Abstract
Isolated and established in culture from the Antarctic in 1988, the nematode Panagrolaimus davidi has proven to be an ideal model for the study of adaptation to the cold. Not only is it the best-documented example of an organism surviving intracellular freezing but it is also able to undergo cryoprotective dehydration. As part of an ongoing effort to develop a molecular understanding of this remarkable organism, we have assembled both a transcriptome and a set of genomic scaffolds. We provide an overview of the transcriptome and a survey of genes involved in temperature stress. We also explore, in silico, the possibility that P. davidi will be susceptible to an environmental RNAi response, important for further functional studies.
Introduction
Panagrolaimus davidi Timm 1971 [1] was first isolated from the McMurdo Sound region of the Antarctic in 1988 and cultured at the University of Otago [2]. Much like its better known temperate cousin, Caenorhabditis elegans, it has, over the years since its early adoption as a model of study, provided surprises that in hindsight made its initial selection, and culturing, very fortunate. Panagrolaimus davidi was found to survive intracellular freezing [3]. Although the cuticle acts as a barrier to ice, at low sub-zero temperatures inoculative freezing can occur through the excretory pore and other orifices, with the ice seeding the body fluid and freezing both intra- and extracellular compartments [3], [4]. Survival of intracellular freezing was first described by Salt in the fat body cells of the goldenrod gall fly [5], [6], [7] but P. davidi still remains the only organism for which it has been described in all compartments of its body. In addition to freezing tolerance, P. davidi has developed other strategies for dealing with low temperatures. At high sub-zero temperatures, and importantly, when the rate of freezing is slow, ice is unable to enter body openings and the nematode supercools. This leads to a vapour pressure difference between the supercooled body fluids and the surrounding ice leading to the transfer of water from the nematode to the surrounding ice; a process termed cryoprotective dehydration [8], [9]. Once P. davidi is cryoprotectively dehydrated, there is not enough water to freeze in either the pseudocoel or other compartments allowing it to sustain low sub-zero temperatures by freeze avoidance. While an increasingly detailed molecular picture is emerging of cryoprotective dehydration using large scale gene expression and proteomic approaches [10], [11], [12], no comparable molecular work has been undertaken on the survival of intracellular freezing.
With the sea change in current sequencing technology, what was previously termed a non-model organism can now become, with a concerted effort and a fraction of the resources that were required even five years ago, a molecular model for a specific physiological trait, which is a powerful change in the way biological systems can now be studied. With this in mind, we have undertaken to sequence not just the transcriptome, which will provide a backbone for expression studies, but also the genome. With the dramatic decrease in cost associated with sequencing genomes, the scaffolds and contigs resulting from a preliminary assembly, even when broken up, are a natural complement to the transcriptome, providing valuable information on gene structure such as intron-exon junctions. To date our only knowledge of the genome comes from a paper by Goldstein and Wharton [13] describing seven synaptonemal complexes, therefore seven chromosome pairs (2n = 14).
The molecular information provided on the scale such as the present study, opens up the exploratory possibilities of gene expression studies. But in order to gain more direct evidence for any such exploratory results, it would be valuable to know whether P. davidi is potentially susceptible to functional genetic methodologies. Since Fire et al. [14] determined the role of RNA interference in C. elegans, this method has provided a clear mechanism by which the role of a pathway or specific genes may be understood, for example, in an organism’s response to environmental stresses [15]. With the two sources of information, the transcripts and their gene structure determined by the genomic sequence, clean RNAi probes for specific targets can be easily developed. Recently, Panagrolaimus superbus was shown to respond to feeding RNAi [16], but a lesson has already been learnt from C. briggsae when, unlike C. elegans for whom it has been a comparative model, it was shown that RNAi was not possible owing to a divergent form of sid-2 [17].
A final word on a mystery that has unfolded in the last few years. In 2009, Lewis et al. published a paper [18] on the phylogenetics of the Panagrolaimus genus. P. davidi (designated CB1, from its presumed origin at Cape Bird, Ross Island) was included in this study, but unexpectedly, two Californian species proved phylogenetically closer to P. davidi CB1 than any other species or strain. Genetic analysis of fresh field material of P. davidi collected during 2005–2007 from Cape Hallett and Gondwana Station on the Victoria Land coast and from Cape Bird showed that the field strain of P. davidi is a different species to P. davidi CB1 [19]. One argument put forward to explain this difference is the possible dominance in culture of a less common strain owing to its parthenogenetic reproductive mode. Questions of invasiveness have also been considered. But the adaptations of P. davidi CB1 to low temperature make this highly unlikely; or highly surprising. Further molecular work should help to resolve this intriguing situation, but in terms of its physiology, the origin of P. davidi CB1 is not of relevance, since its cold tolerance adaptations singles it out as an important organism of study.
Materials and Methods
Culturing, extraction, and library construction for Expressed Sequence Tags
Nematodes were cultured on Escherichia coli strain OP50 on NGM agar plates [20]. RNA was extracted from 580 mg (for a culture grown at 20°C, called PDT) and 730 mg (for a culture grown at 20°C and subsequently brought down to 4°C, called PDF) of P. davidi CB1 (wet weights), respectively. Total RNA (930 µg for PDT and 750 µg for PDF) was prepared with RNagents total RNA Isolation Kit (Promega). Poly(A)+ RNA (2.9 µg for PDT and 5.5 µg for PDF) were isolated with Illustra mRNA Purification Kit (GE healthcare). 2 µg of poly(A)+ RNA was then used to generate a cDNA library with Cloneminer cDNA library Construction Kit (GE healthcare).
Sanger sequencing and quality assurance
Seqclean [21] and Crossmatch [22] was applied to the two (PDT and PDF) P. davidi CB1 Expressed Sequence Tag (EST) libraries, removing the vector and stripping any poly-A tails and poor quality sequence. This left 25,182 reads from the PDT set and 69,958 reads from the PDF set. All of these sequences were added to the post-assembly of the Illumina reads. The ESTs are held in dbEST with accession numbers: JZ585947–JZ681086.
Culturing, extraction, and library construction for Illumina sequencing
The nematodes were cultured in S medium at 20°C for 3 weeks and fed every 3–4 days with E. coli following [23]. Nematodes were extracted using a modified Baermann technique [24]. The worms for the DNA and RNA were then snap frozen with liquid nitrogen in 1.5 ml microcentrifuge tubes and preserved at −80°C until extraction. Seven sets of these worms for the RNA were then subjected to other physiological states to enrich for stress related transcripts. These seven treatments consisted of 1) exposure to cold acclimation at +5°C for 3 days; 2) the previous sample set was then immersed in the bath of a refrigerated circulator where it was cooled from +1°C to −1°C at 0.5°C min−1, and frozen by adding a small ice crystal and maintained at −1°C for 24 h; 4) the previous sample was warmed to +1°C at 0.5°C min−1, and allowed to recover at 20°C for 24 hours; 5) samples from stage 1 were cooled from +1°C to −10°C at 0.5°C min−1; 6) the previous stage was ice nucleated once held at −10°C; 7) the previous sample was warmed to +1°C at 0.5°C min−1. 500 ng (wet weight) was used for the RNA extraction (from each of the 8 different stages, including the culture grown at 20°C) and 500 ng (wet weight) for DNA extraction. The RNA was extracted from whole worms using TRI-sure (Bioline) according to manufacturer’s instructions and purified on Qiagen RNeasy columns. The DNA was extracted from whole worms using the Qiagen DNAeasy Blood and Tissue kit according to manufacturer’s instructions. Both DNA and RNA were checked for purity on standard agarose gels and quantified using a NanoDrop ND-1000 spectrophotometer (Labtech). 8 µg of DNA and 10–15 µg of RNA for each of the stages were used for sequencing.
