Abstract
The Southwest United States, including Arizona and New Mexico, has a diverse climate and is home to many different avian species. We sequenced the hemagglutinin (HA) gene of twenty influenza specimens for the years 2007–2009. This included four from Arizona, and sixteen from New Mexico. We analyzed the sequences and determined the following HA subtypes: H3, H4, H6, H8, and H11. For each subtype, we combined our virus sequences with those from a public database, and inferred phylogeographic models of influenza diffusion.
Statistical phylogeography indicated that overall evolutionary diffusion of avian influenza viruses is geographically structured (p < 0.05). In addition, we found that diffusion to the Southwest was often from nearby states including California.
For H3, H4 and H6, the intra-flyway gene flow rates were significantly (p < 0.001) higher than those of inter-flyway. Such rate difference was also observed in H8 and H11, yet, without statistical significance (p=0.132, p=0.190 respectively). Excluding any one flyway from the calculation generated similar results, suggesting that such barrier effect on gene flow rates is not exclusively produced by any single flyway.
We also calculated the Bayes factor test for the significant non-zero rates between states and identified significant routes both within and across flyways. Such inter-flyway spread of influenza was probably the result of birds from four flyways co-mingling on breeding grounds in northern regions or marshaling on staging areas post breeding in Canada or Alaska, before moving south each fall.
This study provides an initial analysis of evolutionary diffusion of avian influenza virus to and from the Southwest United States. However, more sequences from this region need to be generated to determine the role of host migration and other factors on influenza diffusion.
Keywords: Influenza in Birds, Phylogeography, Southwestern United States
Introduction
The Southwest United States, including Arizona and New Mexico, has a diverse climate (Lenart 2008) and is home to many different avian species (Sharp, 2012). Arizona, in particular is part of the Pacific flyway which starts in Alaska and extends as far as South America (2013b). While there are four migratory flyways in the United States, the Pacific flyway is believed to pose the greatest risk of introduction of avian influenza (AIV) H5N1 virus due to mixing of birds from Asian countries (Hill et al., 2012b). Conversely, New Mexico is in the Central flyway that includes Colorado, Montana, Texas, Oklahoma, Wyoming, Kansas, North and South Dakota, and north into Canada (2013a). Despite the importance of the Southwest for avian migration, there remains uncertainty about its relationship to the diffusion of avian influenza viruses.
Phylogeography is a field that uses sequence data to model geographic diffusion and genetic diversity over time (Avise, 2000). RNA viruses including influenza are often studied because of their short genomes and rapid rate of nucleotide substitutions (Holmes, 2004). Here, geospatial data such as location of the infected host is used in the model to infer diffusion. While there have been many studies that have characterized influenza viruses in migratory birds in the United States (Bahl et al., 2013; Chen and Holmes, 2009; Cross et al., 2013; Dugan et al., 2008; Girard et al., 2012; Henaux et al., 2012; Hill et al., 2012b; Huang et al., 2014; Ip et al., 2008; Jackwood and Stallknecht, 2007; Koehler et al., 2008; Krauss et al., 2004; Lee et al., 2011; Lewis et al., 2013; Pearce et al., 2010; Ramey et al., 2011; Spackman et al., 2005; Suarez et al., 1999; Widjaja et al., 2004; Wille et al., 2011), none have focused on the Southwestern United States. Lam et al. (2012) studied the migration of influenza viruses among a variety of migratory birds in the United States. The authors obtained 100 samples in California, Washington, and Oregon from the United States Geological Survey (USGS) National Wildlife Health Center’s archive, and analyzed with > 1,000 GenBank genome sequences (Lam et al., 2012). Results showed that diffusion of influenza viruses in the United States was isolated by geographic distance and shaped by flyways of the species (Lam et al., 2012). In another study, Girard et al. (2012) examined the phylogeography of influenza among California and Alaskan migratory birds in the Pacific flyway, and found that the geographic origin had the strongest association with virus phylogeny. A study by Hill et al. (2012b) focused on the relationship between migration strategy and avian influenza spread among mallards in the United States, in particular, the Pacific flyway. The authors found that diversity was greater in California, suggesting that wintering states, such as Arizona, might represent an important area for avian influenza reassortment (Hill et al., 2012b). A more recent study by Bahl et al. (2013) found that flyways play a less important role in the long-term persistence of North American avian influenza virus.
The purpose of this study is to understand the phylogeography of avian influenza among migratory birds in the Southwest United States. In addition to the Southwest, we included publically available sequences from North American isolates in order to obtain a complete picture of virus diffusion. We used the combined sequence data sets to identify the association between geography and virus evolution with a particular focus on the Pacific and Central flyways and the Southwest United States.
Materials and Methods
Virus isolation and sequencing
We considered oropharyngeal/cloacal swab samples (N=227) from the USDA National Wildlife Disease Program Avian Tissue Archive. All of the samples originated in Arizona (69/227) or New Mexico (158/227) from 2007 – 2011 and previously tested positive for avian influenza virus by real-time reverse-transcription polymerase chain reaction (rRT-PCR) for the matrix gene at their respective state laboratories. We stored the samples at −80°C in the archive until use for this study. We inoculated each sample into 9–11 day old specific pathogen-free embryonated chicken eggs and incubated at 37°C for 72 hours. We conducted a second passage on all samples. We then harvested amniotic allantoic fluid from each egg and assayed for hemagglutination. We identified thirty-one hemagglutination positive virus isolation samples by rRT-PCR for the matrix gene and extracted RNA using the Ambion MagMAX™ AI/ND Viral RNA Isolation Kit (Life Technologies, Carlsbad, CA).
