Abstract
Efforts to map the Escherichia coli interactome have identified several hundred macromolecular complexes, but direct binary protein-protein interactions (PPIs) have not been surveyed on a large scale. Here we performed yeast two-hybrid screens of 3,305 baits against 3,606 preys (~70% of the E. coli proteome) in duplicate to generate a map of 2,234 interactions, approximately doubling the number of known binary PPIs in E. coli. Integration of binary PPIs and genetic interactions revealed functional dependencies among components involved in cellular processes, including envelope integrity, flagellum assembly and protein quality control. Many of the binary interactions that could be mapped within multi-protein complexes were informative regarding internal topology and indicated that interactions within complexes are significantly more conserved than those interactions connecting different complexes. This resource will be useful for inferring bacterial gene function and provides a draft reference of the basic physical wiring network of this evolutionarily significant model microbe.
Mapping protein-protein interactions (PPIs) is essential to understand biological function. High-throughput tandem affinity purification followed by mass spectrometry (AP-MS) has been used to identify PPIs on a large scale in Escherichia coli 1–3, but this method is unable to distinguish direct interactions when several proteins are purified together. Various high-throughput biochemical approaches have been proposed to examine binary PPIs, including the yeast two-hybrid (Y2H) method which has been successively applied in several eukaryotes 4, 5, prokaryotes 6 and viruses 7. Here we explored the global architecture of the binary interactome in the gram-negative bacterium E. coli by carrying out proteome-scale, high-throughput Y2H screening. We detected network associations for 1,269 E. coli proteins and more than half of the interactions are new. Furthermore, we derived a comprehensive interactome of E. coli by integrating literature curated binary PPIs with Y2H interactions obtained from this study, and analyzed the network structure and function of this E. coli interactome.
RESULTS
Testing the Y2H system for E. coli proteins
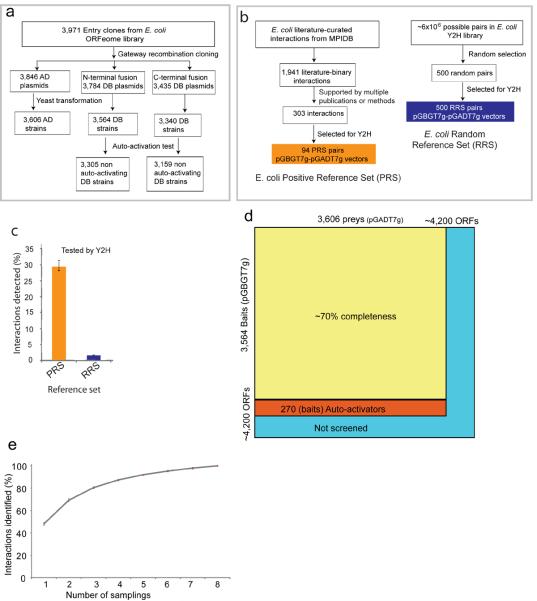
We constructed Y2H libraries by transferring 3,971 Gateway-compatible E. coli entry clones8 into three different Y2H vectors (pGADT7g, pGBGT7g and pGBKCatg) (Fig. 1a, Supplementary Fig. 1a). After the removal of auto-activating baits9, our libraries contained 3,305 baits (in pGBGT7g as DNA binding domain fusions) and 3,606 preys (in pGADT7g as activation domain fusions) (Fig. 1a). To evaluate the efficacy of the Y2H for E. coli proteins, we first benchmarked our system with a reference set of likely (that is, positive) and unlikely (that is, random) PPIs. The positive reference set (PRS) was compiled from manually curated databases10, 11. Approximately 1,941 binary PPIs have been documented for E. coli, most of which have been reported as purified dimers or binary complexes in AP-MS studies. Previous studies have shown that interactions supported by multiple publications are more reliable than those supported by a single publication12, so we selected 303 PPIs that had been described in multiple publications or were characterized by two independent methods.
