Abstract
Purpose of review
Genomic imprinting refers to preferential allele-specific gene expression. DNA methylation-based molecular mechanisms regulate establishment and maintenance of parental imprints during early embryo development and gametogenesis. Because of the coincident timing, a potential association between assisted reproductive technology (ART) procedures and imprinting defects has been investigated in various studies. In this review, we provide an overview of genomic imprinting and present a summary of the relevant clinical data.
Recent findings
ART procedures affect DNA methylation pattern, parental imprinting status, and imprinted gene expression in the mouse embryo. In humans, several case series suggested an association between ART and imprinting disorders, with a three-fold to six-fold higher prevalence of ART use among children born with Beckwith–Wiedemann syndrome compared to the general population. However, more recent studies failed to support these findings and could not demonstrate an association between imprinting disorders and ARTs, independent of subfertility.
Summary
ART procedures may affect methylation status of imprinted regions in the DNA, leading to imprinting disorders. Although the low prevalence of imprinting disorders makes it challenging to perform conclusive clinical trials, further studies in large registries are required to determine the real impact of ARTs on their occurrence.
Keywords: assisted reproductive technologies, genomic imprinting, imprinting disorders
INTRODUCTION
Mammals are biparental diploid organisms that normally have two copies of each autosomal chromosome; one from the mother (maternally derived copy) and one from the father (paternally derived copy). Mammalian genes were initially presumed to be expressed equally from both parental alleles consistent with the fundamental rule of classical Mendelian genetics. This genetic equality assumption was challenged in the mid-1980s using nuclear transplantation and uniparental disomy (UPD) experiments. Initially, haploid parthenogenetic mouse eggs were used as recipients for either a male or a female pronucleus taken from fertilized eggs; eggs receiving a male pronucleus developed to term but those with two female pronuclei developed only poorly after implantation [1] (see Table 1 for definitions of key terms used in this review). A similar study simultaneously showed that diploid mouse embryos with either two female pronuclei (biparental gynogenones) or two male pronuclei (biparental androgenones) were not able to complete normal embryogenesis [2]. Systematic breeding of mice with UPD for individual chromosomes demonstrated that mice with UPD for most chromosomes survive normally; however, UPD for certain chromosomes or chromosomal regions result in anomalous phenotypes suggesting that the parental effect is not associated with the entire genome but is restricted to specific genomic loci [3]. These investigations uncovered the requirement for both maternal and paternal genomes for normal development and were tied to an intriguing biological phenomenon called genomic imprinting.
Table 1.
Brief definitions of biological concepts referred in this review
| Concept | Definition |
|---|---|
| Parthenogenesis | A kind of asexual reproduction in which an egg is developed into an embryo without fertilization. Some species switch between sexual reproduction and parthenogenesis whereas some others reproduce only by parthenogenesis. Artificial parthenogenesis has been achieved by stimulating the egg with the help of various mechanical and chemical agents; usually resulting in incomplete and abnormal development |
| Gynogenone | Diploid embryos with two maternal genomes developed by either transfer of a female pronuclei into the haploid parthenogenetic egg or by replacement of a male pronucleus in the fertilized egg with a female pronucleus from another fertilized egg |
| Androgenone | Diploid embryos with two paternal genomes derived by replacement of the female pronucleus in the fertilized egg with a male pronucleus from another fertilized egg |
| Epigenetic regulation | Heritable changes in gene expression that are controlled by factors other than the DNA sequence, such as DNA methylation and histone acetylation. Epigenetic changes can switch on and off resulting in alternative expression patterns |
| DNA methylation | The biological process in which a methyl group (CH3) is added to a DNA nucleotide. DNA methylation regulates gene expression by turning genes on and off |
| Genomic imprinting | Allele-specific gene expression controlled by parent-of-origin dependent differential methylation of DNA |
| Maternal imprinting | Maternally imprinted genes are methylated and silenced on the maternal copy and expressed from the paternal copy of the chromosome which is inherited from the father |
| Paternal imprinting | Paternally imprinted genes are methylated and silenced on the paternal copy and expressed from the maternal copy of the chromosome which is inherited from the mother |
Imprinting is the epigenetic labeling of certain genes as of paternal or maternal origin, resulting in differential expression depending on whether they are inherited from the mother or the father [4]. Genomic imprinting is required for normal development, and disrupted imprinting is associated with significant pathologies including Angelman syndrome, Prader–Willi syndrome (PWS), and Beckwith–Wiedemann syndrome (BWS) [5–7]. Within the last decade, several studies have raised concerns that assisted reproductive technologies (ARTs) may result in abnormal genomic imprinting, leading to an increased frequency of imprinting-related disorders in children born as a result of infertility treatment (reviewed in [8,9,10■,11]).
In this review, we first present an overview of the molecular mechanisms of genomic imprinting and regulation of imprinting during gametogenesis and early embryo development. Then, we provide a summary of the recent studies investigating the effect of ART procedures on imprinting-related disorders.
MOLECULAR MECHANISM OF GENOMIC IMPRINTING
Investigation of the molecular mechanism responsible for establishment and regulation of genomic imprints have demonstrated that DNA methylation is the main epigenetic modification that distinctly labels maternally and paternally inherited copies of imprinted genomic loci [12,13].
DNA methylation and the role of DNA methyltransferases
DNA methylation refers to the process of addition of a methyl (–CH3) group on cytosines at 5′-cytosineguanine-3′ dinucleotides (CpG) converting DNA base cytosine to 5-methylcytosine (5mC). Specific methylation patterns are observed on guanine-cytosine-rich regions of the genome, called CpG islands, which exhibit high frequencies of CpG sites. Methylation of CpG islands constitutes a key mechanism of epigenetic regulation and plays a central role in the differentiation from embryonic stem cells into specific cells and tissues. Certain CpG islands constitute differentially methylated regions (DMRs) that undergo DNA methylation on only one allele (maternal or paternal). This kind of differential methylation is heritable and results in genomic imprinting.