Amplification of Hemagglutinin Gene
We used Superscript II reverse transcriptase (Life Technologies, Carlsbad, CA) to synthesize cDNA from the RNA extracts using the manufacturer’s protocol. We utilized the Uni12 primer (5’– AGCAAAAGCAGG – 3’) designed by Hoffman et al. (Hoffmann et al., 2001) in the reverse transcription reaction. We then included Hoffman’s (Hoffmann et al., 2001) universal primers Bm-HA-1 (5’– TATTCGTCTCAGGGAGCAAAAGCAGGGG – 3’) and Bm-NS-890R (5’– ATATCGTCTCGTATTAGTAGAAACAAGGGTGTTTT – 3’) to amplify the entire HA region of the influenza genome. We performed the reaction on a C1000 Touch™ thermal cycler (BioRad Carlsbad, CA) and included an initial denaturation phase of 4 minutes at 94°C followed by 30 cycles of: denaturation at 94°C for 20 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 7 minutes. This was followed by a final extension at 72°C for 7 minutes. We ran a 1% agarose gel with GelRed™ stain (Biotium, Hayward, CA) for 45 minutes at 100v and then used a Gel Doc™ XR+ system (BioRad, Hercules, CA) for imaging. We excised bands on a transilluminator (VWR, Radnor, PA) and purified the DNA by using the E.Z.N.A.® Gel Extraction kit (Omega Bio-Tek, Inc., Norcross, GA) according to the manufacturer’s protocol. We used the CloneJET kit (Thermo Scientific, Waltham, MA) to perform blunting and ligation reactions according to the manufacturer’s protocol. We transformed One Shot® TOP10 Chemically Competent E. coli cells (Life Technologies, Carlsbad, CA) and incubated them with 250 μl of SOC media at 37°C and 225rpm for 1 hour. We then plated 25 and 75μl of the cell mixture on pre-warmed LB-amp plates and incubated them overnight at 37°C.
We visually inspected the plates for the presence of colonies, added them to 3.5 ml of LB-amp (in 14 ml culture tubes), and incubated them overnight at 37°C and 225rpm. We screened colonies by performing PCR with the forward and reverse primers provided in the CloneJET kit. We ran the reaction on a C1000 Touch™ thermal cycler and included an initial denaturation phase of 3 minutes at 95°C followed by 25 cycles of: denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 2 minutes. This was followed by a final extension at 72°C for 2 minutes. We ran a 1% agarose gel with GelRed™ stain for 45 minutes at 100v and then used a Gel Doc™ XR+ system (BioRad, Hercules, CA) for imaging. We extracted plasmids using a PureYield™ Plasmid Miniprep kit (Promega, Madison, WI) according to the manufacturer’s protocol. We prepared a sequencing reaction using primers from the CloneJET kit and sent the plasmid template and primers to the sequencing lab at Arizona State University for Sanger sequencing.
We found reliable 5’ and 3’ reads to be about 800–850 bp. Since the hemagglutinin gene is approximately 1778 bp in length, we prepared a third sequencing reaction to capture the gap in the middle of the coding region. Here, we designed a primer on the 5’ sequence using Geneious (Biomatters, Auckland, NZ). We used Geneious to perform assembly and identify contigs and then concatenated the three sequences. We used the National Center for Biotechnology Information (NCBI) Influenza Virus Sequence Annotation Tool (Bao et al., 2007) to perform validity checking of the full-length HA sequence and edited if necessary (including the removal of 5’ and 3’-end vector contamination).
Subtyping
We were able to amplify the HA region of 20 of the 31 isolates across eight different avian species. We determined the subtype by querying NCBI’s BLAST (Altschul et al., 1990) and Influenza Virus Sequence Annotation Tool (Bao et al., 2007). They included: 4 H3, 10 H4, 3 H6, 1 H8, and 2 H11 (Table 1).
Table 1.
GenBank accession and corresponding metadata for the twenty new sequences generated from this work.
| HA | GenBank | Host | State | Date Collected |
|---|---|---|---|---|
| H3 | JX297592 | Anas discors | NM | 2007 |
| H3 | JN864063 | Anas strepera | AZ | 2009 |
| H3 | KF487543 | Anas discors | NM | 2008 |
| H3 | KF511796 | Anas discors | NM | 2007 |
| H4 | CY122409 | Anas platyrhynchos | NM | 2008 |
| H4 | JN673246 | Anas carolinensis | AZ | 2008 |
| H4 | KF511795 | Anas discors | NM | 2007 |
| H4 | KF534792 | Anas discors | NM | 2007 |
| H4 | KF534793 | Anas discors | NM | 2007 |
| H4 | KF537367 | Anas discors | NM | 2007 |
| H4 | KF557768 | Anas cyanoptera | NM | 2008 |
| H4 | KF569945 | Anas discors | NM | 2007 |
| H4 | KF636136 | Anas carolinensis | NM | 2008 |
| H4 | KF636137 | Anas platyrhynchos | NM | 2008 |
| H6 | CY122055 | Anas americana | NM | 2009 |
| H6 | KF569944 | Anas discors | NM | 2008 |
| H6 | KF636135 | Anas discors | NM | 2008 |
| H8 | JN590044 | Anas clypeata | AZ | 2007 |
| H11 | JN798212 | Anas acuta | AZ | 2009 |
| H11 | KF542875 | Anas acuta | NM | 2008 |
Genetic Characterization of North American Wild Bird Lineage Avian Influenza Viruses
We created separate FASTA files for each HA subtype by including reference sequences from the Influenza Research Database (IRD) (Squires et al., 2012). Between September 6–8, 2013, we extracted all available HA sequences of any hosts, geography, collection time, and minimal lengths of 1,659bp with no duplicate sequences. This resulted into five subtype data sets containing 7,301 H3, 888 H4, 1,169 H6, 120 H8, and 422 H11 sequences. We aligned each set using Mafft (Katoh et al., 2002). Then, we used the program RaXML v.7.3 (Stamatakis et al., 2005) to create initial phylogenies by specifying a GTRGAMMA model of nucleotide substitution and a rapid hill climbing approach.
We visually inspected initial phylogenies using FigTree (Rambaut, 2013). For each tree, we identified the lineages that included our sequences and created a new sequence file of only those lineages. We excluded sequences with unknown residues, those outside of the US or Canada, or hosts that were not migratory birds. This resulted into data sets containing 490 H3, 194 H4, 411 H6, 92 H8, and 210 H11 sequences.