Figure 1. Systematic identification of binary PPIs in E. coli.
(a) Pipeline used to construct E. coli Y2H bait and prey libraries. The Y2H libraries were constructed by transferring 3,971 Gateway-compatible E. coli entry clones into AD (activation domain, pGADT7g) and DB (DNA binding domain, pGBGT7g and pGBKCatg) Y2H vectors using Gateway recombination cloning. (b) Pipeline used to assemble literature-curated binary E. coli PPIs (a positive reference set (PRS)) and random reference set (RRS) for binary PPIs. (c) Percentage of the PRS and RRS detected by the Y2H system (error bars denote standard error). (d) Coverage of E. coli proteome (a total of ~4200 ORFs) and the search space of pair-wise protein combinations tested. (e) Sampling sensitivity of Y2H screens measured by screening 92 baits multiple times (error bars denote standard error). Based on this, the mean sampling sensitivity was estimated to be 69% (+/− 2.5%) for the two screens performed on the full search-space.
Among these, our pGBGT7g-pGADT7g library contains both bait and prey clones for 212 PPIs (Supplementary Table 1a). From these we randomly selected 94 PPIs as a PRS (Supplementary Table 1b) and tested them using the pGBGT7g-pGADT7g Y2H vector systems (Fig. 1b, Online Methods). We also tested 100 PRS interactions using the pGBKCatg-pGADT7g vector system (Supplementary Table 1c, Online Methods). Approximately 29% (27 of 94) of the PRS interactions were detected by the pGBGT7g-pGADT7g system, a sensitivity that is comparable to yeast and human Y2H array screening12, 13. We assayed 500 randomly chosen protein pairs comprising the random reference set and detected an interaction for only four, giving a specificity of ~99% (Fig. 1c and Supplementary Table 1d). However, this number cannot be directly extrapolated to the whole data set, given that most false positives arise through a small set of promiscuous baits and preys. We have removed such non-specific interactions from our Y2H dataset (Online Methods).
High-throughput Y2H screening
The Y2H screening was conducted with pools of 3 baits, and positive yeast colonies were selected on synthetic media plates (Supplementary Fig. 1c and Online Methods). The interacting preys were identified by yeast colony PCR, followed by DNA sequencing. The resulting interactions were retested in quadruplicate for each bait-prey combination using fresh yeast bait and prey cultures from archival stocks, and interactions that were either non-reproducible or auto-activated were eliminated (Supplementary Fig. 1d)9, 14. By this approach, we systematically screened 3,305 baits against 3,606 preys, thus covering ~70% of E. coli interactome space (Fig. 1d and Supplementary Fig. 2).
When screening millions of protein pairs, the search space needs to be sampled multiple times to detect all the interactions that are detectable by the assay used 5, 15. Therefore, to estimate the sampling sensitivity of our Y2H system, we performed eight independent sampling screens using 92 baits against the entire prey library (3,606 proteins), resulting in saturation in the number of detectable interactions (Fig. 1e). Based on this analysis, we estimated a mean sampling sensitivity of 69% (+/− 2.5%) for a combination of two screens; at least five-fold sampling would be required to obtain 90% saturation (Fig. 1e). We sampled the entire search space twice independently, resulting in 2,234 high-quality PPIs among 1,269 proteins (Supplementary Table 2). Nearly two-thirds (1,830 of 2,234) of these interactions have not been reported previously. Evaluation against a PRS of 212 interacting pairs indicated a sensitivity of ~21% (44 of 212) in the pGBGT7g-pGADT7g vector system (Supplementary Table 1a), which is comparable to the sensitivity attained in Y2H screening in yeast 5.
To assess independently the quality of these PPIs, we evaluated 114 randomly selected PPIs by two different methods: co-immunoprecipitation (Co-IP) and LUMIER 16 assays (Online Methods). About 86% (99/114) of the Y2H interactions were confirmed by at least one of these biochemical methods (Fig. 2a, b, c and Supplementary Table 3), consistent with previously published Y2H validation rates 4, 5.