Genome-wide methylation pattern is erased and re-established every generation during gameto-genesis and early embryo development. Once established, the pattern of methylated and unmethylated CpGs on the genome tends to be copied through cell divisions to reproduce methylation patterns. This process is called maintenance methylation. The members of the DNA methyltransferase (Dnmt) family, DNMT1, DNMT3A/B, and DNMT3L are the major components of the complex molecular mechanism for acquisition and propagation of methylation across cell divisions. DNMT1 is demonstrated to be the primary maintenance methyltransferase, since targeted mutation of Dnmt1 in mice results in genome-wide DNA demethylation and embryonic lethality [14].
Establishment of DNA methylation, on the other hand, requires de-novo methyltransferases DNMT3A and DNMT3B for epigenetic reprogramming in the embryo and for imprint acquisition in the gametes. Experimental evidence shows that inactivation of both genes blocks de-novo methylation in embryonic stem cells and early embryos, but does not disrupt maintenance of imprinted methylation patterns [15]. Both DNMT3A and DNMT3B are also actively expressed in male and female germ lines [16]. DNMT3L cooperates with DNMT3A and DNMT3B to stimulate their methyltransferase activities for de-novo methylation of maternally imprinted genes in oocytes [17]. Targeted disruption of Dnmt3L does not affect genome-wide methylation levels, but prevents methylation of maternally imprinted sequences resulting in sterility in males and maternal lethality in females [18].
Regulation of imprinting during early embryo development and gametogenesis
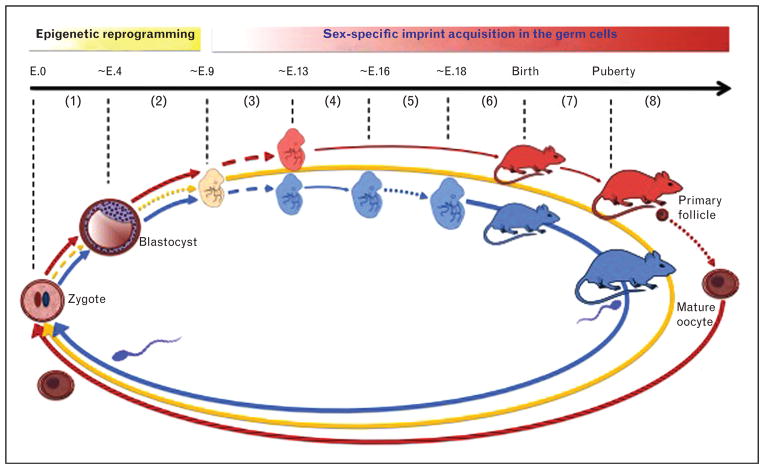
DNA methylation related to genomic imprinting and epigenetic reprogramming is regulated by two major waves of genome-wide demethylation and remethylation: first, biparental genetic totipotency (i.e. cell differentiation) is epigenetically established following fertilization, and second, biparental methylation pattern in the DMRs is eliminated and imprinted methylation is re-established in the germ line for the next generation (Fig. 1).
FIGURE 1.
Regulation of paternally imprinted (blue), maternally imprinted (red), and nonimprinted (yellow) DNA methylation during early embryo development and gametogenesis. Dashed arrows, dotted arrows, and thin arrows indicate demethylation, de-novo methylation, and unmethylated status, respectively. (1) Sex-specific imprints are established in the mature oocyte and in the sperm. Shortly after fertilization, genome-wide demethylation occurs at nonimprinted genomic loci. (2) De-novo methylation at nonimprinted regions on DNA. This epigenetic reprogramming enables cell differentiation by turning different genes on and off in different cells. After that stage, maternal and paternal imprints and nonimprinted methylation are maintained in somatic cells of the embryo throughout life (not shown in the figure). (3) Another wave of demethylation initiates at the imprinted DMRs in primordial germ cells of the embryo. (4) Biparental imprints are erased in the germ cells. (5) Paternal imprint acquisition starts in the spermatogonia of the male fetus, whereas DMRs remain unmethylated in the oocytes of the female fetus. (6) Paternal imprint acquisition is completed in the germ cells of the male fetus and maintained throughout life. (7) Imprinted DMRs remain unmethylated in the primordial oocytes in female embryo. (8) After puberty, maternal imprints are established during oocyte growth.
Imprinted genes are maternally marked in the mature oocyte and paternally marked in the sperm. Shortly after fertilization, before the first cell division, the paternally derived genome undergoes active demethylation by dioxygenase TET3-mediated oxidation of 5mC, changing 5mC into 5-hydroxymethylcytosine [19–21]. In contrast, the maternally derived genome remains methylated during the first DNA replication cycle, but initiates a passive, replication-dependent demethylation process in which 5mC levels gradually decrease through successive cell divisions until the blastocyst stage [19,20]. Genome-wide de-novo methylation occurs around implantation [22]. This epigenetic reprogramming is required for erasure of the inherited epigenetic features and to enable totipotency of the newly formed embryo.
Imprinted DMRs are not affected at this first wave of genome-wide DNA demethylation, and parental imprints are maintained in the somatic tissues of the embryo throughout life. DMRs preserve methylation in the presence of DNMT1 and DNMT1o during preimplantation embryo development [23,24]. ZFP57 and TRIM28 have also been identified as factors contributing to maintenance of methylation within imprinted DMRs [25,26].
Precursor primordial germ cells are biparentally imprinted at early gametogenesis as they are derived from the somatic cells of the embryo. A new germ-line-specific demethylation initiates around mouse embryonic day 8.0–9.0, whereas the primordial germ cells migrate toward the genital ridge in the embryo [27]. This second demethylation occurs at the DMRs and erases the biparental imprinting in the gametes.
Methylation acquisition in the paternal DMRs in male germ cells begins around embryonic day 16.5 and is mostly complete in prospermatogonia by embryonic day 18.5 [28■]. In oocytes, methylation of maternally imprinted DMRs starts only after sexual maturation, around primary to antral stage follicles and is mostly complete in metaphase II (MII) mature oocytes [29] (Fig. 1).
