shift the immune response from an inflammasome-dependent pro-inflammatory cytokine production to anti-inflammatory IFN-β production, thereby further blunting IL-1β and IL-18 signaling through upregulation of IL-1RA and IL-18BP35,36. A similar mechanism has been proposed for LRRFIP2, which inhibits NLRP3 inflammasome activation by recruiting the pseudo caspase-1 substrate Flighthless-I49, but also functions as a cytosolic DNA sensor, which promotes type I IFN production50. Significantly, the IFN-β-inducible expression pattern of POP3 as an early and late response gene was similarly observed in CD68-POP3 TG mice. Thus, our humanized mouse is a valuable model to study inflammasome regulation in vivo, which likely reflects POP3 functions in humans. Thus, there is the prospect that POP3 could also contribute to ameliorating DNA-driven autoimmune disease. Although, POP3 is lacking from mice, macrophage-specific TG expression revealed that POP3 is nevertheless functional in mice and thus, our study represents the first POP in vivo study. Although inflammasomes have been reported in multiple cell types, their relative importance for IL-1β and IL-18-dependent host responses in vivo have not been studied. Our study therefore also represents the first conditional inflammasome mouse model, emphasizing the key role of macrophages in DNA virus-induced inflammasome activation.
ONLINE METHODS
Mice
pCD68-POP3 was generated by replacing CAT in pCAT-Basic containing the human CD68 promoter and the macrophage-specific IVS-1 enhancer37,38 with POP3 and flanking the cassette with AatII restriction sites. The AatII fragment was excised, purified and B6.TgN(CD68-POP3) TG mice were generated by pronuclear injection into C57BL/6 embryos. Two lines were initially analyzed and subsequently a single line was used for most experiments and genotyping was outsourced to Transnetyx. B6.TgN(UbiC-hCAR) TG mice were generated by pronuclear injection of a BglII fragment from pUBI containing the ubiquitin C promoter/intron41 and the human coxsackie and adenovirus receptor (hCAR) with deleted cytoplasmic domain (hCARΔcyt)40. Mice were housed in a specific pathogen-free animal facility and all experiments were performed on age and gender-matched 8–12 weeks old mice conducted according to procedures approved by the Northwestern University Committee on Use and Care of Animals.
Macrophage isolation, culture and transfection
hMΦ were isolated from healthy donor blood after obtaining informed consent under a protocol approved by Northwestern University Institutional Review Board by Ficoll-Hypaque centrifugation (Sigma) and countercurrent centrifugal elutriation in the presence of 10 μg/ml polymyxin B using a JE-6B rotor (Beckman Coulter), as described51 and transfected in 24-well dishes (2.5×105 cells) with 120 nM siRNA duplexes (F2/virofect; Targeting Systems) and analyzed 72 hr posttransfection (POP3 stealth siRNA sense strand: 5′-CAUGGCAUUUCUGGGAAUGCAUGUU-3′, POP3 siRNA#2 sense strand: 5′-GAGCAGGAAACGGUAUAUGUGGGA-3′, and Ctrl stealth siRNA, Invitrogen), as described51. BMDM were flushed from femurs and tibia and differentiated in L929-conditioned medium (25%) in DMEM medium supplemented with 10% heat inactivated FCS (Invitrogen) and analyzed after 7 days. Resting or elicited peritoneal macrophages (PM) were isolated by peritoneal lavage before or 3–5 days after i.p. injection of 1 ml 4% aged thioglycollate medium. THP-1 cells were obtained from ATCC and were routinely tested for mycoplasma contamination. THP-1 cells were stably transduced with pLEX-based lentiviral particles. hMΦ, THP-1 cells, BMDM and PM were treated for the indicated times with 600 ng/mL E. coli LPS (0111:B4, Sigma) or pre-treated with ultra-pure E. coli LPS (0111:B4; Invivogen) (100 ng/mL), MSU (400 ng/mL; Invivogen); mouse and human IFN-β (1500 U/mL; Millipore), MG132 (10 μM; Calbiochem), mIL-1Ra (100 ng/mL; R&D Systems) or recombinant IL-1Ra (anakinra, 10 mg/mL, Amgen). Cells were transfected with poly(dA:dT) (2 ng/mL; Sigma), MDP (20 ug/mL; Invivogen), Salmonella thyphimurium flagellin (140 ng/mL; Invivogen) and Bacillus anthracis Lethal toxin and protective antigen (1 ug/mL; List Biological Laboratories) using Lipofectamine 2000 (Invitrogen). Where indicated, cells were pulsed for 20 min with ATP (5 mM; Sigma) or treated for 45 min with nigericin (5 μM).