Figure 2. Quality assessment and comparison of Y2H interactions with data from the literature.
(a) Immunoblots of 6 of the 114 randomly selected PPIs identified by Y2H assay that were tested by co-immunoprecipitation. The interacting pairs and negative controls (vector) were co-transformed into E. coli and protein binding was detected by immunoblotting (Online Methods). (b) The same 114 randomly selected interacting protein pairs were tested by LUMIER assay 16. Interactions were scored as positive when exhibiting a luminescence intensity ratio (LIR) value > 3 and a p-value <0.05. These two values are plotted against each other in the graph in logarithmic scale. The LIR and p-value thresholds are indicated by dashed lines: (log (LIR) >0.477; log (p) <−1.30). (c) Number of PPIs validated by Co-IP, LUMIER or both assays. (d) Overlap between interactions detected in this large-scale Y2H (pink) and in the literature. The manually curated literature-binary PPIs (purple) are compiled from the microbial protein interaction database (MPIDB; http://jcvi.org/mpidb/about.php) which consists of 1,941 manually curated binary PPIs. The combined-AP-MS PPIs (beige) were predicted from large scale AP-MS studies1–3 and consists of 20,425 PPIs. (e) Pipeline used to detect direct interactions within co-complexes by Y2H matrix screening.
Of the 2,234 PPIs reported in this study, 239 are supported by manually curated PPIs from the literature10, 11, and another 165 are supported by AP-MS studies 1–3 (Fig. 2d). The limited overlap with the literature data is mainly due to low Y2H assay sensitivity (that is, many false-negatives) rather than low specificity (that is, false-positives) (Fig. 1c), consistent with the low overlap observed in the yeast interactome5.
We have shown previously that the Y2H system can be used effectively to analyze direct interactions among proteins within a complex 17. Assessing our Y2H interactions against structures of E. coli protein complexes from the Protein Data Bank 18 (Supplementary Fig. 3a, b, c, d and Supplementary Table 4) revealed that >90% (101/110) of interactions among complex subunits corresponded to direct PPIs (Supplementary Fig. 3 and Supplementary Table 4) 1–3. We then tested 227 putative E. coli protein complexes containing three or more protein subunits 3 by pair-wise Y2H assays of the subunits. To enhance assay sensitivity, the Y2H screening was conducted using both N-terminal and C-terminal fusion bait proteins 13. By this method, we identified a total of 458 PPIs between the subunits of the complexes (Fig. 2e, Supplementary Fig. 4 and Online Methods).
High-quality binary interaction network
To obtain a more comprehensive binary E. coli interactome, we combined the high-quality Y2H data set from this study with binary interactions that we manually curated from the literature 10, 11, hereinafter referred to as “combined-binary” dataset. This dataset contains 3,946 binary interactions among 2,048 proteins (Supplementary Table 5, Fig. 3). Based on our precision, completeness, assay- and sampling-sensitivity measures, we estimated that the total size of the E. coli interactome is on the order of ~10,000 PPIs (Supplementary Results). The combined-binary dataset described in this study comprising ~39% of the whole E. coli binary interactome. We estimate that approximately half of the interactions in the combined-binary interaction dataset likely to be between the components of multimeric complexes (that is, complexes inferred from AP-MS data3 and complexes from EcoCyc http://ecocyc.org/, and half are transient binary interactions (Supplementary Results).
Figure 3. Structural analysis of the E. coli binary interactome.