Phylogeography
We created a FASTA file of sequences for each subtype and annotated each sequence’s definition line (i.e. starting with “>”) to include the latitude and longitude of the state in which the sample was collected. This was identified from the GenBank record (Benson et al., 2011) and is usually designated by the statement “country=”. We used Geonames.org to identify the centroid latitude and longitude of each state.
We submitted the FASTA files to ZooPhy, a phylogeography system developed by one of the authors (Scotch et al., 2010). ZooPhy acts as a pipeline for automatically connecting a series of bioinformatics applications including ClustalW (Larkin et al., 2007) for nucleotide alignment, jModeltest (Posada, 2008) for evaluation of substitution models, and BEAST (Drummond et al., 2012) for generation of Bayesian discrete phylogeographic trees. The output of ZooPhy is a single Maximum Clade Credibility (MCC) tree. By default, ZooPhy currently implements a strict molecular clock. However, we also manipulated the input files to BEAST to assume an uncorrelated lognormal relaxed molecular clock. When necessary, models were run on Arizona State University’s high performance computing environment to improve performance time. We used Tracer (Rambaut et al., 2013) to examine the log files of each Markov-chain Monte Carlo (MCMC) run. Specifically, we focused on the effective sample size of the model parameters to determine if the length of the run was sufficient.
For model comparison, we used Tracer to generate a Bayes factor analysis of the log marginal likelihoods (Table S1). For each subtype, the relaxed clock model was superior and thus was chosen for our analysis. The MCMC chain length for these five data sets (H3, H4, H6, H8, H11) ranged from 10–70 × 106, sub-sampling every 1,000 steps. For H3, we combined two runs in order to increase the effective sample size. We used TreeAnnotator (Drummond et al., 2012) to produce an MCC tree for each gene. Here, we specified a 10% burn-in to disregard the initial steps in the MCMC. We then used FigTree to color-code the branches of the MCC by their most probable geographic state and to time-scale it by years.
In our discrete phylogeography model we utilized a Bayesian stochastic search variable selection (BSSVS) that enables us to compute a Bayes factor test for identifying the most parsimonious diffusion routes (Lemey et al., 2009). In addition, we specified a non-reversible model for considering directionality (i.e. A→ B vs. B→A) which produced K(K-1) transmission rates, where K is the number of discrete locations, as well as their Boolean indicator (0 or 1) for inclusion in the matrix. We calculated the product of each rate-indicator combination and used these numbers to create a new matrix by flyway rather than location (i.e. A and B changed to Flyway 1 and Flyway 2). We then calculated the overall means and differences of 4/3 inter vs. intra-flyway gene flow.
We used additional software for geographic mapping and statistical phylogeography. We used the program SPREAD (Bielejec et al., 2011) to calculate the Bayes factor test for the significant nonzero rates between discrete states. SPREAD also produced a Keyhole Markup Language (KML) file for viewing in Google Earth (Google, Mountain View, CA). In addition to SPREAD, we also used the program BaTS to calculate the Association Index (AI) and the Parsimony Score (PS) which test the hypothesis that tips in the tree are no more likely to share the same trait (e.g. time, host, geography) with adjoining taxa than by chance alone (Parker et al., 2008). For our work, we specified the sample locations as the trait. This enabled us to determine if the evolutionary diffusion of avian influenza viruses was geographically structured. Our final statistical test was the Kullback-Leibler (KL) divergence test (Kullback and Leibler, 1951) to examine the difference between the prior and posterior most recent state (location) of our new viruses. In discrete phylogeography, this enables us to determine the statistical power of a state within the virus’s lineage. We used a fixed prior 1/K for each tree. For this study, we used the statistical software R (R Core Team, 2014) to calculate the KL divergence.
Results
We show the statistical phylogeography results including the KL divergence test for the most recent state before our new viruses from New Mexico and Arizona (Table 2). This provides insight into the states that drove recent diffusion of influenza into the Southwest. For H3, there was variation as the states ranged from DE (Atlantic flyway) to Oregon (Pacific flyway). One virus from New Mexico (GenBank KF511796) had its own state as the most recent. All KL values were < 1 suggesting weak statistical support. The most recent states of all ten H4 viruses are within the Central flyway suggesting that it is a major influence on recent H4 diffusion to the Southwest. In addition, seven of the ten most recent states were New Mexico. While the KL values for these viruses varies, two of these are > 5 suggesting strong support. Meanwhile the results for the three new H6 viruses from New Mexico suggest California is a recent driver of diffusion to this state with very strong support for two of these viruses (KL > 2). We also found a similar result for our single H8 virus. Here, we calculated a low KL value (< 1) for California suggesting weak support. Finally, we identified Washington as the most recent state for our two H11 viruses with weak support (KL < 1).
Table 2.
Most recent ancestor to southwest states New Mexico or Arizona. Statistical measure of certainty measured by the Kullback-Leibler (KL) divergence test.
| HA | GenBank | State | Most Recent State (Highest posterior probability) | Kullback- Leibler divergence |
|---|---|---|---|---|
| H3 | JX297592 | NM | DE (0.45) | < 1 |
| H3 | JN864063 | AZ | OR (0.35) | < 1 |
| H3 | KF487543 | NM | OR (0.23) | < 1 |
| H3 | KF511796 | NM | NM (0.47) | < 1 |
| H4 | CY122409 | NM | NM (0.54) | < 1 |
| H4 | JN673246 | AZ | NM (0.54) | < 1 |
| H4 | KF511795 | NM | NM (0.42) | < 1 |
| H4 | KF534792 | NM | TX (0.57) | < 1 |
| H4 | KF534793 | NM | NM (0.70) | < 1 |
| H4 | KF537367 | NM | AB (0.80) | > 1 |
| H4 | KF557768 | NM | NM (0.99) | > 5 |
| H4 | KF569945 | NM | AB (0.82) | > 1 |
| H4 | KF636136 | NM | NM (0.77) | > 1 |
| H4 | KF636137 | NM | NM (0.99) | > 5 |
| H6 | CY122055 | NM | CA (0.63) | < 1 |
| H6 | KF569944 | NM | CA (0.91) | > 2 |
| H6 | KF636135 | NM | CA (0.94) | > 2 |
| H8 | JN590044 | AZ | CA (0.63) | < 1 |
| H11 | JN798212 | AZ | WA (0.33) | < 1 |
| H11 | KF542875 | NM | WA (0.48) | < 1 |
We show the AI and PS values as well as the upper and lower bounds of the 95% confidence interval (Table 3). All subtypes have vastly different observed and expected values resulting in statistically significant AI and PS. This indicates that the overall evolutionary diffusion of avian influenza viruses among these subtypes are geographically structured in North America.