Identification of imprinted loci
Currently, around 150 imprinted genes [30] and 24 DMRs (three paternally and 21 maternally methylated) have been identified in the mouse genome [31]. The majority of the imprinted genes are located in clusters in approximately 1 Mb sequence throughout the genome. Each of these clusters is under the control of specific DMRs, which are known as imprinting control regions (ICRs). Deletion of ICRs results in loss-of-imprinting [32] and mutations in Dnmt genes cause loss of methylation at the ICRs resulting in bi-allelic expression of the imprinted genes [33].
Imprinted genes on human chromosome 11p15.5 are regulated by two ICRs: KCNQ1OT1 and IGF2/H19. SNRPN ICR controls the imprinting cluster on chromosomal region 15q11-q13 [4]. Imprinted genes in these clusters were identified at the earliest imprinting experiments. In an attempt to identify novel imprinted loci, DNA methylation-based methods were used to detect DMRs and then genes located close to those DMRs were examined to assess their imprinting status [34,35]. More comprehensive search was conducted using expression arrays on parthenogenetic and androgenetic embryos [36,37] or UPD mice models [38,39]. Recently, RNA-Seq analysis of reciprocal mouse crossings enabled transcriptome-wide imprinting detection based on the analysis of strain-specific single nucleotide polymorphisms. Using this novel approach, a total of 1330 imprinted loci were identified in the mouse embryonic and adult brain [40] and 251 candidate imprinted genes in the mouse placenta [41].
Although various analytical approaches have been successfully used to detect imprinted loci, how methylation machinery recognizes and differentially modifies the imprinted regions still remains unknown. In addition to high-density CGIs, tandem-repeat patterns and retrotransposons have been suggested as potential discriminative characteristics of imprinted DMRs. On the other hand, there is evidence that tandem repeats or retrotransposons alone are unlikely predictors of imprint status [42,43]. More recent studies have focused on determining a sequence signature that could distinguish imprinted regions from the rest of the genome, and computational prediction of mouse imprinted genes based on common features such as CpG islands and repeats yielded over 600 candidates [44]. Further studies combining molecular and computational biology approaches are needed to identify the full spectrum of imprinted genes and to determine whether mechanisms other than methylation contribute to genomic imprinting.
IMPACT OF ASSISTED REPRODUCTIVE TECHNOLOGIES ON IMPRINTING AND IMPRINTING DISORDERS
ART-related manipulations to oocytes and embryos, such as follicular stimulation, intracytoplasmic sperm injection (ICSI), and embryo culture, coincide with the timing of epigenetic reprogramming and sex-specific imprint acquisition. Genome-wide and imprinting-related DNA methylation levels change considerably during this period. Consequently, stability of DNA methylation and expression of imprinted genes in oocytes and embryos have been investigated widely in association with ART treatment. Specific imprinted genes are commonly investigated within the context of ART. Characteristics of these genes are presented in Table 2.
Table 2.
The list of imprinted genes in humans, which are commonly investigated in relation to assisted reproductive technology
| Gene symbol | Gene name | Expressed parental allele | Human chromosomal region | Mouse ortholog chromosomal region |
|---|---|---|---|---|
| KCNQ1OT1 (LIT1, KvDMR1) | KCNQ1 overlapping transcript 1 | Paternal expression | 11p15.5 | Distal 7 |
| IGF2 | Insulin-like growth factor 2 | Paternal expression | 11p15.5 | Distal 7 |
| H19 | H19 | Maternal expression | 11p15.5 | Distal 7 |
| PHLDA2 | Pleckstrin homology-like domain family A member 2 | Maternal expression | 11p15.5 | Dist 7 |
| SNRPN | Small nuclear ribonucleoprotein polypeptide N | Paternal expression | 15q11-q13 | Central 7 |
| UBE3A | Ubiquitin-protein ligase E3A | Maternal expression in brain | 15q11-q13 | Central 7 |
| MEST (PEG1) | Mesoderm-specific transcript homolog (paternally expressed gene 1) | Paternal expression | 7q32.2 | Prox 6 |
| PEG10 | Paternally expressed gene 10 | Paternal expression | 7q21.3 | Prox 6 |
| PEG3 | Paternally expressed gene 3 | Paternal expression | 19q13.43 | Prox 7 |
| L3MBTL | Lethal(3) malignant brain tumor-like protein | Paternal expression | 20q13.12 | Chr 2 (not imprinted in mice) |
Imprinting data gathered from [45].
DNA methylation and imprinted gene expression following assisted reproductive technology procedures: animal models
In one of the early studies, disrupted methylation was observed at H19, Mest/Peg1, and Igf2r DMRs in mouse oocytes produced by in-vitro folliculogenesis [46]. Conversely, normal DNA methylation was observed at the imprinted H19, Igf2r, and Snrpn regulatory sequences in MII oocytes grown in a long-term preantral follicle culture system and matured in vitro [47]. The same group also reported that high levels of ammonia and mineral oil overlay during follicle culture did not alter the methylation status of these DNA regions in MII oocytes [48]. A recent systematic literature review reported that mouse ovarian tissue culture and in-vitro follicle culture studies collectively suggest correct imprinted DNA methylation establishment in oocytes [49■].
Investigation of the effect of follicle-stimulating hormone therapy, that is, superovulation, revealed that maternal imprint acquisition was unaffected at Snrpn, Kcnq1ot1, Peg3, and H19 genes in individual mouse oocytes derived from females subjected to low and high hormone treatments [50]. Cryotop vitrification of preantral follicles also did not affect imprinting in vitrified oocytes [51].