Illumina sequencing and quality assurance
The RNA was sequenced on an Illumina HiSeq 2000 resulting in 143,223,606 paired-end reads of 100 bp, after quality control. Quality control consisted of removing adaptors from the sequence, removing reads where the number of unresolved nucleotides exceeded 5%, and reads where the number of nucleotides with phred quality less than or equal to 10 was over 20%. The reads can be downloaded from the SRA repository under the accession number SRP041973.
Genomic DNA was randomly fragmented, with insert sizes of between 500–800 selected through gel electrophoresis, with the fragments gel purified with adapters ligated. After sequencing, quality control consisted of adaptor removal and removing reads in which more than 50% of the bases had phred scores of less than 5. Two runs of 95 and 138 million paired end reads of 100 bp respectively remained after quality control. The sequence data can be downloaded from the SRA repository under the accession number SRP041572.
Transcriptome assembly and annotation
The transcriptome was assembled from 143,223,606 paired-end Illumina reads of 100 bp, and 95,140 sanger sequences. The Illumina reads were assembled first with Soapdenovo [25] using a number of different kmer sizes. Illumina datasets assemble differently depending on the kmer size chosen, with any given dataset having an optimal kmer for numbers and lengths of contigs. In the case of P. davidi CB1, by selecting a number of odd values from 19 to 93, the optimal kmer lay around 69 (see File S1). In order to enrich the assembly as much as possible, all contigs greater than 200 bp from all the different kmer size assemblies were selected (a total of 1,107,215 contigs) and, jointly with the 95,140 sanger sequences, they were assembled together using Newbler [26] and CAP3 [27] consecutively, to eliminate redundancy. The resulting assembly was compared using Blast [28] against the nr database [29] as well as Caenorhabditis elegans [30], Plectus murrayi [31] and Panagrolaimus superbus [32] nematode databases. In addition, a separate blast against the entire WS242 wormbase [30] was carried out. The final assembly set was reduced to those transcripts that were either 500 bp long, or annotated at an e-value less than 1e-10.
Functional groupings of the transcriptome was carried out through Clusters of Orthologous Genes [33], Kegg Orthology [34], and SEED subsystems [35]. Separate annotation of the ESTs was also carried out and they were used in examining specific genes, particularly the LEA-like and HSP-70 genes, owing to their longer lengths combined with their more easily resolved reading frame, where the consensus sequences in the contigs proved more complicated.
DAPI staining of nuclear DNA
C. elegans N2 and P. davidi CB1 were washed out from a 5 cm Nematode Growth Media (NGM) plate and fixed in 2 ml Carnoy’s solution (60% Ethanol, 30% Chloroform, 10% Acetic acid) overnight at room temperature. The fixed worms were then transferred to a watch glass with 50 µl phosphate buffered saline with 0.2% Triton X-100 and 100 ng/ml DAPI (4′,6-diamidino-2-phenylindole), and incubated in a humid chamber in the dark for 30 mins. The worms were mounted on an agar pad slide. Quantification of the intensity of nuclei in ventral nerve cord was carried out with AQUACOSMOS software [36] on fluorescent microscopy pictures with an Axioplan 2 microscope (Carl Zeiss). Estimation of P. davidi CB1 DNA amounts by proportion to C. elegans DNA was done for 10 worms from each species.
Assembling genomic sequence
As with the transcriptome assembly, Soapdenovo, with varying kmer sizes, was used to assemble the DNA paired-end reads. This led to an optimal kmer size of 89. Since it has often been noted that too much sequence coverage can lead to erroneous and more broken assemblies, a second, confirmatory round of assemblies were carried out with only 50x coverage, and then at 50x increments, to see whether the assembly at any lower coverage was better. However, this resulted in confirmation that the more data, the better the assembly. Finally, Blast was used to check and remove any contigs or scaffolds that may have been an assembly of bacterial contaminant.
Results and Discussion
The transcriptome, consisting of 25,875 transcripts, had an average length of 1,163 bp with the GC content at 32.5% (the raw Illumina reads were 35.97%). Against the nr database, 15,748 (61%) of the transcripts were annotated at an e-value of 1e-10 or less. When compared to the C. elegans protein database using blastx, 14,372 (54%) P. davidi CB1 (hereafter referred to simply as P. davidi) transcripts matched 14,395 (57%) of the C. elegans proteins. The P. davidi transcripts matched 62% of the Panagrolaimus superbus EST transcripts [32], the closest nematode for which there is any molecular data, and they matched 52% of the Plectus murrayi ESTs [31], the only other Antarctic nematode for which any molecular data has been published. When compared to all nematode databases currently housed at wormbase (WS242), 68.5% of the P. davidi transcripts matched at an e-value of 1e-10 or lower. The remaining transcripts that had no annotation, and in particular, those that matched no nematode are obviously of interest in that they may well contain potential clues to the unique physiological adaptations of P. davidi. File S2 lists all the transcripts that match against nr at an e-value of 1e-10 or lower, along with their corresponding description.