Virus preparation
Recombinant adenovirus was generated by cloning GFP or GFP-POP3 into pShuttle, recombination with pAdEasy in E.coli BJ5183 and purification from HEK293N cells on a caesium chloride gradient. Lentiviral particles were generated in HEK293T-Lenti cells (Clontech) transfected with pLEX containing GFP or GFP-POP3 and the packaging plasmids pMD.2G and psPAX2 (Addgene plasmids 12259 and 12260). Murine cytomegalovirus (MCMV, Smith strain, ATCC #VR-1399) was obtained from the American Type Culture Collection (ATCC) and propagated in mouse embryo fibroblast SG-1 cells (ATCC #CRL-1404) for cell-based experiments and passaged twice for 2 weeks each, in the salivary glands of 6–8 week-old BALB/c mice after i.p. injection of 1.5 × 105 PFU/mL MCMV. Mice were euthanized and salivary glands collected, homogenized in HBSS, clarified and the viral titer determined by plaque formation assay and a Taqman qPCR assay based on MCMV iE and glycoprotein B (Invitrogen), using a MCMV standard curve and stored in aliquots at −80°C. Non-infected clarified salivary gland homogenates were used for mock infection. 2.5×105 macrophages were infected with 1×105 PFU/well in 24-well plates. Vaccinia virus (MVA, modified Vaccinia virus Ankara, ATCC # VR-1508) was obtained from ATCC and amplified in hamster fibroblast BHK-21 cells (ATCC #CCL-10). MVA titer was determined by a plaque-forming assay using BHK-21 cells. 2.5×105 macrophages were infected with 1×106 PFU/well in 24-well plates. Kaposi’s Sarcoma-associated herpes virus (KSHV) lytic cycle was induced from BCBL-1 cells by supplementing media with TPA (20 ng/mL). KSHV-containing culture SN was collected after 96 h, clarified by centrifugation (330xg for 5 min followed by centrifugation at 1540×g for 30min) and filtered through 0.45 μm pore size filters. KSHV was subsequently concentrated by ultracentrifugation at 20,000 rpm for 90 min (SW28 rotor, 4°C). Viral pellets were resuspended in EBM2 medium (Lonza), 0.45 μm filtered, and titered on the endothelial cell line iHMVEC52. KSHV was used to infect 2.5×105 macrophages at 1.2×105 IU/24-well.
Plasmids
pCDNA3-based expression constructs for ASC, POP1 and POP2 were described earlier17–19,51,53,54. POP3 (Acc. No.: KF562078), AIM2, AIM2-PYD, IFI16, IFI16-PYD, IFIX, IFIX-PYD, MNDA, MNDA-PYD, were generated by standard PCR from cDNAs and expressed sequence tags (EST) (Open Biosystems) and cloned in pcDNA3, pLEX or pShuttle with N-terminal myc, HA, Flag, GFP or RFP tags. All expression constructs were sequence verified.