Nodes (representing proteins) are colored according to the availability of structural data. Edges (representing PPIs) are colored according to their source (literature, our Y2H experiment or both). The sub-networks (enlarged parts) are based on a previous AP-MS study 3 and for which Y2H and literature binary interactions provided the topology for a sub-complex of at least three components. Protein structural data is overlaid. For example, protein complex 42 (bottom left) contains seven proteins; the Y2H binary interactions suggest a topology for five subunits in the complex, and all the subunits have either a complete structure or a model. Furthermore, for two interactions between three different components we have structures of the binary sub-complexes. For complexes 72, 100, and 103 the Y2H binary interactions identified in our screens suggest an almost complete topology.
Like other biological networks 19, both the combined-binary interactions and the combined AP-MS data sets2, 3 are scale-free; that is, many interacting proteins have low connectivity and few have high connectivity (Supplementary Fig. 5a). As with the yeast binary interactome 5, we found that AP-MS interactions were enriched (p = 1.8 × 10−31) in essential protein pairs (Supplementary Fig. 5b), and essential proteins were better connected (Fig. 4a) in the AP-MS network than in the combined-binary interaction network. Moreover, the path length between proteins in the combined-binary interaction network was longer (> 5) compared to the combined AP-MS network (Fig. 4b), reflecting the tendency of Y2H to detect more interactions outside of protein complexes than AP-MS5.
Figure 4. Comparison of the properties of binary and AP-MS interaction networks.
(a) The degree of a node in a network (degree distribution) involving essential E. coli protein pairs from the combined-binary (data from this study and literature binary interactions10) and combined AP-MS networks2, 3. (b) Frequency distribution involving protein pairs from the combined-binary and combined AP-MS PPI networks at different path lengths. For panels A and B, the data were sampled by considering the same number of interactions among the same number of nodes in the two datasets. (c) Distribution of the Pearson correlation coefficient (PCC) between the GI profiles for gene pairs encoding interacting proteins derived from combined-binary or combined-AP-MS network versus random gene pairs. (d) Distribution of co-expression and (e) condition-dependent phenotypic correlation profiles with corresponding interacting proteins, shown as in (c). The p-values for panels (c,d,e) were computed (i.e., AP-MS vs. random (blue) and combined-binary vs. random (brown)) using the Student's t- test. (f) Average semantic similarity of the combined-binary and combined-AP-MS PPI networks is shown for Gene Ontology (GO) categories.
As in yeast 20, we found that physically interacting proteins from the combined-binary dataset and from the combined AP-MS dataset were more likely to have highly correlated genetic interactions (GI) profiles (p ≤ 0.05) than randomly selected protein pairs (Fig. 4c). Physically interacting protein pairs in both datasets also showed positively correlated co-expression (p ≤ 0.05) and phenotypic (p ≤ 0.05) profiles (Fig. 4d, e) 5. Moreover, the average semantic similarity 21 of Gene Ontology (GO) annotations in the combined-binary PPI network was comparable to that of the combined AP-MS network, suggesting that the binary PPIs are as functionally coherent as the AP-MS-derived PPIs (Fig. 4f).
Protein complex topology of the E. coli interactome
AP-MS-derived complexes do not provide information about the internal topology of multi-protein assemblies 22, 23. We therefore assessed whether Y2H experiments could be used to detect direct physical interactions within complexes identified by large-scale AP-MS experiments. We compiled a list of 227 E. coli protein complexes that have three components or more as identified in a large-scale AP-MS screen 3. Next, we identified interactions between subunits of these complexes in our Y2H data set and among literature-curated binary interactions (Supplementary Table 6 and Online Methods). We mapped 745 binary interactions within 203 complexes (Supplementary Fig. 6), of which 319 (43%) interactions were found in this study and 426 (57%) were obtained from the literature (108 of which were recapitulated in our Y2H study). We could deduce a putative complete internal topology for 15 multi-protein complexes; for these 15 complexes the binary interactions connected all components of the complex, therefore providing a hypothesis on its internal connectivity. For another 45 complexes we determined the putative internal topology of a sub-complex with at least three subunits, and for 46 complexes we determined the internal connections between pairs of subunits (Supplementary Fig. 6). For an additional 97 complexes we could only map homomeric interactions (that is interactions between multiple copies of the same protein). We found in the literature a structure (or a homology model) for 1,097 proteins and 202 interactions (Supplementary Fig. 7) within the complexes. For several complexes, we found a sub-complex of at least three components for which complete topology was revealed by experimental binary interaction data and structural data was available for all its subunits (Fig. 3 and Supplementary Fig. 8).