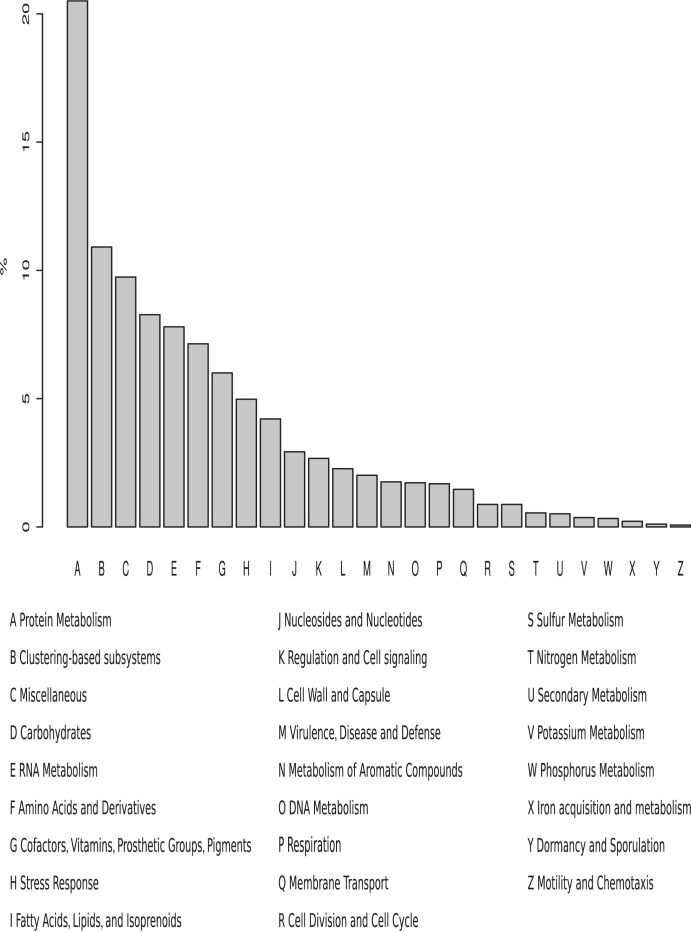
Functional classifications were able to be determined for 55% of the whole transcriptome. Figure 1 shows the functional spread provided by SEED subsystems. Although not shown, both the Clusters of Orthologous Genes and KEGG Orthology analyses, while broader in their categories, provided the same breakdown. The highest proportion of the transcriptome is clearly involved in protein metabolism, with a high proportion of clustering-based subsystems, a designation of genes based on their co-localisation across many genomes. Other highly represented subsystem groups were the carbohydrates, amino acids and derivatives, and RNA metabolism. A separate analysis was done on the two EST libraries with the breakdowns showing similar percentage patterns (File S3) with, like the transcriptome as a whole, protein metabolism having the strongest representation.
Figure 1. SEED subsystem analyses of the transcriptome.
The y-axis indicates the percentage of the total annotated set represented by the specific category.
That the transcriptome is characterised so strongly by genes involved in protein turnover is a clear indication of activity and change which is hardly surprising given the fact that different physiological states were mixed together, where such activity might be expected. With the similar proportions also apparent in the two EST libraries this seems to imply that such activity and change is occuring even within each stage. This is probably reflective of the need to be highly responsive to changes in the environmental conditions.
A natural complement to the transcriptome, even in preliminary form, is the genome sequence. Prior to sequencing however, it is useful to know the size of the genome, even approximately. DAPI staining and comparison to the C. elegans nuclear DNA was carried out with the result that P. davidi is estimated to be roughly ∼90 Mb, slightly smaller than C. elegans at ∼97 Mb. We then sequenced two short-insert paired-end libraries (<1,000 bp) of nuclear DNA, which according to the genome size by DAPI staining, would provide a sequence depth of coverage of roughly 517. Assembly resulted in 86% inclusion of the incorporated genomic reads. The scaffold N50 is 6,352 bp, with an average size of 4,150 bp, and a total size of 195 Mb. File S4 shows the size distribution of the scaffolds greater than the N50 with the largest at 73,240 bp (the N50 of the contigs (those joined together to form the scaffolds) is 1,873 bp with the largest being 21,613 bp with the total size being 93 Mb).
The total size of the scaffolds indicates an observation that had already been noted with the EST libraries, namely that while P. davidi is a parthenogenetic species, which should give rise to a homozygous line, evidence suggests that it is in fact heterozygous. Generation of the ESTs was constructed from a single animal with >10x generations to produce an isogenic line, yet many of the ESTs showed a heterozygosity. Examining the LEA ESTs for example, provided evidence that is best explained by heterozygous descendent sequences in which there is crossing over. This was followed up by examining the LEA loci on the genome, as well as examples in the cDNA of two other genes, glutamine synthetase and 6-phosphogluconate dehydrogenase, all of which led to the same conclusion (see File S5).
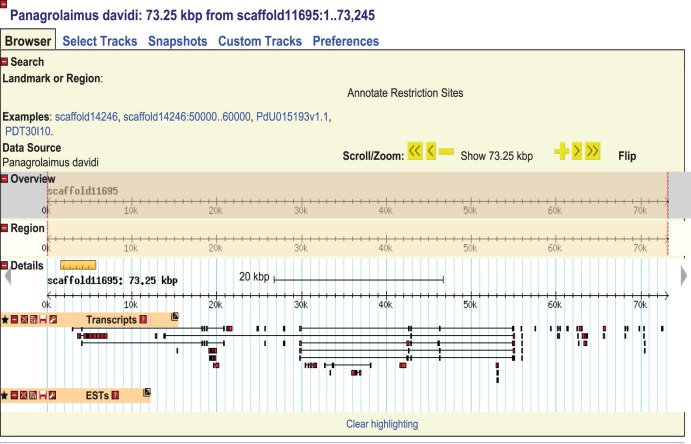
Mapping of the transcriptome onto the scaffolds resulted in 94% of the transcripts finding a match at an e-value of 1e-10 or lower, indicating that while broken up, the genome sequence is reasonably complete. Further work is being carried out to build the assembly into a more contiguous form (see Figure 2). Subsequent work on building up a well-annotated genome should also be made easier with close comparative models, such as other Panagrolaimus species.
Figure 2. Browser of the scaffolds and their transcript alignments, housed at genomes.bas.ac.uk.
The browser will be updated during the continued development of the genome assembly.
The genomic scaffolds that have been assembled have already provided valuable information for the development of PCR probes, allowing one to examine the splice sites for many of the transcripts, as well as for current work in developing probes for RNAi. A small example resulting from the assembly that is redolent of the early work on the C. elegans genome providing information on gene structure is the intron size that was found to have a peak at 47 bp [37], and in C. briggsae at 54 bp [38]. For P. davidi, 139 random intronic regions were examined in closer detail across 37 scaffolds showing a clear peak at 49 bp (see File S6).
One of the key reasons for deep sequencing and undertaking a large-scale genomics approach in P davidi, is to aid in isolating key genes involved in cold tolerance. One class of protein that has proven elusive so far, despite a previously reported but inconclusive result [39] are the ice-active proteins (IAP): ice nucleating proteins (INPs), antifreeze proteins (AFPs) and recrystallization-inhibiting proteins (RIPs) [39]. Adhikari et al. [31] has reported a type II antifreeze protein in Plectus murrayi, and it was expected that such a find would also be made in the current P. davidi transcript set. However, neither searches of the annotation or homology matches with the Plectus murrayi EST has identified such a gene, except for a very weak match to transcript PdU008960v1.1, a c-type lectin carbohydrate-binding protein. To date all effort to find even one ice-active protein has failed. In 2009, a newly discovered antifreeze molecule, xylomannan, was isolated from the freeze tolerant Alaskan Beetle, Upis ceramboides [40]. Since this is not a protein, but a combination of saccharide and fatty acid, such an avenue may prove useful with P. davidi, but has yet to be undertaken.