Immunoblot analysis, immunoprecipitation and immunohistochemistry
Rabbit polyclonal and mouse monoclonal POP3 antibodies were custom raised (KLH-conjugated-CGSPSSARSVSQSRL), rabbit polyclonal antibody to ASC (Chemicon clone 2EI-7 and custom), mouse monoclonal antibody to ASC (custom), mouse polyclonal antibody to caspase-1 (Santa Cruz Biotech clone M-20), mouse monoclonal antibody to hCAR (Santa Cruz Biotech clone Mab.E[mh1]), mouse monoclonal antibody to GFP (Santa Cruz Biotech clone B-2), mouse monoclonal antibody to dsRED (Santa Cruz Biotech clone F-9), mouse monoclonal antibody to myc (Roche and Santa Cruz Biotech clone 9E10), mouse monoclonal antibody to the N-terminus of IFI16 (Santa Cruz Biotech clone 1G7), mouse monoclonal antibody to the C-terminus of IFI16 (Abcam clone ab104409), rabbit polyclonal antibody to the C-terminus of AIM2 (Cell Signaling Technology clone 8055), rabbit polyclonal antibodies to IκBα (clone 44D4)/p-IκBα (clone 14D4), JNK (clone 9252)/p-JNK (9251), p38 (clone 9212)/p-p38 (clone 12F8), p42/44 (clone 9102)/p-p42/44 (9101), IRF3 (clone D83B9), p-IRF3 (clone 4D4G) (all Cell Signaling Technology) and mouse monoclonal antibody to β-tubulin (Santa Cruz Biotech clone TU-02), mouse monoclonal antibody to GST (Santa Cruz Biotech clone B-14) and mouse monoclonal antibody to NLRP3 (Adipogen clone Cryo-2) were used for immunoblot. For co-immunoprecipitations (IP), HEK293 cells were transfected with GFP-POP3, HA-ASC, RFP-AIM2 or empty plasmid in 100 mm dishes (Lipofectamine 2000, Invitrogen). Cells were lysed (50 mM Hepes pH 7.4, 150 mM NaCl, 10% Glycerol, 2 mM EDTA, 0.5% Triton X-100, supplemented with protease inhibitors) 36 hrs post transfection. Cleared lysates were subjected to IP by incubating with immobilized antibodies as indicated for 16 hrs at 4°C, followed by extensive washing with lysis buffer. Bound proteins were separated by SDS-PAGE, transferred to PVDF membranes and analyzed by immunoblotting with indicated antibodies and HRP-conjugated secondary antibodies, ECL detection (Pierce), and image acquisition (Ultralum). TCL (5%) were also analyzed where indicated.
Endogenous NLRP3 and AIM2 inflammasome complexes were similarly purified from ultrapure LPS-primed (16 hrs, 100 ng/mL) THP-1 cells following nigericin treatment (45 min, 5 μM) or MVA-infection (90 min), respectively. For GST pull down experiments, POP3 was cloned into pGEX-4T1 and affinity purified as a GST fusion protein from E. coli BL21. Proteins were either prepared by in vitro transcription/translation (TNT Quick Coupled Transcription/Translation, Promega), or TCL were prepared from IFN-β-treated (16 hrs) BMDM or THP-1 cells by lysis (50 mM Hepes pH 7.4, 120 mM NaCl, 10% Glycerol, 2 mM EDTA, 0.5% Triton X-100, supplemented with protease inhibitors) as a source of endogenous proteins, and cleared lysates were incubated with immobilized GST-POP3 or GST control for16 hrs at 4°C, followed by extensive washing with lysis buffer and analysis as above. For ASC cross-linking, 4×106 BMDM were seeded in 60 mm plates and subjected to cross-linking as described55. Briefly, cells were transfected with 1 μg/ml poly (dA:dT) for 5 hrs, supernatants were removed, cells rinsed with ice-cold PBS and lysed (20 mM Hepes pH 7.4, 100 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate, supplemented with protease inhibitors) and further lysed by shearing. Cleared lysates were stored for immunoblot analysis and the insoluble pellets were resuspended in 500 μl PBS, supplemented with 2 mM disuccinimydyl suberate (DSS, Pierce) and incubated with rotation at room temperature for 30 min. Samples were centrifuged at 5,000 rpm for 10 min at 4°C and the cross-linked pellets were resuspended in 50 μl Laemmli sample buffer and analyzed by immunoblot. Human lung tissue was embedded in paraffin, cut into 3 μm sections, mounted, deparaffinized and immunostained with mouse monoclonal CD68 (Dako) and rabbit polyclonal POP3 and peroxidase (HRP)/DAB+ and alkaline phosphatase (AP)/Fast Red enzyme/chromogen combinations (Dako) and specific isotype controls (Dako) and haemotoxylin counterstaining of nuclei.