In contrast to the oocyte, disrupted DNA methylation was consistently observed in ART-conceived mouse embryos. Aberrant DNA methylation at Igf2-H19 imprinted region was observed in in-vitro conceived mouse morulas and blastocysts [52]. Analysis of methylation status and expression level of H19 in individual blastocysts revealed that epigenetic alterations of the H19 were affected in in-vitro fertilization (IVF), whereas superovulation resulted in disrupted H19 expression [53]. The disruptive effect of superovulation on imprinting status was also reported in terms of loss of methylation in maternally methylated Snrpn, Peg3, and Kcnq1ot1 and gain of methylation at the maternal allele in paternally methylated H19 imprinted regions [54]. Notably, this disruption was dose-dependent, with aberrant imprinted methylation more frequent at the high hormone dosage. The same group also showed that embryo culture in five different commercial media systems resulted in loss of imprinted methylation at different levels and that combined treatment of superovulation and embryo culture resulted in increased perturbation of genomic imprinting [55]. In addition, the effect of maternal age on DNA methylation was investigated in blastocysts and mid-gestation embryos and interestingly, no age-related differences were detected in DMRs of Snrpn, U2af1rs1, Kcnq1ot1, Igfr2, Peg1, and H19 imprinted genes [56].
Altered expression of imprinted genes is an expected consequence of disrupted methylation in DMRs. Aberrant expression of silent paternal H19 allele in the mouse oocyte was observed in one of the two commercial embryo culture media tested [57]; and preimplantation embryo culture in the presence of serum resulted in differential expression of imprinted H19, Igfr2, and Grb10 genes, leading to aberrant fetal growth and development in mice [58]. A series of studies indicated abnormal imprinted gene expression in the placental tissues of the ART conceptuses [59–61]. These studies were based on the fact that the trophectoderm is directly in contact with the artificial culture medium, which can potentially impact placental development [61].
Imprinting disorders in assisted reproductive technology-conceived children
In addition to the animal studies described above, aberrant DNA methylation was observed at imprinted DMRs of in-vitro cultured human oocytes and ART-conceived human embryos [62,63], and DNA methylation-associated gene expression differences were detected in in-vitro conceived children [64]. These observations led to the investigation of the potential impact of ART on imprinting-related human disorders.
BWS, Angelman syndrome, and PWS are the most common imprinting disorders investigated within the context of ART. BWS is an overgrowth disorder resulting from genetic or epigenetic defects affecting KCNQ1OT1 and H19 imprinting domains [5]. The incidence of BWS is estimated to be 1 of 13 700 [65]. Angelman syndrome is a neurological disorder characterized by mental retardation, severe learning difficulties, subtle dysmorphic facial features, and a happy, sociable disposition [6]. The syndrome is caused by downregulation of maternal UBE3A expression in the SNRPN imprinting cluster. PWS is characterized by hyperphagia, stubbornness, and compulsive traits [7]. PWS also arises from imprinting defects at the SNRPN DMR. Both Angel-man syndrome and PWS are relatively rare diseases with a frequency of one of 10 000 to one of 15 000 [7].
Initially in 2002, a case report by Cox et al. [66] observed sporadic loss of methylation in maternally imprinted SNRPN locus in two ICSI children who developed Angelman syndrome and pointed out the possibility of disrupted maternal imprinting as a consequence of ICSI. Orstavik et al. [67] also reported an ICSI child with Angelman syndrome having sporadic imprinting defects in SNRPN locus. Ludwig et al. [68] investigated the incidence of ART in a larger cohort including 79 Angelman syndrome patients of which 16 were born to subfertile families. Imprinting defects were detected at SNRPN region in four out of these 16 individuals: one ICSI child, one child born after hormonal therapy, and two children born without ART. The authors [68] suggested that subfertility and imprinting defects may have a common cause.
The first evidence on association of ART with BWS was published in 2003 reporting altered DNA methylation at LIT1 (KCNQ1OT1) or H19 imprinted DMRs in five of the seven BWS children born after ART [69]. The prevalence of ART in the BWS registry assessed in this study was 4.6% (3/65), which represented a six-fold increase compared to 0.76% live births resulting from ART in the United States in 1999 [69].
These initial observations were followed by population- and cohort-based analyses from the United Kingdom, France, and Australia, indicating significantly increased prevalence of BWS among ART-conceived children compared with children conceived naturally. In the study by Maher et al., [70] six of 149 (4%) BWS children were born after ART in the United Kingdom. As ART accounts for only approximately 1.2% of births in the general population in the United Kingdom, this corresponded to an approximately three-fold increase in the incidence of exposure to ART in children with BWS [70]. Two out of six cases were assessed for KvDMR1 methylation and both showed loss of methylation on the maternal allele [70]. In another cohort of 149 BWS cases from France, Gicquel et al. [71] identified six post-ART children with isolated demethylation at KvDMR1. The prevalence of ART treatment in children with BWS (4%) in this study was three times higher than the general population in France, where ART accounts for 1.3% of births [71]. Halliday et al. [72] estimated the risk of BWS in the IVF population as approximately one out of 4000 comparing the occurrence ratio of 37 BWS cases in the general population to four post-ART BWS cases in the IVF population. Three out of four post-ART BWS children were tested for KvDMR1 methylation, and loss of methylation was detected in all three cases [72]. Sutcliffe et al. [73] conducted a survey including 79 BWS, 75 Angelman syndrome, 163 PWS, and 23 transient neonatal diabetes mellitus cases, and a significantly increased frequency of ART use was confirmed only in the BWS population (2.9% versus 0.8% expected). Altered methylation was observed at KvDMR1 region in all of the eight BWS cases and at SNRPN region in one of the three Angelman syndrome cases examined [73].
Although loss of methylation in the maternally imprinted KvDMR1 was investigated as the main imprinting defect associated with post-ART BWS cases, abnormal methylation was observed at multiple genomic loci in both ART-conceived and naturally conceived BWS patients [74,75].
With the aim of testing the hypothesis that the culture media could be implicated as a common factor among post-ART BWS children, Chang et al. [76] conducted an analysis on 341 patients with BWS that were recruited to the BWS registries in the United States between 1994 and 2003. Nineteen of the 341 individuals were born after ART. Analysis of the available medical records for 12 of these 19 ART-BWS cases revealed that all 12 women had received some type of ovarian stimulation medication; however, investigations were not able to identify any specific ART procedure or in-vitro culture media as being associated with BWS in this patient group [76].