Much is already known of the molecular processes involved in cryoprotective dehydration [10], [11], [41] and Table 1 lists a number of the key genes which have been identified in previous expression profiling studies. These include genes in the trehalose synthesis pathway, the aquaporins, chaperones and oxidoreductase genes associated with cell stress, and desaturase genes involved in membrane fluidity.
Table 1. Associated P. davidi transcripts matching genes significant for cold tolerance.
| Function | C elegans homolog | P davidi contig | E-value |
| Trehalose -6-phosphate synthase | tps-2 | PdU054346v1.1 | 0 |
| tps-2 | PdU054198v1.1 | e-133 | |
| Trehalase | tre1 | PdU000858v1.1 | 0 |
| tre-2 | PdU000867v1.1 | 0 | |
| tre-3 isoform a | PdU002260v1.1 | e-165 | |
| tre-3 isoform b | PdU001362v1.1 | e-165 | |
| Trehalose -6-phosphate phosphatase | gob-1 | PdU001633v1.1 | e-145 |
| Aquaporin | aqp-2 | PdU054401v1.1 | e-067 |
| aqp-3 | PdU008003v1.1 | e-082 | |
| aqp-4 | PdU055737v1.1 | e-059 | |
| aqp-7 | PdU008968v1.1 | e-89 | |
| aqp-7 | PdU011262v1.1 | e-99 | |
| aqp-8 | PdU054717v1.1 | e-076 | |
| Desaturase | fat-2 | PdU004098v1.1 | e-136 |
| fat-7 | PdU004305v1.1 | e-116 | |
| fat-7 | PdU004197v1.1 | e-115 | |
| Superoxide dismutase | sod-1 | PdU057258v1.1 | e-051 |
| Catalase | ctl-1 | PdU002908v1.1 | 0 |
| Peroxyredoxin | prdx-2 | PdU058248v1.1 | e-090 |
| Glutathione peroxidase | gpx-1 | PdU020041v1.1 | e-060 |
| Glutaredoxin | glrx-5 | PdU019570v1.1 | e-040 |
| Glutathione S-transferase | gsto-1 | PdU008618v1.1 | e-037 |
| gst-21 | PdU016688v1.1 | e-030 | |
| gst-1 | PdU015378v1.1 | e-022 |
Trehalose is commonly synthesised from glycogen and has been shown to act as an anhydroprotectant [42] by preserving the functionality of biomolecules and acting as a water replacement in terms of a compatible osmolyte [43], and by glass formation and chemical stability [44]. Two enzymes are directly involved in the synthesis of trehalose: trehalose-6-phosphate synthase (tps) and trehalose 6-phosphate phosphatase (gob), with trehalase (tre) involved in the breakdown of this sugar. Previous work on desiccation has identified a duplication of the tps gene in other species: Megaphorura arctica [11], C. elegans [45] and Brachionus plicatilis [46]. Transcripts with sequence similarity to trehalose-6-phosphate synthase (tps) were identified in the P. davidi dataset, but these appeared to be two non-overlapping portions of the same gene, namely tps-2. However searching the genomic scaffolds indicate that they may come from different regions of the genome, and therefore a potential duplication. TPS-1 was not identified in this dataset. However, in line with the previous studies on duplicated tps genes, potential duplicates of the trehalase gene were also identified in P. davidi. Homologs of the remaining two tre genes, tre-4 and −5 that are present in C. elegans, were not found.
Two membrane function gene families have been identified as being significantly expressed during cryoprotective dehydration, the aquaporins (associated with solute transport across membranes) [47] and the Δ9-acyl-CoA desaturases (involved in changing membrane fluidity via fatty acid composition) (eg [48]). To date 12 aquaporin (aqp) and 7 desaturase (fat) genes have been identified in the genome of C. elegans. In the P. davidi dataset 6 aqp and 3 fat genes were identified. Of particular note was the potential duplication of both aqp-7 and fat-7, which would be specific to P. davidi. Searching the genomic scaffolds, both fat-7 transcripts aligned onto the same scaffold implying different regions of the same gene. However, the two transcripts of aqp-7 aligned to different scaffolds, which indicate a potential duplication. It is worth noting that aqp-7, as well as aqp-3, are the aquaglyceroporin genes, the glycerol-permeable homologs of the classical aquaporins for water transport.
Much of the recent molecular interest in desiccation survival has involved the study of the late embryogenesis abundant (LEA) protein family. These were initially found during the embryogenesis of cottonseed in 1981 [49], [50] and are hydrophilic, intrinsically disordered proteins. They have received a great deal of attention over the last decade or so, since they were found to play a role not only in the desiccation of plants, but also in animals. The verdict is still out in terms of both the classification system that should define the types – the plant types do not so easily translate to the animal types – but also in terms of all the possible functions the LEAs might play [51]. This paper will not attempt to weigh into the debate on the types, as there has been some good work to date focussed on this issue [52], [53], [54], [55]. Owing to the inconclusive designations many researchers prefer to refer to certain LEAs found in animals as LEA-like (i.e. [56]) with all LEA-related proteins found in animals to date most similar to type 3 LEA, with the exception of two type 1 LEA sequences from Artemia franciscana [57]. More work also needs to be done on differentiating the functional differences of the separate types before too much is made of the syntax, even though it is likely that any syntactic differences will be reflected functionally. As Tunnacliffe and Wise [51] have phrased it, the LEAs remain a conundrum.
Among the many functional properties attributed to the LEA (and LEA-like) proteins are as a molecular shield inhibiting aggregation of denaturing proteins, as an antioxidant, to provide protection to membranes preventing damaging phase transitions during freezing, as hydration buffers, slowing water loss - among others (see [51], [58], [56]). In nematodes trehalose is an important constituent of desiccation survival [59], unlike in tardigrades and bdelloid rotifers where trehalose is not accumulated, or even present during desiccation [60], [61], [62]. Yet Gal et al. [63] found that silencing the C. elegans lea-1 gene significantly reduced survival during induction of desiccation as well as of osmotic and heat stress. However, the more remarkable of recent studies is one conducted on human hepatoma cell lines in which two type 3 LEA proteins from Artemia fransiscana were transfected, with 98% of cells retaining membrane integrity after rehydration from low water content, compared to 0% without transfection [64]. When the same was attempted without the trehalose and only one of the transfected LEAs, 94% of the cells retained membrane integrity. Within the current P. davidi dataset, both the contigs and the EST reads were searched for potential LEAs with at least 26 individual reads or contigs found. These were checked for the ability to compose an amphiphilic α-motif [65], [51], and whether they were natively unfolded using Foldit [66]. These ESTs and contigs were then clustered using Clustal [67] (see Figure 3) as well as checked for homology to the genomic scaffolds (see Table 2). The colouring scheme found in Figure 3 depict those sequences found on the same scaffold (in Table 2). As can be seen, the colouring matches perfectly the independent clustering based on Clustal which provides evidence that there are possibly up to 9 different LEA-type genes. However, translation to amino acid, combined with Cd-hit [68] at a relatively low stringency threshold of 0.8, resulted in 13 separate clusters. File S7 contains the cd-hit clustering and the resulting representative sequences.