Immunofluorescence Microscopy
hMΦ were grown on cover slips and either IFN-β treated for 16 hrs or infected with GFP, GFP-POP3 expressing adenovirus, MVA for 2 hrs or KSHV for 8 hrs, fixed, permeabilized, and immunostained with AIM2 (Cell Signaling clone 8055), IFI16 (Santa Cruz Biotech clone 1G7) and POP3 (custom raised) antibodies and secondary Alexa Fluor 546-conjugated antibodies and DAPI (Invitrogen)54. Images were acquired by fluorescence microscopy on a Nikon TE2000E2-PFS with a 100x oil objective and image deconvolution (Nikon Elements).
Cytokine and caspase-1 measurement
IL-1β, IL-18, TNFα, IL-6, IFN-β and IFN-γ secretion was quantified from clarified culture SN obtained from hMΦ, BMDMs, PM and from mouse serum by ELISA (BD Biosciences, eBiosciences, Invitrogen). Samples were analyzed in triplicates and repeated at least three times, showing a representative result. Active caspase-1 p10 was detected by immunoblot in TCA-precipitated serum-free culture supernatants 4 hrs after treatment51.
mRNA analysis
mRNA expression of target genes was quantified by RT-PCR or in vivo by using gold nanoparticles conjugated to specific oligonucleotides duplexed with Cy5-labeled reporter strands, which are non-toxic and are endocytosed by live cells (SmartFlares; Millipore). Subsequent analysis was performed by flow cytometry in combination with linage specific markers. Briefly, 7–12 weeks old mice received an i.p. injection of MCMV (105 PFU) for 6 hrs. Mice were euthanized and peritoneal cells were obtained by lavage. Blood was obtained by retro-orbital bleeding, collected in EDTA-containing tubes, and incubated for 16 hrs with control or POP3 specific SmartFlares (1:1000 dilution in HBSS). Subsequently, cells were blocked with Fc-Block (2.4G2, BD), stained with fluorochrome-conjugated antibodies (see below), fixed and depleted of red blood cells using BD FACS Lysing solution (BD Biosciences) and analyzed on a BD LSR II flow cytometer. Data were compensated and evaluated using FlowJo software (Tree Star, Ashland, OR, USA). Doublets and debris were excluded and leukocytes were identified using the pan-hematopoietic marker CD45 (30-F11, BD). Leukocyte subsets were identified as following: CD4 T cells as CD4+ (RM4-5, BD), CD8 T cells as CD8+ (53-6.7, BD), B cells as B220+ (RA3-6B2, BD), NK cells as NK1.1+ (PK-136, BD), neutrophils as CD11b+ (M1/70, eBioscience) and Ly6G+ (1A8, BD), monocytes as CD11b+ (M1/70, eBioscience) and Ly-6C+ (AL-21, BD) and macrophages as CD11b+ (M1/70, eBioscience) and F4/80+ (BM8, eBioscience). Monocytes were further subdivided into Ly-6Chi classical or inflammatory and Ly-6Clo/med non-classical or resident monocytes. Total RNA was isolated from hMΦ, BMDM or mouse blood using Trizol (Invitrogen) or the mouse RiboPure-blood RNA isolation kit (Invitrogen), treated with DNase I, reverse transcribed with GoScript (Promega) and analyzed by TaqMan Real-time gene expression system using predesigned FAM labeled primer/probes on an ABI 7300 Real time PCR machine (Applied Biosystems) and displayed as relative expression compared to GAPDH or β-actin. The POP3 TaqMan assay was custom designed: POP3-Fwd: 5′-AGCACGAGTAGCCAACTTGATT-3′, POP3-Rev: 5′-GGTCTTCCTCACTGCAGACA-3′ and POP3-FAM probe: 5′-CCATGCCAGCGTTTTA-3′. The RT-PCR primers for POP3 were: POP3-Fwd: 5′-ATGGAGAGTAAATATAAGGAG-3′, POP3-Rev: 5′-TCAACATGCATTCCCAGAAAT-3′.