The earlier reports suggesting an association between ART and imprinting disorders were not supported by the findings of subsequent case series and cohort studies. Analysis of Danish registry data including 6052 IVF children born in 1995–2001 did not suggest an increased risk of imprinting diseases after IVF [77]. In a survey of both clinically diagnosed and previously unrecognized BWS and Angel-man syndrome cases, the absolute risk of imprinting disorders in ART-conceived babies was estimated to be less than 1% [78]. According to a Dutch study, 6.4% of the 220 children with one of the Angelman syndrome, BWS, or PWS were born after any form of ART, whereas 6.8% of the affected children were born to subfertile couples without ART [79]. Because the relative risks of assessed syndromes were the same in subfertile families with or without ART, the increased prevalence of imprinting disorders was attributed to fertility problems rather than ART procedures. The infertility factor was further discussed in a case report of a family having two children diagnosed with BWS, one through spontaneous conception and the other one born after ART [80]. A summary of studies investigating the association between ART and imprinting disorders is given in Table 3.
Table 3.
Summary of human studies investigating the association between assisted reproductive technology and imprinting disorders
| Study | Study design |
Aim | Study population | Treatment | Frequency of imprinting defect in analyzed samples |
Affected DMR – gene |
Prevalence and risk estimation |
Country | Period |
|---|---|---|---|---|---|---|---|---|---|
| Ludwig et al. [68] | Case series (survey) | Determine whether subfertility is associated with Angelman syndrome | 79 Angelman syndrome cases; 16/79 born to subfertile families | ICSI/ovarian stimulation | 4/16 (ICSI: 1/16; ovarian stimulation: 1/16; no ART: 2/16) | SNRPN | Imprinting defect in Angelman syndrome children born to subfertile families: 4/16 (25%); Imprinting defect in Angelman syndrome population: 4%. RR: 6.25-fold increase in subfertile couples | Germany | NA |
| DeBaun et al. [69] | Case series | Determine whether ART is associated with BWS | 65 BWS cases; 3/65 born after ART + 4 ART-BWS from a previous registry | IVF/ICSI | 5/6 | H19 KvDMR1 (KCNQ1OT1) | ART in BWS population: 3/65 (4.6%). ART in general population: 0.76%. Relative increase: approximately six-fold | USA | BWS registries 2001–2002 and 1994–2000 |
| Maher et al. [70] | Case series | Determine whether ART is associated with BWS | 149 BWS cases; 6/149 born after ART | IVF/ICSI | 2/2 | KvDMR1 (KCNQ1OT1) | ART in BWS population: 6/149 (4%). ART in general population: 1.2%. Relative increase: approximately 3.3-fold | UK | BWS: 1989–2002. ART: 1995–2000 |
| Gicquel et al. [71] | Case series | Determine whether ART is associated with BWS | 149 BWS cases; 6/149 born after ART | IVF/ICSI/fresh or frozen cycles | 6/6 | KvDMR1 (KCNQ1OT1) | ART in BWS population: 6/149 (4%). ART in general population: 1.3%. Relative increase: approximately 3.1-fold | France | NA |
| Halliday et al. [72] | Case control | Determine whether ART is associated with BWS | 37 BWS cases; 4/37 born after ART | IVF/ICSI/fresh or frozen cycles | 3/3 | KvDMR1 (KCNQ1OT1) | Overall BWS prevalence: 37/1 316 500 (~1/35 580). The risk of BWS in ART population: 4/14 894 (~1/4000). Relative increase: approximately nine-fold | State of Victoria, Australia | 1983–2003 |
| Sutcliffe et al. [73] | Case series (survey) | Determine whether ART is associated with BWS, Angelman syndrome, PWS, or TNDM | 79 BWS cases, 11/79 ART, 75 Angelman syndrome cases, 3/75 ART, 163 PWS cases, 9/163 ART 23 TNDM cases, and 1/23 ART | IVF/ICSI/ovarian stimulation | 8/8 BWS; 1/3 Angelman syndrome | KvDMR1 (KCNQ1OT1) in BWS; SNRPN in Angelman syndrome | ART in BWS population: 2.9% (corrected rate). ART in general population: 0.8%. Relative increase: 3.5-fold | UK and Ireland | Syndromes: 1987–2001. ART: 1999–2002 |
| Rossignol et al. [74] | Cohort | Assess the occurrence of abnormal imprinting in loci other than KvDMR1 in BWS conceived with ART versus those conceived without ART | 11 post-ART BWS cases; 29 non-ART BWS cases | IVF/ICSI/cryo preservation | 40/40 | KvDMR1 (KCNQ1OT1) IGF2 SNRPN | Abnormal methylation at a locus other than KvDMR1: In post-ART BWS cases: 3/11 (27%); In non-ART BWS cases: 7/29 (24%) | France | NA |
| Lim et al. [75] | Cohort | Assess the occurrence of abnormal imprinting in loci other than KvDMR1 in BWS conceived with ART versus those conceived without ART | 25 post-ART BWS cases; 87 non-ART BWS cases | IVF/ICSI | 24/25 | KvDMR1 (KCNQ1OT1) additional DMRs at 6q24, 7q32, and 15q13 | Abnormal methylation at a locus other than KvDMR1: In post-ART BWS cases: 37.5%; In non-ART BWS cases: 6.4% | UK | NA |
| Chang et al. [76] | Case series | Determine whether the type of culture media used in ART is associated with BWS | 341 BWS cases; 19/341 born after ART; 12/19 medical records | IVF/ICSI/ovarian stimulation | NA | NA | No specific ART method, specific in-vitro culture media, or timing of embryo transfer was associated with BWS | USA | BWS registry 1994–2003 |
| Lidegaard et al. [77] | Cohort | Determine whether IVF is associated with imprinting disorders (BWS, PWS, kidney cancer, and retinoblastoma) in singleton children | 6052 singleton ART children; 442 349 singleton non-ART children | IVF/ICSI | NA | NA | ART was not associated with any imprinting disorder | Denmark | 1995–2001 |
| Bowdin et al. [78] | Case series (survey) | Determine whether ART is associated with BWS or Angelman syndrome | 1/1524 BWS in ART children; 0/1524 Angelman syndrome in ART children | IVF/ICSI | 1/47 | KvDMR1 (KCNQ1OT1) | The absolute risk of imprinting disorders in children conceived by ART is small (<1%) | UK and Ireland | 1997–2003. 1989–2002 Ireland |
| Doornbos et al. [79] | Case series (survey) | Determine whether ART is associated with BWS, Angelman syndrome, or PWS | 71 BWS; 63 Angelman syndrome; 86 PWS; 220 total affected children; 14/220 post-ART; 15/220 born to subfertile families without ART | IVF/ICSI/ovarian stimulation/IUI/donor insemination | NA | NA | ART in affected group: 14/220 (6.4%). ART in general population: 2.1%. Non-ART children of subfertile families in affected group: 15/220 (6.8%). Non-ART children of subfertile families in general population: 3.5% | Netherlands | 1983–2003 |
ARTs, assisted reproductive technologies; BWS, Beckwith–Wiedemann syndrome; ICSI, intracytoplasmic sperm injection; IVF, in-vitro fertilization; IUI, intrauterine insemination; NA, not available; PWS, Prader–Willi syndrome; TNDM, transient neonatal diabetes mellitus.