Figure 3. UPGMA clustering of the LEA transcripts and ESTs from Table 2 .
The colouring represents those transcripts and ESTs that aligned on the same scaffolds, as shown in Table 2. The close clustering and independent scaffold alignment provides evidence of distinct LEA genes.
Table 2. P. davidi LEA-like sequences.
| Transcript/EST | Length (aa) | Unfoldability* | Charge* | Phobic* | Genomic Scaffold‡ |
| PdU001599v1.1 | 339 | −0.078 | 0.006 | 0.388 | n/a |
| PDT20F06 | 594 | −0.023 | 0.015 | 0.41 | n/a |
| PDF06F09 | 288 | −0.189 | 0.031 | 0.356 | Scaffold19015 |
| PDF82L15 | 201 | −0.18 | 0.06 | 0.37 | Scaffold19015 |
| PdU055885v1.1 | 307 | −0.146 | 0.023 | 0.369 | Scaffold19015 |
| PDF40F19 | 317 | −0.189 | 0.028 | 0.356 | Scaffold19015 |
| PDF16M17 | 306 | −0.194 | 0.029 | 0.354 | Scaffold19015 |
| PDF96E03 | 209 | −0.225 | 0.115 | 0.374 | Scaffold19015 |
| PDT56F11 | 172 | −0.128 | 0.029 | 0.378 | Scaffold19015 |
| PdU055084v1.1 | 235 | −0.203 | 0.047 | 0.357 | Scaffold51708 |
| PDT10D20 | 235 | −0.206 | 0.047 | 0.356 | Scaffold51708 |
| PdU013001v1.1 | 169 | −0.109 | 0.065 | 0.398 | Scaffold10457 |
| PDF39E07 | 90 | −0.076 | 0.022 | 0.394 | Scaffold18176 |
| PDT45G14 | 98 | −0.084 | 0.041 | 0.398 | Scaffold18176 |
| PdU014555v1.1 | 150 | −0.049 | 0.033 | 0.408 | Scaffold1422 |
| PdU002998v1.1 | 502 | −0.181 | 0.008 | 0.351 | Scaffold12112 |
| PDT19L04 | 1002 | −0.151 | 0.009 | 0.362 | Scaffold12112 |
| PdU017016v1.1 | 200 | −0.179 | 0 | 0.349 | Scaffold12112 |
| PdU053999v1.1 | 593 | −0.148 | 0.007 | 0.363 | Scaffold12112 |
| PdU010487v1.1 | 272 | −0.171 | 0.011 | 0.356 | Scaffold12112 |
| PDT30I10 | 593 | −0.129 | 0.002 | 0.368 | Scaffold12112 |
| PDT06A14 | 320 | −0.06 | 0.034 | 0.404 | Scaffold38706 |
| PdU017506v1.1 | 119 | −0.175 | 0.05 | 0.369 | Scaffold38706 |
| PDF80K07 | 282 | −0.049 | 0.032 | 0.407 | Scaffold38706 |
| PdU015612v1.1 | 234 | −0.076 | 0.017 | 0.392 | Scaffold38706 |
| PDF77N04 | 78 | −0.359 | 0.103 | 0.321 | Scaffold59957 |
Although too many to be included in Table 1, another important class of temperature stress proteins involved in protein stability are the numerous heat shock proteins. C. elegans has 12 types of HSP-70 genes, with two pairs (F44E5.4/F44E5.5 and HSP-3/4) almost identical, suggesting they have been raised by gene duplication. In P. davidi, analysis to date has indicated 7 HSP-70 homologs, with 20 HSP-70-like genes. The P. davidi sequence provides homologs of HSP-1, HSP-3/4, HSP-6, HSP-70, HSP-110, F44E5.4/.5, and T14G8.3, with HSP-1, HSP-3/4 and HSP-6 indicating the presence of orthologs. File S8 provides the P. davidi sequences of the different groups.
For the cold tolerant process of intracellular freezing, very little is as yet known of the mechanisms that allow this to occur, with no molecular work done on any organism to date. However it would be surprising if many of the above mentioned genes were not in some way involved, either mechanistically, or in terms of stress response.
From the beginning of the molecular focus on P. davidi, a vital question has been whether it is susceptible to environmental RNA interference. If so, it would provide a method of functionally investigating survival of intracellular freezing. We have provided an in silico search for RNAi specific genes (following [69]). As with the IAP, an inability to find a key gene does not preclude there being one. However, so far there has been no in silico evidence of sid-2, even though a number of other associated genes are present (see Table 3). As pointed out in the introduction, without sid-2, even if other associated RNAi genes were present, it is considered unlikely that P. davidi would have an environmental response to RNAi [17]. The results could potentially mirror the relationship between C. elegans and C. briggsae, since Panagrolaimus superbus has been shown to have an RNAi response [16]. Work in determining whether there is an environmental response is being done in Otago, but so far the results have been inconclusive (A. Seybold, per. comm.). If the information hinted at in the in silico search is correct, it would be disappointing, since any lack of response would imply similar difficulties with the soaking method [70]. While microinjection is still a possibility in providing an RNAi response [14], it is unlikely to be of help in understanding an environmental response where large numbers of nematodes are needed to provide a statistical indication of survival.
Table 3. An in silico search of the P. davidi transcriptome for the genes associated with RNAi (following [69]).