In vivo virus infection and intracellular IFN-γ staining
8–10 week-old age and gender matched wild type (WT) and CD68-POP3 mice were randomly infected with 1×105 to 1×106 PFU by i.p. injection of MCMV or mock salivary gland homogenates and euthanized after 36 hrs. Spleens were digested with collagenase type D (Roche) (1 mg/mL) and DNase I (Roche) (0.1 mg/mL) in HBSS at 37°C for 15 min, passed through 40 μm nylon cell strainers (BD Biosciences), after which red cells were lysed using 1x BD Pharm Lyse buffer (BD Biosciences), and washed with complete RPMI medium (RPMI 1640 with 10% FCS, 2 mM glutamine, 100 U penicillin/0.1 mg streptomycin/mL, 10 mM HEPES buffer, and 1 mM sodium pyruvate). Splenocytes were counted (Countess cell counter; Invitrogen) and 3×106 splenocytes were directly stained for IFN-γ expression, and an additional 3×106 splenocytes were first suspended in complete RPMI medium and stimulated for 4 hrs in the presence of leukocyte activation cocktail (2 μL/mL, BD Biosciences) before staining6. Splenocytes were pre-incubated with mouse Fc block, and labeled with pre-titrated fluorescent antibodies to B220, CD4, CD8, CD11b, CD69, NK1.1, as described above and Ly49H (3D10, eBioscience). Intracellular staining for IFN-γ (XMG1.2, eBioscience) was accomplished using a BD Cytofix/Cytoperm Kit according to the manufacturer’s specifications (BD Biosciences) and dead cells were excluded using Aqua live/dead staining (Invitrogen). At least 400,000 events per sample were acquired on a BD LSRII instrument and data were analyzed with FlowJo software (TreeStar, Inc).
MSU-induced peritonitis
10–12 week-old age and gender matched WT and CD68-POP3 mice were randomly i.p. injected with either PBS (0.5 mL/mouse) or MSU crystals in PBS (10 mg in 0.5 mL PBS/mouse). 5 hrs after MSU injection, mice were i.p. administered the luminescent Xenolight Rediject Inflammation probe (200 mg/kg, PerkinElmer)56. Images were exposed for 5 min (IVIS Spectrum, PerkinElmer) and luminescence quantified with Living Image (PerkinElmer). Mice were also euthanized 7 hrs after MSU injection and peritoneal cavities were flushed with 2 mL of ice-cold PBS/10% FBS, clarified by centrifugation, and analyzed for IL-1β by ELISA.