Table 3.
Statistical phylogeography results including association index (AI), parsimony score (PS). Numbers in parenthesis are the upper and lower bounds of the 95% confidence interval.
| HA | AI | PS | ||
|---|---|---|---|---|
| Observed Mean | Null Mean | Observed Mean | Null Mean | |
| H3 | 6.17 (5.60, 6.78) | 41.67 (39.91, 43.33) | 85.23 (84.00, 87.00) | 263.13 (254.68, 268.80) |
| H4 | 2.41 (1.83, 3.02) | 14.62 (13.35, 15.67) | 39.08 (37.00, 41.00) | 91.27 (88.99, 93.25) |
| H6 | 6.42 (5.74, 7.13) | 39.97 (38.63, 41.23) | 91.86 (89.00, 95.00) | 274.59 (268.48, 280.44) |
| H8 | 0.65 (0.55, 0.91) | 5.83 (4.98, 6.71) | 14.97 (15.00, 15.00) | 35.29 (33.34, 37.01) |
| H11 | 3.73 (3.11, 4.37) | 18.82 (17.81, 19.93) | 50.09 (49.00, 51.00) | 129.32 (125.57, 132.54) |
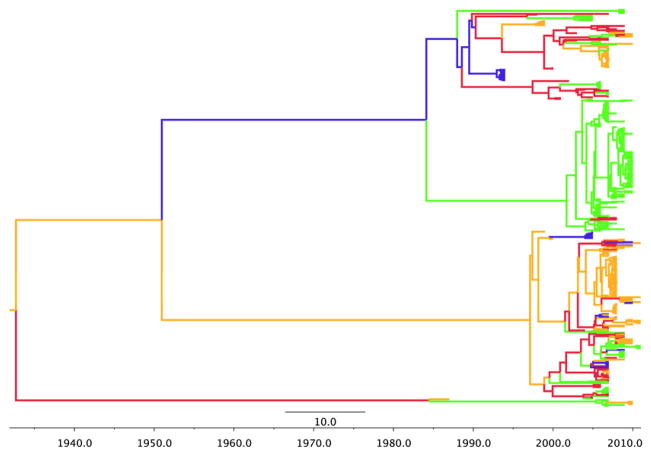
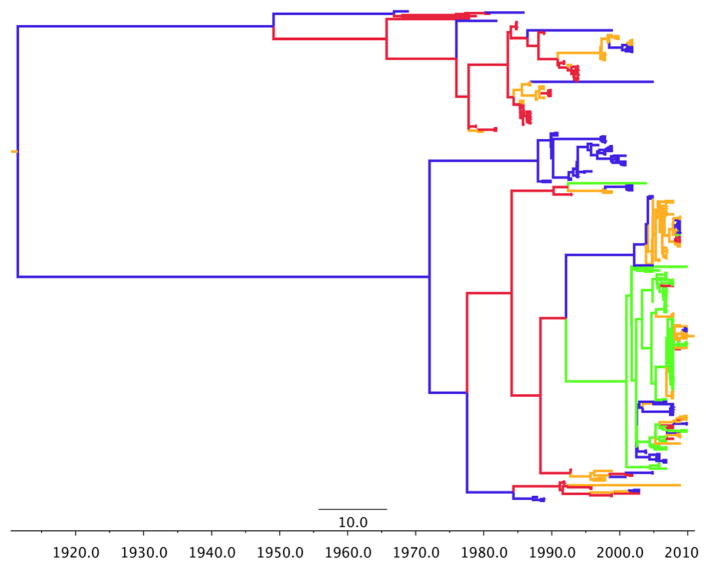
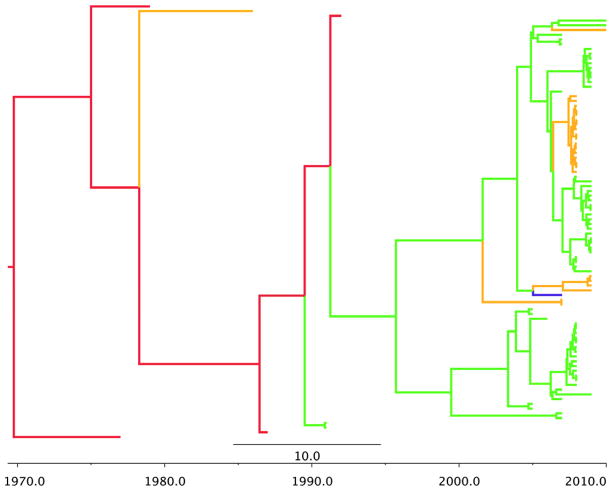
We also provide the phylogeographic trees for each model in Figures 1–5. In order to obtain a clear depiction of the diffusion process between flyways, we modified our trees by converting discrete states from individual state names to the name of their respective flyway. For example, “Arizona” and “Oregon” were changed to “Pacific”, while “New Mexico” and “North Dakota” were changed to “Central”. The branch colors represent these four US flyways: green=Pacific flyway, red=Central, gold=Mississippi, blue=Atlantic. For H3 (Figure 1) the root is deep as initial divergence between Mississippi and Central flyways occurs in 1932. The Pacific flyway clade has remained relatively homogenous since 1984. Conversely, the other three flyways show spatial admixture. For H4, (Figure 2), the root is much shorter and the tree is dominated by the Central and Pacific flyways. H6 (Figure 3) resembles an evolutionary structure more consistent to H3 as the root is very deep, however the clade for the Pacific flyway does contain more spatial admixture. This suggests that recent mixing of the virus from avian species in other flyways has resulted in transmission to birds that are found in Western states.