| Function | C elegans homologs | P davidi contigs | E-value* |
| Small RNA biosynthetic proteins | drh-3 | PdU000268v1.1 | e-83 |
| drsh-1 | PdU006095v1.1 | e-73 | |
| xpo-1 | PdU054092v1.1 | 0 | |
| xpo-2 | PdU003765v1.1 | e-31 | |
| dcr-1 | PdU000401v1.1 | e-139 | |
| drh-1 | PdU000369v1.1 | e-63 | |
| pash-1 | PdU009300v1.1 | e-16 | |
| rde-4 | PdU000474v1.1 | e-47 | |
| xpo-3 | - | ||
| dsRNA uptake and spreading,and siRNA amplification effectors | smg-2 | PdU000228v1.1 | 0 |
| smg-6 | - | ||
| ego-1 | PdU000148v1.1 | e-177 | |
| rrf-3 | PdU000148v1.1 | e-122 | |
| rrf-1 | PdU000148v1.1 | e-175 | |
| smg-5 | - | ||
| rsd-2 | - | ||
| rsd-3 | PdU001889v1.1 | e-39 | |
| sid-1 | - | ||
| rsd-6 | - | ||
| sid-2 | - | ||
| Argonautes (AGOs) andRNA-induced SilencingComplex (RISC) components | alg-1 | PdU000474v1.1 | 0 |
| R06C7.1 (wago-1) | PdU000507v1.1 | e-102 | |
| C04f12.1 | PdU000451v1.1 | e-52 | |
| F58G1.1 (wago-4) | PdU000507v1.1 | e-105 | |
| alg-4 | PdU000474v1.1 | e-135 | |
| rde-1 | PdU000474v1.1 | e-47 | |
| C16C10.3 (wago-9) | PdU055266v1.1 | e-54 | |
| ppw-1 | PdU000507v1.1 | e-53 | |
| csr-1 | PdU000451v1.1 | e-50 | |
| ppw-2 | PdU000507v1.1 | e-97 | |
| sago-1 | PdU000453v1.1 | e-39 | |
| T22B3.2 | PdU000474v1.1 | e-128 | |
| T22H9.3 (wago-10) | - | ||
| alg-2 | PdU000474v1.1 | 0 | |
| ergo-1 | PdU000474v1.1 | e-49 | |
| prg-1 | PdU000474v1.1 | e-32 | |
| F55A12.1 (wago-2) | PdU000453v1.1 | e-72 | |
| T23D8.7 (hpo-24) | PdU000474v1.1 | e-170 | |
| nrde-3 | PdU055266v1.1 | e-38 | |
| sago-2 | PdU000507v1.1 | e-53 | |
| T23B3.2 | - | ||
| Y49F6A.1 (wago-11) | PdU055266v1.1 | e-49 | |
| ZK1248.7 (wago-5) | PdU000507v1.1 | e-107 | |
| prg-2 | PdU000474v1.1 | e-29 | |
| C06A1.4 | PdU000507v1.1 | e-88 | |
| C14B1.7 | PdU055266v1.1 | e-46 | |
| tsn-1 | PdU002202v1.1 | e-131 | |
| ain-1 | - | ||
| vig-1 | PdU004590v1.1 | e-13 | |
| ain-2 | - | ||
| RNAi inhibitors | eri-1 | PdU006594v1.1 | e-29 |
| xrn-2 | PdU056342v1.1 | e-129 | |
| adr-2 | PdU002126v1.1 | e-24 | |
| xrn-1 | PdU056342v1.1 | e-64 | |
| adr-1 | - | ||
| lin-15b | - | ||
| eri-5 | - | ||
| eri-6/7 | PdU001476v1.1 | e-45 | |
| eri-3 | - | ||
| Nuclear RNAi effectors | mut-7 | PdU057227v1.1 | e-15 |
| cid-1 | PdU001764v1.1 | e-26 | |
| ekl-1 | PdU010210v1.1 | e-12 | |
| gfl-1 | PdU010550v1.1 | e-32 | |
| mes-2 | PdU000971v1.1 | e-57 | |
| rha-1 | PdU053907v1.1 | e-148 | |
| ekl-6 | PdU000462v1.1 | e-27 | |
| zfp-1 | PdU000768v1.1 | e-27 | |
| mut-2 | PdU001764v1.1 | e-22 | |
| ekl-5 | - | ||
| mes-3 | - | ||
| mut-16 | - | ||
| rde-2 | - |
*Homologs not found at the e-value cutoff of 1e-10 are shown as absent with a hyphen.
With this, the first large scale molecular work done to date on P. davidi, we now have the information to begin exploring the physiological adaptations of this extraordinary nematode in greater depth.
Supporting Information
The resulting number of contigs of certain lengths resulting from the choice of different kmer sizes on the Illumina data for the transcriptome by Soapdenovo.
(PDF)
A listing of the transcriptome transcripts constructed from the EST and Illumina data with their corresponding match in the nr database at an e-value of 1e-10 or below. Only those transcripts with a match are represented.
(TXT)
SEED subsystem analysis of the two EST libraries PDT (20°C) and PDF (4°C). The colouring scheme for the legend is read in a counterclockwise manner.
(PDF)
Distribution of the genomic scaffold sizes above the N50 value.
(PDF)
Some evidence of heterozygosity in P. davidi . Pg 1 are the amino acid sequences of a LEA subfamily. Blue: silent substitution, Red: amino acid substitution, Gray italic: missing in LEA1.1 and LEA1.3. Consensus sequences for LEA1 subfamily and general LEA proteins are indicated under the amino acid sequences (shown in blue and magenta, respectively). Pg 2 are the cDNA sequences of the same LEA1 subfamily: Red: single nucleotide variation (SNV) sites. Pg 3 is the trace of the LEA1 gene locus, amplified by PCR from a single worm and directly sequenced from the DNA. The SNV sites found by cDNA analysis (indicated by asterisks and arrow heads) appear as double bands, confirming these sites. Pg 4 are the LEA1 genomic sequences, corresponding to the cDNA sequences. Pg 5 and 6 are cDNA sequences of two other genes, gln-5 and T25B9.9 homologs in P. davidi, which also show SNV patterns (the SNV sites are shown in Black/Blue – except for position 628 in the T25B9.9 homolog, which is a sequencing error). Similar SNV patterns have been observed in many other genes including other LEAs, 18s RNA, 28s RNA, and members of HSP-70.
(PDF)
Intron size distribution in P. davidi . The peak around 49 is similar to the intron peak found in C. elegans.
(PDF)
Cd-hit clustering of the 26 LEA transcripts and the resulting 13 represented amino acid sequences.
(PDF)
Sequences of the HSP-70 and HSP-70-like genes found in P. davidi.
(PDF)
Acknowledgments
The authors would like to thank Yuji Kohara, Hironori Niki, Yohei Minakuchi and Tadasu Shin-i at the National Institute of Genetics in Japan for the EST sequencing and support, and the University of Otago Biochemistry Department and the Beijing Genomics Institute in Shenzhen for conducting the Illumina sequencing. We would also like to thank Karen Judge at the University of Otago, and Jeremy Robst at the British Antarctic Survey for technical support. The authors would further like to thank two anonymous reviewers for comments on the manuscript, and Mark Blaxter for some enlightening comments on assembly issues that unfortunately came too late to be adopted.