Statistics
Graphs represent the mean +/− s.e.m. A standard two-tailed unpaired t-test was used for statistical analysis of two groups with all data points showing a normal distribution, and values of p < 0.05 were considered significant and listed in the figure legends (Prism 5, GraphPad). The investigators were not blinded to the genotype of the mice/cells. Sample sizes were selected on the basis of preliminary results to ensure a power of 80% with 95% confidence between populations.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM071723, HL097183, AI092490, AI082406, AI099009 and AR064349 to C.S., AR057532 to A.D., AR050250, AR054796, AI092490 and HL108795 to H.P. and AR064313 to C.M.C.), a Cancer Center Support Grant (CA060553), the Skin Disease Research Center (AR057216) and the American Heart Association (12GRNT12080035) to C.S.. S.K. was an Arthritis Foundation fellow (AF161715), L.d.A. was supported by the American Heart Association (11POST585000) and the NIH (T32AR007611) and H.P. was supported by funds provided by the Solovy/Arthritis Research Society Professor. The ubiquitin C promoter was kindly provided by Brian C. Schaefer (Uniformed Services University of the Health Sciences), pLXS-hCARΔcyt by James DeGregori (University of Colorado Health Sciences Center), Plasmids pMD2.G and psPAX2 by Didier Trono (École Polytechnique Fédérale de Lausanne) and Aim2−/− mice by Katherine A Fitzgerald (University of Massachusetts, Worcester). Transgenic mice were generated with the assistance of the Northwestern University Transgenic and Targeted Mutagenesis Laboratory. We thank Alexander D. Radian for isolation of human macrophages.
Footnotes
Author Contributions. C.S. and A.D. designed the research; S.K., R.A.R., L.d.A., C.M.C., S.L.R., A.V.M., M.C.W. and A.G. performed experiments; E.F., E.G., D.R.G., H.P. and J.C.R. provided essential reagents, expertise and advice; S.K., R.A.R., L.d.A., C.M.C., A.V.M., H.P., A.D. and C.S. analyzed results; S.K., A.D. and C.S. wrote the paper; A.D. and C.S. conceived the study, designed the experiments and provided overall direction.
Competing Financial Interests. The authors declare no competing financial interests.
Database accession numbers. POP3: KF562078
References
- 1.Martinon F, Burns K, Tschopp J. The Inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-1β. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 2.Stehlik C, Lee SH, Dorfleutner A, Stassinopoulos A, Sagara J, Reed JC. Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. J Immunol. 2003;171:6154–63. doi: 10.4049/jimmunol.171.11.6154. [DOI] [PubMed] [Google Scholar]
- 3.Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for Caspase-1. J Biol Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- 4.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Ann Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–6. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerur N, et al. IFI16 Acts as a Nuclear Pathogen Sensor to Induce the Inflammasome in Response to Kaposi Sarcoma-Associated Herpesvirus Infection. Cell Host Microbe. 2011;9:363–75. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–87. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 10.Jin T, et al. Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 15.Schattgen SA, Fitzgerald KA. The PYHIN protein family as mediators of host defenses. Immunol Rev. 2011;243:109–18. doi: 10.1111/j.1600-065X.2011.01053.x. [DOI] [PubMed] [Google Scholar]
- 16.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Dorfleutner A, et al. A Shope Fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes. 2007;35:685–694. doi: 10.1007/s11262-007-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorfleutner A, et al. Cellular pyrin domain-only protein 2 is a candidate regulator of inflammasome activation. Infect Immun. 2007;75:1484–1492. doi: 10.1128/IAI.01315-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated NF-κB and pro-Caspase-1 regulation. Biochem J. 2003;373:101–113. doi: 10.1042/BJ20030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stehlik C, Dorfleutner A. COPs and POPs: Modulators of Inflammasome Activity. J Immunol. 2007;179:7993–8. doi: 10.4049/jimmunol.179.12.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedoya F, Sandler LL, Harton JA. Pyrin-only protein 2 modulates NF-kappaB and disrupts ASC:CLR interactions. J Immunol. 2007;178:3837–45. doi: 10.4049/jimmunol.178.6.3837. [DOI] [PubMed] [Google Scholar]