Figure 1.
H3 phylogeographic maximum clade credibility (MCC) tree. Discrete state names were reassigned to their respective flyways. Branch colors represent US flyways: green=Pacific flyway, red=Central, gold=Mississippi, blue=Atlantic.
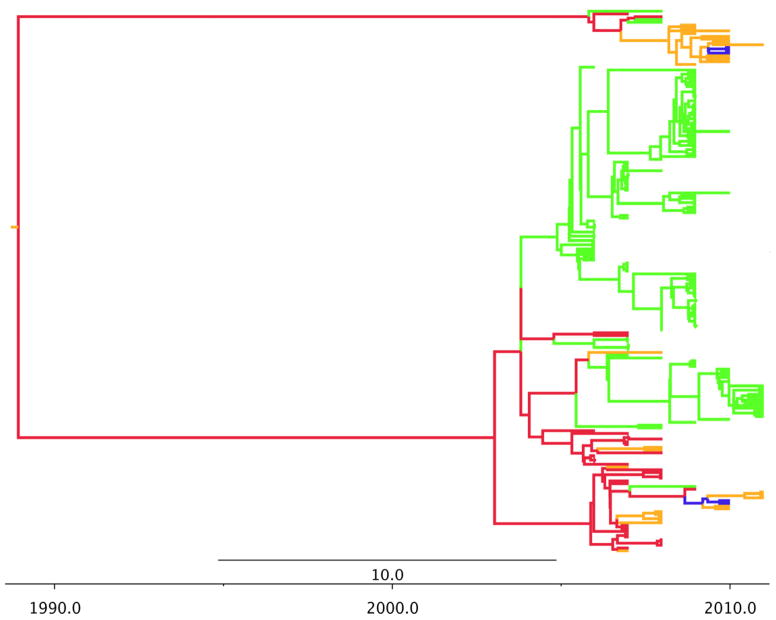
Figure 5.
H11 phylogeographic MCC tree. Branch colors represent US flyways: green=Pacific flyway, red=Central, gold=Mississippi, blue=Atlantic.
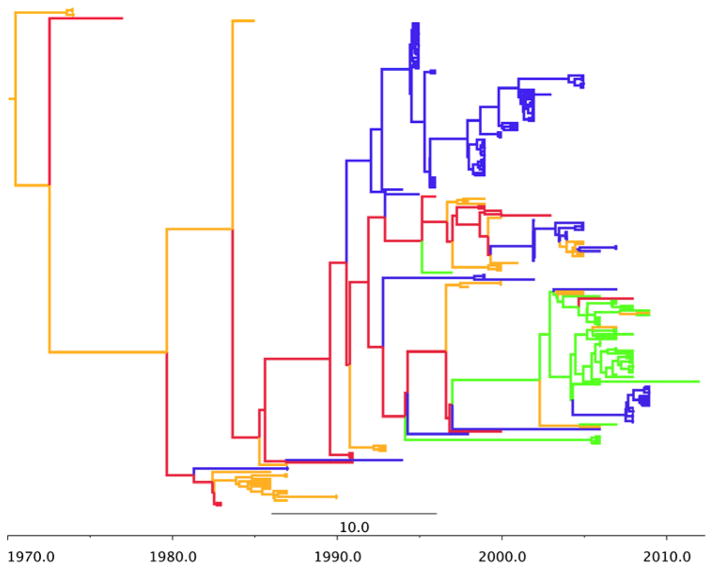
Figure 2.
H4 phylogeographic MCC tree. Branch colors represent US flyways: green=Pacific flyway, red=Central, gold=Mississippi, blue=Atlantic.
Figure 3.
H6 phylogeographic MCC tree. Branch colors represent US flyways: green=Pacific flyway, red=Central, gold=Mississippi, blue=Atlantic.
The phylogeography of H8 (Figure 4) resembles H4 with a more recent origin and an evolutionary history that is driven by the Central and Pacific flyways with little admixture. Finally H11 (Figure 5) also has a more recent origin with little admixture. However, there appears to be more evolutionary contribution from all four flyways.
Figure 4.
H8 phylogeographic MCC tree. Branch colors represent US flyways: green=Pacific flyway, red=Central, gold=Mississippi, blue=Atlantic.
We show the results of the Bayes factor (BF) test to identify significant non-zero geographic state transition rates (Figures S1-S5). We used a BF cutoff of 3 that is consistent with the literature (Baele et al., 2013; Lemey et al., 2009; Li and Drummond, 2012). We show a complete list of the rates in Table S2. Since we used a nonreversible diffusion model, we calculate separate rates for each origin and destination (i.e. A→B vs. B→A). In table 4, we summarize gene flow between intra and inter-flyway routes. For example, we considered a route from California to Arizona as intra-flyway (Pacific) and Iowa to Arizona as an inter-flyway route. For H3, H4 and H6 gene, the intra-flyway gene flow rates are significantly (p < 0.001 using two-sample T-test) higher than inter-flyway rates. Such rate difference was also observed in H8 and H11, yet, without statistical significance (p=0.132, p=0.190 respectively). Excluding any one flyway from the calculation produced similar results suggesting that such barrier effect on gene flow rates is not exclusively produced by any single flyway.
Table 4.