Funding Statement
MAST and MSC were supported by NERC core funding to the British Antarctic Survey. HK was supported by grant-in-aid for scientific research (No. 23510239) from the Japan Society for the Promotion of Science. Further support came from the Transdisciplinary Research Integration Center (TRIC), the Research Organisation of Information and Systems (ROIS), and the Departments of Zoology and Biochemistry at the University of Otago. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Timm RW (1971) Antarctic Soil and Freshwater Nematodes from the McMurdo Sound Region. Proc Helm Soc Wash 38: 42–52. [Google Scholar]
- 2. Wharton DA, Brown IM (1989) A survey of the terrestrial nematodes from the McMurdo Sound region, Antarctica. New Zeal J Zooloogy 16: 467–470. [Google Scholar]
- 3. Wharton DA, Ferns DJ (1995) Survival of intracellular freezing by the Antarctic nematode Panagrolaimus davidi. . J Exp Biol 198: 1381–1387. [DOI] [PubMed] [Google Scholar]
- 4.Wharton DA (2011) Cold tolerance. In: Perry RN, Wharton DA, editors. Molecular and Physiological Basis of Nematode Survival. Wallingford: Centre for Agriculture and Biosciences International. 182–204. [Google Scholar]
- 5. Salt RW (1957) Natural occurrence of glycerol in insects and its relation to their ability to survive freezing. Can Entomol 89: 491–494. [Google Scholar]
- 6. Salt RW (1959) Survival of frozen fat body cells in an Insect. Nature 184: 1426.14402719 [Google Scholar]
- 7. Salt RW (1962) Intracellular freezing in insects. Nature 193: 1207–1208. [Google Scholar]
- 8. Wharton DA, Goodall G, Marshall CJ (2003) Freezing survival and cryoprotective dehydration as cold tolerance mechanisms in the Antarctic nematode Panagrolaimus davidi. . J Exp Biol 206: 215–221. [DOI] [PubMed] [Google Scholar]
- 9. Wharton DA, Downes MF, Goodall G, Marshall CJ (2005) Freezing and cryoprotective dehydration in an Antarctic nematode (Panagrolaimus davidi) visualised using a freeze substitution technique. Cryobiology 50: 21–28. [DOI] [PubMed] [Google Scholar]
- 10. Clark MS, Thorne MAS, Purać J, Grubor-Lajšić G, Kube M, et al. (2007) Surviving extreme polar winters by desiccation: clues from Arctic springtail (Onychiurus arcticus) EST libraries. BMC Genomics 8: 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark MS, Thorne MAS, Purać J, Burns G, Hillyard G, et al. (2009) Surviving the cold: molecular analyses of insect cryoprotective dehydration in the Arctic springtail Megaphorura arctica (Tullberg). BMC Genomics 10: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thorne MAS, Worland M, Feret R, Deery M, Lilley K, et al. (2011) Proteomics of cryoprotective dehydration in Megaphorura arctica Tullberg 1876 (Onychiuridae: Collembola). Insect Mol Biol 20: 303–310. [DOI] [PubMed] [Google Scholar]
- 13. Goldstein P, Wharton DA (1996) The synaptonemal complexes of the meiotic parthenogenetic Antarctic nematode Panagrolaimus davidi: Karyotype analysis and three-dimensional reconstruction of pachytene nuclei. Cytobios 85: 81–90. [PubMed] [Google Scholar]
- 14. Timmons L, Fire A (1998) Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- 15. Boutros M, Ahringer J (2008) The art and design of genetic screens: RNA interference. Nat Rev Genet 9: 554. [DOI] [PubMed] [Google Scholar]
- 16. Shannon AJ, Tyson T, Dix I, Boyd J, Burnell AM (2008) Systemic RNAi mediated gene silencing in the anhydrobiotic nematode Panagrolaimus superbus . BMC Mol Biol 9: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP (2007) Caenorhabditis elegans sid-2 is required for environmental RNA interference. Proc Natl Acad Sci U S A 104: 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lewis SC, Dyal LA, Hilburn CF, Weitz S, Liau W-S, et al. (2009) Molecular evolution in Panagrolaimus nematodes: origins of parthenogenesis, hermaphroditism and the Antarctic species P. davidi. . BMC Evol Biol 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raymond MR, Wharton DA, Marshall CJ (2013) Nematodes from the Victoria Land coast, Antarctica and comparisons with cultured Panagrolaimus davidi . Antarct Sci doi:10.1017/S0954102013000230 [Google Scholar]
- 20. Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SeqClean download website. Available: https://sourceforge.net/projects/seqclean/. Accessed Feb 15 2014.
- 22.Phil Green’s Phred/Phrap/Consed website. Available: http://www.phrap.org/phredphrapconsed.html. Accessed Feb 15 2014.
- 23. Raymond MR, Wharton DA (2013) The ability of the Antarctic nematode Panagrolaimus davidi to survive intracellular freezing is dependent upon nutritional status. J Comp Physiol B 183: 181–188. [DOI] [PubMed] [Google Scholar]
- 24.Hooper DJ (1986) Extraction of free-living stages from soil. In: Southey JF, editor. Laboratory Methods for Work with Plant and Soil Nematodes. London: HMSO. 5–30. [Google Scholar]
- 25.Short Oligonucleotide Analysis Package website. Available: soap.genomics.org.cn. Accessed Feb 15 2014.
- 26.454 Life Sciences website. Available: www.my454.com. Accessed Feb 15 2014.
- 27. Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 29.Genbank Blast database ftp site. Available: http://ftp.ncbi.nlm.nih.gov/blast/db/. Accessed Feb 15 2014.
- 30.Wormbase website. Available: www.wormbase.org. Accessed Feb 15 2014.
- 31. Adhikari BN, Wall DH, Adams BJ (2009) Desiccation survival in an Antarctic nematode: Molecular analysis using expressed sequenced tags. BMC Genomics 10: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tyson T, Zamora G, Wong S, Skelton M, Daly B, et al. (2012) A molecular analysis of desiccation tolerance mechanisms in the anhydrobiotic nematode Panagrolaimus superbus using expressed sequenced tags. BMC Research Notes 5: 68–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278: 631–637. [DOI] [PubMed] [Google Scholar]
- 34. Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Overbeek R, Begley T, Butler R, Choudhuri J, Chuang H-Y, et al. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Research 33: 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamamatsu website. Available: www.hamamatsu.com. Accessed Feb 15 2014.