- 22.Johnston JB, et al. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–98. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Hiller S, et al. NMR Structure of the Apoptosis- and Inflammation-Related NALP1 Pyrin Domain. Structure. 2003;11:1199–205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the Absent in Melanoma 2 (AIM2) Pyrin Domain Provides Insights into the Mechanisms of AIM2 Autoinhibition and Inflammasome Assembly. J Biol Chem. 2013;288:13225–35. doi: 10.1074/jbc.M113.468033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–4. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 26.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–24. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 27.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 28.Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U S A. 2008;105:9035–40. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate Immune Activation Through Nalp3 Inflammasome Sensing of Asbestos and Silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–56. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–82. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 32.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–75. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 33.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guarda G, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–23. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Kaser A, et al. Interferon-alpha induces interleukin-18 binding protein in chronic hepatitis C patients. Clin Exp Immunol. 2002;129:332–8. doi: 10.1046/j.1365-2249.2002.01911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sciacca FL, Canal N, Grimaldi LM. Induction of IL-1 receptor antagonist by interferon beta: implication for the treatment of multiple sclerosis. J Neurovirol. 2000;6 (Suppl 2):S33–7. [PubMed] [Google Scholar]
- 37.Gough PJ, Gordon S, Greaves DR. The use of human CD68 transcriptional regulatory sequences to direct high-level expression of class A scavenger receptor in macrophages in vitro and in vivo. Immunology. 2001;103:351–61. doi: 10.1046/j.1365-2567.2001.01256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greaves DR, Quinn CM, Seldin MF, Gordon S. Functional comparison of the murine macrosialin and human CD68 promoters in macrophage and nonmacrophage cell lines. Genomics. 1998;54:165–8. doi: 10.1006/geno.1998.5546. [DOI] [PubMed] [Google Scholar]
- 39.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 40.Leon RP, et al. Adenoviral-mediated gene transfer in lymphocytes. Proc Natl Acad Sci U S A. 1998;95:13159–64. doi: 10.1073/pnas.95.22.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell Immunol. 2001;214:110–22. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- 42.Pien GC, Satoskar AR, Takeda K, Akira S, Biron CA. Cutting edge: selective IL-18 requirements for induction of compartmental IFN-gamma responses during viral infection. J Immunol. 2000;165:4787–91. doi: 10.4049/jimmunol.165.9.4787. [DOI] [PubMed] [Google Scholar]
- 43.Smith HRC, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–31. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews DM, Scalzo AA, Yokoyama WM, Smyth MJ, Degli-Esposti MA. Functional interactions between dendritic cells and NK cells during viral infection. Nat Immunol. 2003;4:175–81. doi: 10.1038/ni880. [DOI] [PubMed] [Google Scholar]
- 45.McDonald B, et al. Intravascular Danger Signals Guide Neutrophils to Sites of Sterile Inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 46.Pisetsky DS. The role of innate immunity in the induction of autoimmunity. Autoimmunity Rev. 2008;8:69. doi: 10.1016/j.autrev.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 47.Cridland JA, et al. The mammalian PYHIN gene family: Phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.González-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12:125–35. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin J, et al. LRRFIP2 negatively regulates NLRP3 inflammasome activation in macrophages by promoting Flightless-I-mediated caspase-1 inhibition. Nat Commun. 2013;4:2075. doi: 10.1038/ncomms3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang P, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–94. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 51.Khare S, et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 2012;36:464–76. doi: 10.1016/j.immuni.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao R, Guo X. Human microvascular endothelial cells immortalized with human telomerase catalytic protein: a model for the study of in vitro angiogenesis. Biochem Biophys Res Commun. 2004;321:788–794. doi: 10.1016/j.bbrc.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 53.Bryan NB, Dorfleutner A, Kramer SJ, Yun C, Rojanasakul Y, Stehlik C. Differential splicing of the apoptosis-associated speck like protein containing a caspase recruitment domain (ASC) regulates inflammasomes. J Inflamm (Lond) 2010;7:23. doi: 10.1186/1476-9255-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryan NB, Dorfleutner A, Rojanasakul Y, Stehlik C. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J Immunol. 2009;182:3173–82. doi: 10.4049/jimmunol.0802367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gross S, et al. Bioluminescence imaging of myeloperoxidase activity in vivo. Nat Med. 2009;15:455–61. doi: 10.1038/nm.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.