Integration of PPI and GI networks
Identification of physical interactions among proteins does not necessarily imply that all interacting partners belong to the same functional pathway24, 25. By contrast, genetic screens can elucidate functional relationships between genes26, 27. Analogous to the yeast synthetic genetic array approach 28, we have previously developed a high-throughput synthetic genetic array screening technology, termed eSGA 29 to map GI networks in E. coli to elucidate pathway-level relationships. Integration of this GI data with the PPI networks should provide information that is not attainable from either study alone29. Using a Markov clustering algorithm 30, we identified physically related groupings in the binary interaction network and overlaid with the currently available large-scale E. coli GIs datasets 24, 29, 31 (publically accessible at http://ecoli.med.utoronto.ca/esga; Supplementary Table 7) for the corresponding protein interactions to determine the functional relationships of individual proteins and multi-protein complexes in various pathways and processes (Fig. 5, Supplementary Tables 7, 8).
Figure 5. Integrative analyses on the PPI and GI data.
Examples of sub-networks showing the physical and genetic connectivity among the components of various bioprocesses. Sub-networks containing Y2H physical interactions (shown as grey edges) among 1,269 proteins are derived using a Markov clustering approach. The GIs (red edges for negative interactions and green for positive interactions) from the published large scale eSGA surveys24, 29, 31 were overlaid on the PPI network. Sub-networks with positive and/or negative interactions are highlighted with shaded ovals: (a) secretion components (cluster ID: 5); (b) flagellum or motility components (cluster ID: 9); (c) subunits of ATP-dependent protease complexes (cluster ID: 14); and (d) ycfM and pbpG (cluster ID: 8). For details on GIs in each of the sub-networks and clusters IDs see Supplementary Tables 7 and 8. Large nodes in each sub-network indicated that genetic associations with the indicated protein subunits are known; small nodes indicate an absence of GIs.
This integrative analysis captured interactions that were missed by either one of the methods. For example, in the secretory pathway the inner membrane protein SecA is peripherally associated with the multi-subunit translocation apparatus SecYEG 32; Y2H alone captured the physical interaction between SecA and SecY and not by eSGA (Fig. 5a); however interactions between other Sec components missed by Y2H were detected as GIs by eSGA. Also, for components of the flagellum (e.g., fliN-cheR and fliA-fliI) (Fig. 5b), physical interactions among the proteins are overlaid by a negative phenotype in the GI network, consistent with their joint participation in flagellum assembly and motility33. Another example involves a sub-network of physically connected subunits of the ATP-dependent protease complex (ClpAX) known for their cooperative function in protein quality control 34. These had a positive GI phenotype (Fig. 5c), suggesting that they operate in the same pathway 28, 29, 35. Similarly, the Y2H interaction between the recently characterized outer membrane lipoprotein YcfM/LpoB and penicillin binding protein PbpG showed a strong positive interaction (Fig. 5d), consistent with a recently proposed regulatory role of YcfM in peptidoglycan synthesis 36
Conservation of binary PPI
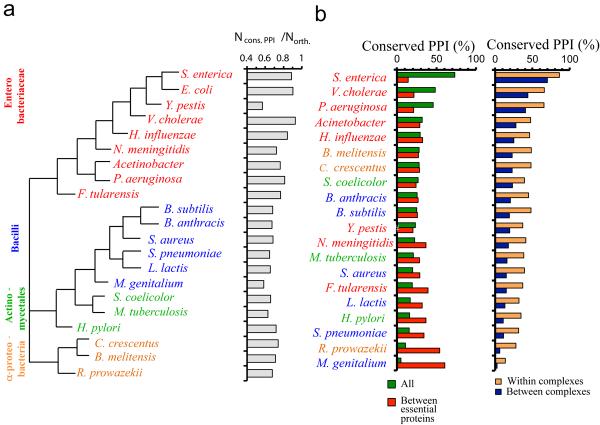