- 37.Spieth J, Lawson D (2006) Overview of gene structure. In: The C. elegans Research Community, editors. Wormbook www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 38. Stein L, Bao Z, Blasiar D, Blumenthal T, Brent M, et al. (2003) The Genome Sequence of Caenorhabditis briggsae: A Platform for Comparative Genomics. PLoS Biol 1: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wharton DA, Barrett J, Goodall G, Marshall CJ, Ramløv H (2005) Ice-active proteins from the Antarctic nematode Panagrolaimus davidi . Cryobiology 51: 198–207. [DOI] [PubMed] [Google Scholar]
- 40. Walters KR Jr, Serianni AS, Sformo T, Barnes BM, Duman JG (2009) A nonprotein thermal hysteresis-producing xylomannan antifreeze in the freeze-tolerant Alaskan beetle Upis ceramboides . Proc Natl Acad Sci U S A 106: 20210–20215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bahrndorff S, Tunnacliffe A, Wise M, McGee B, Holmstrup M, et al. (2009) Bioinformatics and protein expression analyses implicate LEA proteins in the drought response of Collembola. J Insect Physiol 55: 210–217. [DOI] [PubMed] [Google Scholar]
- 42. Ring RA, Danks HV (1998) The role of trehalose in cold-hardiness and desiccation. Cryo-Lett 19: 275–282. [Google Scholar]
- 43. Yancey PH (2005) Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol 208: 2819–2830. [DOI] [PubMed] [Google Scholar]
- 44. Crowe JH, Crowe LM, Chapman D (1984) Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science 223: 701–703. [DOI] [PubMed] [Google Scholar]
- 45. Pellerone F, Archer S, Behm C, Grant WN, Lacey MJ, et al. (2003) Trehalose metabolism genes in Caenorhabditis elegans and filarial nematodes. Int J Parasitol 33: 1195–1206. [DOI] [PubMed] [Google Scholar]
- 46. Denekamp NY, Thorne MAS, Clark MS, Kube M, Reinhardt R, et al. (2009) Discovering genes associated with dormancy in the monogonont rotifer Brachionus plicatilis . BMC Genomics 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kruse E, Uehlein N, Kaldenhoff R (2006) The aquaporins. Genome Biol 7: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kayukawa T, Chen B, Hoshizaki S, Ishikawa Y (2007) Upregulation of a desaturase is associated with the enhancement of cold hardiness in the onion maggot, Delia antiqua. . Insect Biochem Mol Biol 37: 1160–1167. [DOI] [PubMed] [Google Scholar]
- 49. Dure III L, Greenway SC, Galau GA (1981) Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 20: 4162–4168. [DOI] [PubMed] [Google Scholar]
- 50. Galau G, Dure III L (1981) Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by reciprocal heterologous complementary deoxyribonucleic acid-messenger ribonucleic acid hybridization. Biochemistry 20: 4169–4178. [DOI] [PubMed] [Google Scholar]
- 51. Tunnacliffe A, Wise M (2007) The continuing conundrum of the LEA proteins. Naturwissenschaften 94: 791–812. [DOI] [PubMed] [Google Scholar]
- 52. Wise M (2002) The POPPs: clustering and searching using peptide probability profiles. Bioinformatics Supp. 1: S38-S45. [DOI] [PubMed] [Google Scholar]
- 53. Wise M (2003) LEAping to conclusions: A computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hunault G, Jaspard E (2010) LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genomics 11: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jaspard E, Macherel D, Hunault G (2012) Computational and Statistical Analysis of Amino acid usage and physico-chemical properties of the twelve late embryogenesis abundant protein classes. PLoS ONE 7: e36968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hand S, Menze M, Toner M, Boswell L, Moore D (2011) LEA proteins during water stress: Not just for plants anymore. Annu Rev Physiol 73: 115–134. [DOI] [PubMed] [Google Scholar]
- 57. Sharon M, Kozarova A, Clegg J, Vacratsis P, Warner A (2009) Characterization of a group 1 late embryogenesis abundant protein in encysted embryos of the brine shrimp Artemia fransiscana. . Biochem Cell Biol 87: 415–430. [DOI] [PubMed] [Google Scholar]
- 58. Chakrabortee S, Tripathi R, Watson M, Schierle G, Kurniawan D, et al. (2012) Intrinsically disordered proteins as molecular shields. Mol Biosyst 8: 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Madin K, Crowe J (1975) Anhydrobiosis in nematodes: carbohydrate and lipid metabolism during dehydration. J Exp Zool 193: 335–342. [Google Scholar]
- 60. Tunnacliffe A, Lapinski J (2003) Resurrecting Van Leeuwenhoek’s rotifers: a reappraisal of the role of disaccharidesin anhydrobiosis. Philos T Roy Soc B 358: 1755–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tunnacliffe A, Lapinski J, McGee B (2005) A putative LEA protein, but no trehalose, is present in anhydrobiotic bdelloid rotifers. Hydrobiologia 546: 315–321. [Google Scholar]
- 62. Hengherr S, Heyer A, Kohler H-R, Schill R (2008) Trehalose and anhydrobiosis in tardigrades: evidence for divergence in response to dehydration. FEBS 275: 281–288. [DOI] [PubMed] [Google Scholar]
- 63. Gal TZ, Glazer I, Koltai H (2004) An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett 577: 21–26. [DOI] [PubMed] [Google Scholar]
- 64. Li S, Chakraborty N, Borcar A, Menze MA, Toner M, et al. (2012) Late embryogenesis abundant proteins protect human hepatoma cells during acute desiccation. Proc Natl Acad Sci U S A 109: 20859–20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA (2001) Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta 1544: 196–206. [DOI] [PubMed] [Google Scholar]
- 66. Prilusky J, Felder C, Zeev-Ben-Mordehai T, Rydberg E, Man O, et al. (2005) FoldIndex©: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21: 3435–3438. [DOI] [PubMed] [Google Scholar]
- 67. Higgins DG, Sharp PM (1988) CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene 73: 237–244. [DOI] [PubMed] [Google Scholar]
- 68. Weizhong L, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 69. Dalzell JJ, McVeigh P, Warnock ND, Mitreva M, Mck Bird D, et al. (2011) RNAi effector diversity in nematodes. PLoS Neglect Trop D 5: e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tabara H, Grishok A, Mello CC (1998) RNAi in C. elegans: soaking in the genome sequence. Science 282: 430–431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The resulting number of contigs of certain lengths resulting from the choice of different kmer sizes on the Illumina data for the transcriptome by Soapdenovo.
(PDF)
A listing of the transcriptome transcripts constructed from the EST and Illumina data with their corresponding match in the nr database at an e-value of 1e-10 or below. Only those transcripts with a match are represented.
(TXT)
SEED subsystem analysis of the two EST libraries PDT (20°C) and PDF (4°C). The colouring scheme for the legend is read in a counterclockwise manner.
(PDF)
Distribution of the genomic scaffold sizes above the N50 value.
(PDF)
Some evidence of heterozygosity in P. davidi . Pg 1 are the amino acid sequences of a LEA subfamily. Blue: silent substitution, Red: amino acid substitution, Gray italic: missing in LEA1.1 and LEA1.3. Consensus sequences for LEA1 subfamily and general LEA proteins are indicated under the amino acid sequences (shown in blue and magenta, respectively). Pg 2 are the cDNA sequences of the same LEA1 subfamily: Red: single nucleotide variation (SNV) sites. Pg 3 is the trace of the LEA1 gene locus, amplified by PCR from a single worm and directly sequenced from the DNA. The SNV sites found by cDNA analysis (indicated by asterisks and arrow heads) appear as double bands, confirming these sites. Pg 4 are the LEA1 genomic sequences, corresponding to the cDNA sequences. Pg 5 and 6 are cDNA sequences of two other genes, gln-5 and T25B9.9 homologs in P. davidi, which also show SNV patterns (the SNV sites are shown in Black/Blue – except for position 628 in the T25B9.9 homolog, which is a sequencing error). Similar SNV patterns have been observed in many other genes including other LEAs, 18s RNA, 28s RNA, and members of HSP-70.
(PDF)
Intron size distribution in P. davidi . The peak around 49 is similar to the intron peak found in C. elegans.
(PDF)
Cd-hit clustering of the 26 LEA transcripts and the resulting 13 represented amino acid sequences.
(PDF)
Sequences of the HSP-70 and HSP-70-like genes found in P. davidi.
(PDF)