In addition, stability of DNA methylation was examined in in-vitro conceived children independent of the diagnosed syndromes. Normal methylation pattern was observed at SNRPN region in all 92 ICSI children examined [81], whereas hypo-methylation at KvDMR1 region was reported in three out of 18 clinically normal ART-conceived children [82]. Methylation status of the IGF2/H19 ICR in 61 phenotypically normal ART newborns was compared to 30 naturally conceived newborns and showed that overall methylation patterns were similar between the two groups [83]. In a more comprehensive analysis, 10 DMRs were analyzed in 77 ICSI, 35 IVF, and 73 natural conception children, and a slight hypermethylation was observed only at MEST region in IVF children [84]. Comparison of whole-genome gene expression in blood samples obtained from 60 ART-conceived and 60 naturally conceived babies demonstrated similar genomic imprinting expression between the two groups, except for differential expression of PEG10, L3MBTL, and PHLDA2 genes in the ART group and loss of methylation at L3MBTL region in one ICSI case [85]. Examination of DNA methylation at four imprinted loci in 66 IVF-conceived and 69 naturally conceived children revealed no difference at percentage of methylation between the groups [86]. These findings together suggest that ART does not significantly alter the DNA methylation pattern in humans and imprinting defects are not common in the ART population.
CONCLUSION
Genomic imprinting is a specialized epigenetic mechanism that silences one parental allele, while enabling gene expression from the other parental allele. Although imprinting seems to regulate only a limited number of genes, disruption of imprinting may have serious health consequences, leading to significant pathologies including BWS, Angelman syndrome, and PWS.
As ART-related manipulations of oocytes and embryos coincide with the timing of epigenetic reprogramming and sex-specific imprint acquisition, stability of DNA methylation and imprinted gene expression in oocytes and embryos have been investigated in association with ART. Animal studies suggested an effect of ART on genomic imprinting in embryos. In addition, several case series studies reported an approximately three-fold to six-fold increase in the occurrence of BWS in association with ART, compared to the general population. However, abnormal DNA methylation could not be consistently identified in IVF children, and an effect of ART independent of infertility could not be demonstrated.
At present, our understanding of the impact of ART on genomic imprinting and imprinting disorders is limited as the syndromes associated with aberrant imprinting are quite rare, making clinical cohort studies extremely difficult and randomized prospective trials impossible. A combined effort from clinical and basic researchers is needed to determine the actual impact of ART on imprinting disorders and to delineate how different types of ART related interventions may affect genomic imprinting.
KEY POINTS.
Manipulation of oocytes and embryos in ART coincides with the timing of epigenetic reprogramming and sex-specific imprint acquisition.
Several studies reported an estimated three-fold to sixfold increase in the occurrence of BWS in association with ART, compared to the general population.
More recent reports suggest that infertility might underlie disrupted imprinting because the relative risks for the assessed syndromes seem to be similar in subfertile families with or without ART treatment.
Further comprehensive surveys and analyses are required to clarify the relationship between ART and imprinting disorders.
Acknowledgments
E.S. is supported by award no. R01HD059909 from the National Institutes of Health (NIH).