To investigate the evolutionary significance of PPIs detected in E. coli with respect to other microbes, we examined the presence of PPI orthologs in other bacteria (Fig. 6a) by using the combined-binary interactions of E. coli and binary interactions reported for other bacteria in literature curated database10 and from the large scale bacterial interactome studies (Online Methods). Previous investigations have shown that interactions in yeast are conserved in worms37 and metazoan interactions are able to predict homologous connections in yeast 38. We found that interacting protein pairs from our study are highly conserved in bacteria that are closely related to E. coli but less so with increasing distance from E. coli in the phylogenetic tree (Fig. 6a). However, the opposite trend was observed when interactions between proteins encoded by essential E. coli genes were considered (Fig. 6b). We also found that interactions between the subunits of protein complexes are more conserved than interactions between proteins belonging to different complexes (Fig. 6b).
Figure 6. Conservation of physical interactions between and within protein complexes.
(a) Phylogenetic tree based on the comparison of complete proteomes of 20 different bacteria that are closely related to E. coli. Using bacterial proteins that had orthologs in E. coli, we determined the sets of interologs (that is, conserved interacting homologs, Ncons.PPI) in each organism. Normalizing the number of interologs by the corresponding number of orthologs in each organism (Northo), we observed a declining trend with increasing distance from E. coli in the phylogenetic tree. (b) Bacteria were sorted according to their numbers of predicted interologs and the data suggest that PPIs in E. coli were more conserved in evolutionarily closer organisms. However, we found the opposite trend when we considered the fraction of conserved interactions between essential E. coli proteins (left panel, red). We determined the number of interactions between proteins in other species that are in the same complex or in different complexes compared to protein complexes in E. coli (right panel) and found that interactions are more conserved in species closely related to E. coli, but interactions within complexes are more often conserved than interactions between complexes.
DISCUSSION
Our study substantially expands the available resources for protein complexes and their connections to pathways in the model gram-negative bacterium, E. coli. Y2H and AP-MS studies provide largely complementary information about the interactome and both are essential to obtaining complete protein networks in prokaryotic biology. The current dataset confirms some known binary interactions and contains many novel PPIs within and between complexes.
Binary PPI interaction maps also provide information about the internal topology of multimeric protein complexes, which should allow for their structural modeling, once the stoichiometry of each component is known22. This is illustrated by the structures of two well-characterized E. coli complexes: RecBCD 39 and the F1 portion of the ATP synthase complex 40 containing three copies of each of its subunits (alpha, AtpA and beta, AtpD) (Fig. 3).
Comparison of the binary interactome to complexes found through AP-MS and in the EcoCyc database (Supplementary Tables 9 and 10) suggests that the majority of PPIs (2128) are not in complexes. We suspect that many protein complexes remain undetected and may form only under certain growth conditions; the Y2H assay may detect PPIs irrespective of the growth condition in E. coli as it is carried out in yeast. Furthermore, the conditions used for affinity purification may preclude the detection of less stable interactions that are detectable by Y2H. Our data suggest that there are distinct subsets of interactions found in complexes or as more transient binary interactions. Based on the assessment of the the total number of interactions in each group, we estimate that the combined total is on the order of 9,200 to 11,400 interactions (Supplementary Results). To our knowledge, this study represents the largest experimentally derived catalog to date of the E. coli binary interaction network and thus is a draft reference of the basic physical wiring network of an evolutionarily well conserved model microbe.