Overall means and differences of 4/3 inter vs. intra-flyway gene flow with 95% confidence intervals. Abbreviations: P= Pacific, C=Central, M=Mississippi, A= Atlantic.
| Intra-flyway | Inter-flyway | Intra-flyway – Inter-flyway | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HA | P-C-M | P-C-A | C-M-A | P-M-A | P-C-M | P-C-A | C-M-A | P-M-A | P-C-M | P-C-A | C-M-A | P-M-A | P-C-M-A |
| H3 | 0.083 (0.064, 0.102) | 0.103 (0.071, 0.135) | 0.077 (0.057, 0.098) | 0.078 (0.059, 0.096) | 0.049 (0.041, 0.057) | 0.052 (0.043, 0.061) | 0.048 (0.039, 0.056) | 0.041 (0.034, 0.048) | 0.034 | 0.051 | 0.029 | 0.037 | 0.036* |
| H4 | 0.118 (0.090, 0.145) | 0.105 (0.069, 0.141) | 0.124 (0.094, 0.153) | 0.115 (0.079, 0.150) | 0.050 (0.040, 0.060) | 0.055 (0.042, 0.068) | 0.058 (0.045, 0.07) | 0.049 (0.038, 0.060) | 0.068 | 0.05 | 0.066 | 0.066 | 0.063* |
| H6 | 0.120 (0.067, 0.173) | 0.107 (0.062, 0.152) | 0.113 (0.072, 0.155) | 0.104 (0.060, 0.149) | 0.055 (0.042, 0.067) | 0.043 (0.032, 0.055) | 0.052 (0.041, 0.062) | 0.036 (0.028, 0.044) | 0.065 | 0.064 | 0.061 | 0.068 | 0.064* |
| H8 | 0.246 (0.149, 0.344) | 0.309 (0.134, 0.484) | 0.184 (0.057, 0.311) | 0.246 (0.149, 0.344) | 0.151 (0.106, 0.196) | 0.178 (0.119, 0.236) | 0.179 (0.131, 0.227) | 0.194 (0.147, 0.241) | 0.095 | 0.131 | 0.005 | 0.052 | 0.071 |
| H11 | 0.063 (0.030, 0.0958) | 0.110 (0.061, 0.159) | 0.088 (0.044, 0.131) | 0.093 (0.054, 0.133) | 0.062 (0.044, 0.080) | 0.071 (0.053, 0.089) | 0.071 (0.052, 0.089) | 0.070 (0.053, 0.088) | 0.001 | 0.039 | 0.017 | 0.023 | 0.023 |
p-values < 0.05 as determined by two-sample T-test.
Discussion
The Southwest United States contributes to the Pacific and Central wild waterfowl migratory flyways and includes a variety of states from the Pacific Ocean through the Great Plains. This region represents an important area for AIV transmission especially since research has found that the Pacific flyway represents the most likely route in the US for HPAI H5N1 compared to other flyways (Huang et al., 2014). Our results suggest variation in evolutionary diffusion of influenza subtypes to and from the Southwest United States. We found that diffusion to the Southwest was often from nearby states including California. Meanwhile we found that intra-flyway migration is propelling influenza gene flow, however a barrier effect on gene flow rates is not exclusively produced by any single flyway. However, we did not study host contribution to virus diffusion and thus cannot determine the avian hosts that drive influenza gene flow. For example, a study by Hill et al. (2012a) examined the relationship between influenza gene flow among mallards and migration strategy. The study focused on the wintering areas in California and no difference in influenza prevalence was found between migration strategy: long-distance (Alaska), resident (California), and intermediate-distance (Pacific Northwest Rim such as Washington) (Hill et al., 2012a). However, there was a difference in gene flow depending on migration strategy with migrants introducing viruses such as H1, H4, and N6 from Alaska and N1 and N2 from Washington resulting in limited circulation in wintering populations in California (Hill et al., 2012a). Conversely, resident mallards were year-round reservoirs for a limited number of avian influenza subtypes (Hill et al., 2012a). Our analysis of significant non-zero rates through Bayes Factor (Table S2) included inter-flyway routes. North American migratory flyways are oriented in a north-south direction (vertical). While studies have demonstrated some degree of admigration, or switching between flyways (Guillemain et al., 2005), it is unlikely that horizontal migration across flyways is the primary driver of influenza in North American birds. Rather, it is more probable that birds in all four flyways are co-mingling on breeding grounds in northern regions or marshaling on staging areas post breeding in Canada or Alaska, before moving south each fall. In addition, our study is consistent with the work by Lam et al. (2012) that found that avian influenza in the United States is structured by geography.
The authors recognize several limitations with this work. First, it represents a small number (twenty) of the total samples in GenBank or IRD. We were unable to amplify 11 isolates potentially due to RNA degradation. Thus, more sequences from this region are needed to generate a definitive understanding of influenza diffusion to and from this region. The small size also did not enable us to focus on propagation to the Southwest US by specific hosts. For example, blue-winged teal and cinnamon teal have been associated with the appearance of North American lineages of LPAI and HPAI in South America (Gonzalez-Reiche et al., 2012; Gonzalez-Reicheabc and Perez, 2012). In order to increase our sample size, we will attempt to amplify the remaining isolates and contribute these to future work on phylogeography in this region. In addition, we will sequence the complete virus genome in order to obtain a better understanding of the relationship between virus reassortment and phylogeography. This will test whether the wintering sites including Arizona and New Mexico might promote influenza virus reassortment (Hill et al., 2012b). As another limitation, we did not include geographic distance for diffusion and gene flow thus our analysis cannot address the impact caused by length of migration. Also, the arid climate in the Southwest US finds concentrations of birds in riparian zones and at annual lakes, where there is unusually high species diversity as well as density of birds (Szaro, 1980). We did not sample birds as part of this work thus we cannot determine local persistence of the virus. Finally, many of these birds over-winter in Central America, thereby providing linkage between species which breed in South America in the austral summer as well as winter visitors. In our analysis, we did not consider regions south of the United States that might be important for AIV spread.
This study provides an initial analysis of evolutionary diffusion of influenza to and from the Southwest United States. However, more sequences from this region need to be generated to determine the role of host migration and other factors on influenza diffusion.