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
#x025A0; of special interest
#x025A0;■ of outstanding interest
- 1.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 2.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 3.Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- 4.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 5.Maher ER, Reik W. Beckwith–Wiedemann syndrome: imprinting in clusters revisited. J Clin Invest. 2000;105:247–252. doi: 10.1172/JCI9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40:87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassidy SB, Dykens E, Williams CA. Prader–Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet. 2000;97:136–146. doi: 10.1002/1096-8628(200022)97:2<136::aid-ajmg5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 8.Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23:2826–2834. doi: 10.1093/humrep/den310. [DOI] [PubMed] [Google Scholar]
- 9.Odom LN, Segars J. Imprinting disorders and assisted reproductive technology. Curr Opin Endocrinol Diabetes Obes. 2010;17:517–522. doi: 10.1097/MED.0b013e32834040a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10■.Denomme MM, Mann MR. Genomic imprints as a model for the analysis of epigenetic stability during assisted reproductive technologies. Reproduction. 2012;144:393–409. doi: 10.1530/REP-12-0237. A comprehensive literature review contending that aberrant DNA methylation arising from ART procedures or infertility may not be restricted to the imprinted genes but might be an indicator of more global epigenetic instability. [DOI] [PubMed] [Google Scholar]
- 11.van Montfoort AP, Hanssen LL, de Sutter P, et al. Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update. 2012;18:171–197. doi: 10.1093/humupd/dmr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sapienza C, Peterson AC, Rossant J, Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987;328:251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- 13.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 14.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 15.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 16.Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- 17.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 18.Bourc’his D, Xu GL, Lin CS, et al. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 19.Mayer W, Niveleau A, Walter J, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 20.Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 21.Gu TP, Guo F, Yang H, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 22.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 23.Howell CY, Bestor TH, Ding F, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 24.Hirasawa R, Chiba H, Kaneda M, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Ito M, Zhou F, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messerschmidt DM, de Vries W, Ito M, et al. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 27.Guibert S, Forne T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012;22:633–641. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28■.Kobayashi H, Sakurai T, Miura F, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23:616–627. doi: 10.1101/gr.148023.112. In this study, investigators performed genome-wide DNA methylation analysis in male and female mouse primordial germ cells at embryonic days 10.5, 13.5, and 16.5 by whole-genome shotgun bisulfite sequencing and identified gender-specific differences in CpG methylation at genome-wide and gene-specific levels during fetal germline progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse preimplantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 30.Williamson C, Blake A, Thomas S, et al. World wide web site: mouse imprinting data and references. Oxfordshire: MRC Harwell; 2014. [Google Scholar]
- 31.Macdonald WA, Mann MR. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol Reprod Dev. 2014;81:126–140. doi: 10.1002/mrd.22220. [DOI] [PubMed] [Google Scholar]
- 32.Spahn L, Barlow DP. An ICE pattern crystallizes. Nat Genet. 2003;35:11–12. doi: 10.1038/ng0903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneda M, Okano M, Hata K, et al. Essential role for de novo DNA methyl-transferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 34.Peters J, Wroe SF, Wells CA, et al. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci U S A. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith RJ, Dean W, Konfortova G, Kelsey G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 2003;13:558–569. doi: 10.1101/gr.781503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizuno Y, Sotomaru Y, Katsuzawa Y, et al. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem Biophys Res Commun. 2002;290:1499–1505. doi: 10.1006/bbrc.2002.6370. [DOI] [PubMed] [Google Scholar]
- 37.Sritanaudomchai H, Ma H, Clepper L, et al. Discovery of a novel imprinted gene by transcriptional analysis of parthenogenetic embryonic stem cells. Hum Reprod. 2010;25:1927–1941. doi: 10.1093/humrep/deq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi JD, Underkoffler LA, Collins JN, et al. Microarray expression profiling of tissues from mice with uniparental duplications of chromosomes 7 and 11 to identify imprinted genes. Mamm Genome. 2001;12:758–764. doi: 10.1007/s00335-001-3027-5. [DOI] [PubMed] [Google Scholar]
- 39.Schulz R, Menheniott TR, Woodfine K, et al. Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res. 2006;34:e88. doi: 10.1093/nar/gkl461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregg C, Zhang J, Weissbourd B, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Soloway PD, Clark AG. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics. 2011;189:109–122. doi: 10.1534/genetics.111.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed MR, Riggs AD, Mann JR. Deletion of a direct repeat element has no effect on Igf2 and H19 imprinting. Mamm Genome. 2001;12:873–876. doi: 10.1007/s00335-001-2027-9. [DOI] [PubMed] [Google Scholar]
- 43.Lewis A, Mitsuya K, Constancia M, Reik W. Tandem repeat hypothesis in imprinting: deletion of a conserved direct repeat element upstream of H19 has no effect on imprinting in the Igf2-H19 region. Mol Cell Biol. 2004;24:5650–5656. doi: 10.1128/MCB.24.13.5650-5656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Kerjean A, Couvert P, Heams T, et al. In vitro follicular growth affects oocyte imprinting establishment in mice. Eur J Hum Genet. 2003;11:493–496. doi: 10.1038/sj.ejhg.5200990. [DOI] [PubMed] [Google Scholar]
- 47.Anckaert E, Adriaenssens T, Romero S, et al. Unaltered imprinting establishment of key imprinted genes in mouse oocytes after in vitro follicle culture under variable follicle-stimulating hormone exposure. Int J Dev Biol. 2009;53:541–548. doi: 10.1387/ijdb.082619ea. [DOI] [PubMed] [Google Scholar]
- 48.Anckaert E, Adriaenssens T, Romero S, Smitz J. Ammonium accumulation and use of mineral oil overlay do not alter imprinting establishment at three key imprinted genes in mouse oocytes grown and matured in a long-term follicle culture. Biol Reprod. 2009;81:666–673. doi: 10.1095/biolreprod.109.076810. [DOI] [PubMed] [Google Scholar]
- 49■.Anckaert E, De Rycke M, Smitz J. Culture of oocytes and risk of imprinting defects. Hum Reprod Update. 2013;19:52–66. doi: 10.1093/humupd/dms042. In this systematic review, authors concluded that ovarian tissue culture and in-vitro follicle culture are not associated with abnormal DNA methylation at imprinted loci in mouse oocytes, whereas data on the impact of human IVM on imprinting are lacking. [DOI] [PubMed] [Google Scholar]