Supplementary Material
Acknowledgments
S.V.R and P.U. were supported by the National Institutes of Health grant GM079710. P.U. and P.A. were supported by the Seventh Research Framework Programme of the European Union (AntiPathoGN; EU grant HEALTH-F3-2009-223101). M.B. was supported by the Discovery Grant from the Natural Sciences and Engineering Research Council (DG-20234). We thank Dr. Xavier De Bolle for providing pRH016 and pRH018 vectors.
Footnotes
Author contributions: The project was conceived by S.V.R. and P.U, and directed by S.V.R and R.P. P.K., J.F., R.H., G.S. and S.V.R performed the experiments. S.V.R., A.K., R.M., S.B.P., J.V., R.A., S.P., A.C., S.W., P.A. and M.B performed computational analysis. A.E. provided support for computational analysis tools. S.V.R., P.A. and M.B wrote the manuscript, with input from P.U.
References
- 1.Arifuzzaman M, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 3.Hu P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Titz B, et al. The binary protein interactome of Treponema pallidum--the syphilis spirochete. PLoS ONE. 2008;3:e2292. doi: 10.1371/journal.pone.0002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uetz P, et al. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopala SV, et al. The Escherichia coli K-12 ORFeome: a resource for comparative molecular microbiology. Bmc Genomics. 2010;11:470. doi: 10.1186/1471-2164-11-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopala SV, Uetz P. Analysis of protein-protein interactions using high-throughput yeast two-hybrid screens. Methods Mol Biol. 2011;781:1–29. doi: 10.1007/978-1-61779-276-2_1. [DOI] [PubMed] [Google Scholar]
- 10.Goll J, et al. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24:1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopala SV, et al. MPI-LIT: a literature-curated dataset of microbial binary protein--protein interactions. Bioinformatics. 2008;24:2622–2627. doi: 10.1093/bioinformatics/btn481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun P, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YC, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods. 2010;7:667–668. doi: 10.1038/nmeth0910-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidalain PO, Boxem M, Ge H, Li S, Vidal M. Increasing specificity in high-throughput yeast two-hybrid experiments. Methods. 2004;32:363–370. doi: 10.1016/j.ymeth.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrios-Rodiles M, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopala SVS, P. Caufield JH, Tovchigrechko A, Uetz P. Studying protein complexes by the yeast two-hybrid system. Methods. 2012 doi: 10.1016/j.ymeth.2012.07.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman HM, et al. The Protein Data Bank. Nucleic acids research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barabasi AL. Scale-free networks: a decade and beyond. Science. 2009;325:412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 20.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Du Z, Payattakool R, Yu PS, Chen CJ. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23:1274–1281. doi: 10.1093/bioinformatics/btm087. [DOI] [PubMed] [Google Scholar]
- 22.Aloy P, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 23.Lasker K, et al. Integrative structure modeling of macromolecular assemblies from proteomics data. Mol Cell Proteomics. 2010;9:1689–1702. doi: 10.1074/mcp.R110.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babu M, et al. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS genetics. 2011;7:e1002377. doi: 10.1371/journal.pgen.1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput Biol. 2008;4:e1000065. doi: 10.1371/journal.pcbi.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141:739–745. doi: 10.1016/j.cell.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 28.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butland G, et al. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 30.Brohee S, van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babu M. Quantitative genome-wide genetic interaction screens reveal global epistatic relationships of protein complexes in Escherichia coli. PLoS genetics. 2014 doi: 10.1371/journal.pgen.1004120. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver DB, Beckwith JE. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopala SV, et al. The protein network of bacterial motility. Mol Syst Biol. 2007;3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bershtein S, Mu W, Serohijos AW, Zhou J, Shakhnovich EI. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell. 2013;49:133–144. doi: 10.1016/j.molcel.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 36.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews LR, et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharan R, et al. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 40.Cingolani G, Duncan TM. Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nature structural & molecular biology. 2011;18:701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.