Supplementary Material
We study the phylogeography of the first Southwest US avian influenza sequences.
We sequence twenty hemagglutinin genes from New Mexico and Arizona.
Analysis indicated that the evolutionary diffusion are geographically structured.
The intra-flyway gene flow rates were higher than those of inter-flyway and a barrier effect on gene flow rates is not exclusively produced by any single flyway.
Influenza diffusion to the Southwest was often from nearby states including California.
Acknowledgments
The project described was supported by award number R00LM009825 from the National Library of Medicine and an Arizona State University/Mayo Clinic seed grant to Matthew Scotch. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine, the National Institutes of Health, or Arizona State University and the Mayo Clinic. The authors would like to thank Christina Weller for her work on virus isolation. The authors would also like to thank the Arizona State University Advanced Computing Center (A2C2) for the use of the Saguaro supercomputer.
Abbreviations
- AIV
Avian Influenza Virus
- IRD
Influenza Research Database
- NCBI
National Center for Biotechnology Information
- RNA
Ribonucleic Acid
- cDNA
complimentary Deoxyribonucleic Acid
- PCR
Polymerase Chain Reaction
- USGS
United States Geological Survey
- USDA
United States Department of Agriculture
- KML
Keyhole Markup Language
- AI
Association Index
- PS
Parsimony Score
- KL
Kullback-Leibler
- MCMC
Markov chain Monte Carlo
- MCC
Maximum Clade Credibility
- NIAID
National Institute of Allergy and Infectious Diseases
- HA
Hemagglutinin
- NA
Neuraminidase
- BF
Bayes Factor
- BaTS
Bayesian Tip-Significance Testing
- LPAI
Low Pathogenic Avian Influenza
- HPAI
Highly Pathogenic Avian Influenza
- AK
Alaska
- AB
Alberta
- AZ
Arizona
- BC
British Columbia
- CA
California
- DE
Delaware
- GA
Georgia
- IA
Iowa
- IL
Illinois
- LA
Louisiana
- MB
Manitoba
- MD
Maryland
- MI
Michigan
- MN
Minnesota
- MO
Missouri
- MS
Mississippi
- NE
Nebraska
- NB
New Brunswick
- NL
Newfoundland
- NJ
New Jersey
- NM
New Mexico
- NY
New York
- ND
North Dakota
- NS
Nova Scotia
- OH
Ohio
- ON
Ontario
- OR
Oregon
- PA
Pennsylvania
- QC
Quebec
- SK
Saskatchewan
- SD
South Dakota
- TN
Tennessee
- TX
Texas
- WA
Washington
- WI
Wisconsin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Central Flyway. U.S. Fish and Wildlife Service; 2013a. http://central.flyways.us. [Google Scholar]
- Pacific Flyway. Audubon Society; 2013b. http://conservation.audubon.org/pacific-flyway. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography : the history and formation of species. Harvard University Press; Cambridge, Mass: 2000. [Google Scholar]
- Baele G, Lemey P, Vansteelandt S. Make the most of your samples: Bayes factor estimators for high-dimensional models of sequence evolution. BMC bioinformatics. 2013;14:85. doi: 10.1186/1471-2105-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl J, Krauss S, Kuhnert D, Fourment M, Raven G, Pryor SP, Niles LJ, Danner A, Walker D, Mendenhall IH, Su YC, Dugan VG, Halpin RA, Stockwell TB, Webby RJ, Wentworth DE, Drummond AJ, Smith GJ, Webster RG. Influenza a virus migration and persistence in North American wild birds. PLoS pathogens. 2013;9:e1003570. doi: 10.1371/journal.ppat.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Tatusova T. FLAN: a web server for influenza virus genome annotation. Nucleic acids research. 2007;35:W280–284. doi: 10.1093/nar/gkm354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic acids research. 2011;39:D32–37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27:2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Holmes EC. Frequent inter-species transmission and geographic subdivision in avian influenza viruses from wild birds. Virology. 2009;383:156–161. doi: 10.1016/j.virol.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Arsnoe DM, Minnis RB, King DT, Swafford S, Pedersen K, Owen JC. Prevalence of avian paramyxovirus 1 and avian influenza virus in double-crested cormorants (phalacrocorax auritus) in eastern north america. J Wildl Dis. 2013;49:965–977. doi: 10.7589/2012-06-164. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS pathogens. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard YA, Runstadler JA, Aldehoff F, Boyce W. Genetic structure of Pacific Flyway avian influenza viruses is shaped by geographic location, host species, and sampling period. Virus Genes. 2012;44:415–428. doi: 10.1007/s11262-011-0706-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reiche AS, Morales-Betoulle ME, Alvarez D, Betoulle JL, Muller ML, Sosa SM, Perez DR. Influenza a viruses from wild birds in Guatemala belong to the North American lineage. PloS one. 2012;7:e32873. doi: 10.1371/journal.pone.0032873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reicheabc AS, Perez DR. Where do avian influenza viruses meet in the Americas? Avian diseases. 2012;56:1025–1033. doi: 10.1637/10203-041412-Reg.1. [DOI] [PubMed] [Google Scholar]
- Guillemain M, Sadoul N, Simon G. European flyway permeability and abmigration in Teal Anas crecca, an analysis based on ringing recoveries. Ibis. 2005;147:688–696. [Google Scholar]
- Henaux V, Samuel MD, Dusek RJ, Fleskes JP, Ip HS. Presence of avian influenza viruses in waterfowl and wetlands during summer 2010 in California: are resident birds a potential reservoir? PloS one. 2012;7:e31471. doi: 10.1371/journal.pone.0031471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill NJ, Takekawa JY, Ackerman JT, Hobson KA, Herring G, Cardona CJ, Runstadler JA, Boyce WM. Migration strategy affects avian influenza dynamics in mallards (Anas platyrhynchos) Mol Ecol. 2012a;21:5986–5999. doi: 10.1111/j.1365-294X.2012.05735.x. [DOI] [PubMed] [Google Scholar]