- 50.Denomme MM, Zhang L, Mann MR. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil Steril. 2011;96:734–738. e732. doi: 10.1016/j.fertnstert.2011.06.055. [DOI] [PubMed] [Google Scholar]
- 51.Trapphoff T, El Hajj N, Zechner U, et al. DNA integrity, growth pattern, spindle formation, chromosomal constitution and imprinting patterns of mouse oocytes from vitrified preantral follicles. Hum Reprod. 2010;25:3025–3042. doi: 10.1093/humrep/deq278. [DOI] [PubMed] [Google Scholar]
- 52.Li T, Vu TH, Ulaner GA, et al. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11:631–640. doi: 10.1093/molehr/gah230. [DOI] [PubMed] [Google Scholar]
- 53.Fauque P, Jouannet P, Lesaffre C, et al. Assisted reproductive technology affects developmental kinetics, H19 imprinting control region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol. 2007;7:116. doi: 10.1186/1471-213X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Market-Velker BA, Zhang L, Magri LS, et al. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 55.Market-Velker BA, Fernandes AD, Mann MR. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol Reprod. 2010;83:938–950. doi: 10.1095/biolreprod.110.085480. [DOI] [PubMed] [Google Scholar]
- 56.Lopes FL, Fortier AL, Darricarrere N, et al. Reproductive and epigenetic outcomes associated with aging mouse oocytes. Hum Mol Genet. 2009;18:2032–2044. doi: 10.1093/hmg/ddp127. [DOI] [PubMed] [Google Scholar]
- 57.Doherty AS, Mann MR, Tremblay KD, et al. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- 58.Khosla S, Dean W, Brown D, et al. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- 59.Mann MR, Lee SS, Doherty AS, et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 60.Rivera RM, Stein P, Weaver JR, et al. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17:1–14. doi: 10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
- 61.Fauque P, Mondon F, Letourneur F, et al. In vitro fertilization and embryo culture strongly impact the placental transcriptome in the mouse model. PLoS One. 2010;5:e9218. doi: 10.1371/journal.pone.0009218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khoueiry R, Ibala-Rhomdane S, Mery L, et al. Dynamic CpG methylation of the KCNQ1OT1 gene during maturation of human oocytes. J Med Genet. 2008;45:583–588. doi: 10.1136/jmg.2008.057943. [DOI] [PubMed] [Google Scholar]
- 63.Chen SL, Shi XY, Zheng HY, et al. Aberrant DNA methylation of imprinted H19 gene in human preimplantation embryos. Fertil Steril. 2010;94:2356–2358. 2358.e1. doi: 10.1016/j.fertnstert.2010.01.120. [DOI] [PubMed] [Google Scholar]
- 64.Katari S, Turan N, Bibikova M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18:3769–3778. doi: 10.1093/hmg/ddp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weksberg R, Shuman C, Beckwith JB. Beckwith–Wiedemann syndrome. Eur J Hum Genet. 2010;18:8–14. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cox GF, Burger J, Lip V, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71:162–164. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orstavik KH, Eiklid K, van der Hagen CB, et al. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet. 2003;72:218–219. doi: 10.1086/346030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ludwig M, Katalinic A, Gross S, et al. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet. 2005;42:289–291. doi: 10.1136/jmg.2004.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith–Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maher ER, Brueton LA, Bowdin SC, et al. Beckwith–Wiedemann syndrome and assisted reproduction technology (ART) J Med Genet. 2003;40:62–64. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gicquel C, Gaston V, Mandelbaum J, et al. In vitro fertilization may increase the risk of Beckwith–Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72:1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halliday J, Oke K, Breheny S, et al. Beckwith–Wiedemann syndrome and IVF: a case–control study. Am J Hum Genet. 2004;75:526–528. doi: 10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sutcliffe AG, Peters CJ, Bowdin S, et al. Assisted reproductive therapies and imprinting disorders: a preliminary British survey. Hum Reprod. 2006;21:1009–1011. doi: 10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- 74.Rossignol S, Steunou V, Chalas C, et al. The epigenetic imprinting defect of patients with Beckwith–Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet. 2006;43:902–907. doi: 10.1136/jmg.2006.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lim D, Bowdin SC, Tee L, et al. Clinical and molecular genetic features of Beckwith–Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod. 2009;24:741–747. doi: 10.1093/humrep/den406. [DOI] [PubMed] [Google Scholar]
- 76.Chang AS, Moley KH, Wangler M, et al. Association between Beckwith–Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril. 2005;83:349–354. doi: 10.1016/j.fertnstert.2004.07.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lidegaard O, Pinborg A, Andersen AN. Imprinting diseases and IVF: Danish National IVF cohort study. Hum Reprod. 2005;20:950–954. doi: 10.1093/humrep/deh714. [DOI] [PubMed] [Google Scholar]
- 78.Bowdin S, Allen C, Kirby G, et al. A survey of assisted reproductive technology births and imprinting disorders. Hum Reprod. 2007;22:3237–3240. doi: 10.1093/humrep/dem268. [DOI] [PubMed] [Google Scholar]
- 79.Doornbos ME, Maas SM, McDonnell J, et al. Infertility, assisted reproduction technologies and imprinting disturbances: a Dutch study. Hum Reprod. 2007;22:2476–2480. doi: 10.1093/humrep/dem172. [DOI] [PubMed] [Google Scholar]
- 80.Strawn EY, Jr, Bick D, Swanson A. Is it the patient or the IVF? Beckwith–Wiedemann syndrome in both spontaneous and assisted reproductive conceptions. Fertil Steril. 2010;94:754.e1–754.e2. doi: 10.1016/j.fertnstert.2010.01.067. [DOI] [PubMed] [Google Scholar]
- 81.Manning M, Lissens W, Bonduelle M, et al. Study of DNA-methylation patterns at chromosome 15q11-q13 in children born after ICSI reveals no imprinting defects. Mol Hum Reprod. 2000;6:1049–1053. doi: 10.1093/molehr/6.11.1049. [DOI] [PubMed] [Google Scholar]
- 82.Gomes MV, Huber J, Ferriani RA, et al. Abnormal methylation at the KvDMR1 imprinting control region in clinically normal children conceived by assisted reproductive technologies. Mol Hum Reprod. 2009;15:471–477. doi: 10.1093/molehr/gap038. [DOI] [PubMed] [Google Scholar]
- 83.Shi X, Ni Y, Zheng H, et al. Abnormal methylation patterns at the IGF2/H19 imprinting control region in phenotypically normal babies conceived by assisted reproductive technologies. Eur J Obstet Gynecol Reprod Biol. 2011;158:52–55. doi: 10.1016/j.ejogrb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 84.Tierling S, Souren NY, Gries J, et al. Assisted reproductive technologies do not enhance the variability of DNA methylation imprints in human. J Med Genet. 2010;47:371–376. doi: 10.1136/jmg.2009.073189. [DOI] [PubMed] [Google Scholar]
- 85.Feng C, Tian S, Zhang Y, et al. General imprinting status is stable in assisted reproduction-conceived offspring. Fertil Steril. 2011;96:1417–1423. e1419. doi: 10.1016/j.fertnstert.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 86.Oliver VF, Miles HL, Cutfield WS, et al. Defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population. Fertil Steril. 2012;97:147–153.e7. doi: 10.1016/j.fertnstert.2011.10.027. [DOI] [PubMed] [Google Scholar]