- Hill NJ, Takekawa JY, Cardona CJ, Meixell BW, Ackerman JT, Runstadler JA, Boyce WM. Cross-seasonal patterns of avian influenza virus in breeding and wintering migratory birds: a flyway perspective. Vector borne and zoonotic diseases. 2012b;12:243–253. doi: 10.1089/vbz.2010.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13:745–756. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wille M, Dobbin A, Walzthoni NM, Robertson GJ, Ojkic D, Whitney H, Lang AS. Genetic structure of avian influenza viruses from ducks of the atlantic flyway of north america. PloS one. 2014;9:e86999. doi: 10.1371/journal.pone.0086999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip HS, Flint PL, Franson JC, Dusek RJ, Derksen DV, Gill RE, Jr, Ely CR, Pearce JM, Lanctot RB, Matsuoka SM, Irons DB, Fischer JB, Oates RM, Petersen MR, Fondell TF, Rocque DA, Pedersen JC, Rothe TC. Prevalence of Influenza A viruses in wild migratory birds in Alaska: patterns of variation in detection at a crossroads of intercontinental flyways. Virol J. 2008;5:71. doi: 10.1186/1743-422X-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood MW, Stallknecht DE. Molecular epidemiologic studies on North American H9 avian influenza virus isolates from waterfowl and shorebirds. Avian diseases. 2007;51:448–450. doi: 10.1637/7536-032706R.1. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler AV, Pearce JM, Flint PL, Franson JC, Ip HS. Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta) Mol Ecol. 2008;17:4754–4762. doi: 10.1111/j.1365-294X.2008.03953.x. [DOI] [PubMed] [Google Scholar]
- Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 2004;4:177–189. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- Kullback S, Leibler RA. On Information and Sufficiency. The Annals of Mathematical Statistics. 1951;22:79–86. [Google Scholar]
- Lam TT, Ip HS, Ghedin E, Wentworth DE, Halpin RA, Stockwell TB, Spiro DJ, Dusek RJ, Bortner JB, Hoskins J, Bales BD, Yparraguirre DR, Holmes EC. Migratory flyway and geographical distance are barriers to the gene flow of influenza virus among North American birds. Ecol Lett. 2012;15:24–33. doi: 10.1111/j.1461-0248.2011.01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee DH, Lee HJ, Lee YN, Park JK, Lim TH, Kim MS, Youn HN, Lee JB, Park SY, Choi IS, Song CS. Evidence of intercontinental transfer of North American lineage avian influenza virus into Korea. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11:232–236. doi: 10.1016/j.meegid.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart M. Climate of the Southwest. Southwest Climate Change Network; 2008. [Google Scholar]
- Lewis NS, Javakhishvili Z, Russell CA, Machablishvili A, Lexmond P, Verhagen JH, Vuong O, Onashvili T, Donduashvili M, Smith DJ, Fouchier RA. Avian influenza virus surveillance in wild birds in Georgia: 2009–2011. PloS one. 2013;8:e58534. doi: 10.1371/journal.pone.0058534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WL, Drummond AJ. Model averaging and Bayes factor calculation of relaxed molecular clocks in Bayesian phylogenetics. Molecular biology and evolution. 2012;29:751–761. doi: 10.1093/molbev/msr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2008;8:239–246. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Pearce JM, Ramey AM, Ip HS, Gill RE., Jr Limited evidence of trans-hemispheric movement of avian influenza viruses among contemporary North American shorebird isolates. Virus Res. 2010;148:44–50. doi: 10.1016/j.virusres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular biology and evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- Rambaut A. FigTree. 2013 http://tree.bio.ed.ac.uk/software/figtree/
- Rambaut A, Suchard MA, Drummond AJ. Tracer. 2013 http://tree.bio.ed.ac.uk/software/tracer/
- Ramey AM, Pearce JM, Reeves AB, Franson JC, Petersen MR, Ip HS. Evidence for limited exchange of avian influenza viruses between seaducks and dabbling ducks at Alaska Peninsula coastal lagoons. Arch Virol. 2011;156:1813–1821. doi: 10.1007/s00705-011-1059-z. [DOI] [PubMed] [Google Scholar]
- Scotch M, Mei C, Brandt C, Sarkar IN, Cheung K. At the intersection of public-health informatics and bioinformatics: using advanced Web technologies for phylogeography. Epidemiology. 2010;21:764–768. doi: 10.1097/EDE.0b013e3181f534dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JW. Follow the Water. 2012. Looking for Birds in the Southwest? [Google Scholar]
- Spackman E, Stallknecht DE, Slemons RD, Winker K, Suarez DL, Scott M, Swayne DE. Phylogenetic analyses of type A influenza genes in natural reservoir species in North America reveals genetic variation. Virus Res. 2005;114:89–100. doi: 10.1016/j.virusres.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Squires RB, Noronha J, Hunt V, Garcia-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respi Viruses. 2012;6:404–416. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihoodbased inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Suarez DL, Garcia M, Latimer J, Senne D, Perdue M. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. Journal of virology. 1999;73:3567–3573. doi: 10.1128/jvi.73.5.3567-3573.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaro RC. Forest Service Gen Tech Report. 1980. Factors influencing bird populations in southwestern riparian forests, Management of western forests and grasslands for non-game birds USDA; pp. 403–418. [Google Scholar]
- Widjaja L, Krauss SL, Webby RJ, Xie T, Webster RG. Matrix gene of influenza a viruses isolated from wild aquatic birds: ecology and emergence of influenza a viruses. Journal of virology. 2004;78:8771–8779. doi: 10.1128/JVI.78.16.8771-8779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M, Robertson GJ, Whitney H, Bishop MA, Runstadler JA, Lang AS. Extensive geographic mosaicism in avian influenza viruses from gulls in the northern hemisphere. PloS one. 2011;6:e20664. doi: 10.1371/journal.pone.0020664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.