Abstract
Gene 6 protein of bacteriophage T7 has 5′-3′-exonuclease activity specific for duplex DNA. We have found that gene 6 protein also has flap endonuclease activity. The flap endonuclease activity is considerably weaker than the exonuclease activity. Unlike the human homolog of gene 6 protein, the flap endonuclease activity of gene 6 protein is dependent on the length of the 5′-flap. This dependency of activity on the length of the 5′-flap may result from the structured helical gateway region of gene 6 protein which differs from that of human flap endonuclease 1. The flap endonuclease activity provides a mechanism by which RNA-terminated Okazaki fragments, displaced by the lagging strand DNA polymerase, are processed. 3′-extensions generated during degradation of duplex DNA by the exonuclease activity of gene 6 protein are inhibitory to further degradation of the 5′-terminus by the exonuclease activity of gene 6 protein. The single-stranded DNA binding protein of T7 overcomes this inhibition.
Keywords: flap endonuclease, gene 6 protein, bacteriophage T7, Okazaki fragment, DNA replication
Introduction
Bacteriophage T7 gene 6 protein (gp6), a double-strand specific 5′-3′-exonuclease, is essential for phage growth.1-4 The exonuclease activity has been studied extensively and has been shown to initiate hydrolysis at the 5′-termini of duplex DNA, hydrolyzing the DNA in a non-processive manner to release nucleoside 5′-monophosphates.1,2 We recently found that gp6 also has a structure-specific 5′-flap endonuclease activity.5 The endonuclease activity depends on the length of 5′-flaps: the activity decreases as the length increases. T7 single-strand DNA binding protein (gp2.5) encoded by gene 2.5 stimulates both the flap endonuclease activity and the exonuclease activity of gp6. The question arises as to the role of the flap endonuclease activity in phage-infected cells.
The exonuclease activity of gp6 plays roles in several DNA metabolic processes in phage-infected cells. In replication, gp6 participates in the degradation of the bacterial DNA to provide nucleoside 5′-monophosphate precursors for the synthesis of T7 progeny DNA.6 In addition, the maturation of Okazaki fragments requires the removal of the 5′-terminal RNA primer, a process performed by gp6.7 Recombination in T7-infected cells is extremely efficient even in the host lacking RecA pathway.8 However, no recombinant molecules are generated when Escherichia coli is infected with bacteriophage T7 deficient in gp6.9 Most likely gp6 plays an important role in creating single-stranded regions in T7 in preparation for recombination.
We recently characterized the flap endonuclease activity of gp6.5 The major position of cleavage occurs one nucleotide into the duplex region adjacent to double-stranded/single-stranded DNA junction. Unlike eukaryotic flap endonucleases, the efficiency of flap cleavage decreases with increasing length of the 5′-overhang. A comparison of known structures from homologs of flap endonuclease provides a model as to the effect of the length of the 5′-overhang on the flap endonuclease activity of gp6.
Flap endonucleases play a variety of roles in all life forms including replication, recombination, and repair pathways. While the role of the flap endonuclease activity of gp6 in T7-infected cells is not known, one can speculate on its role based on the known role of homologous flap endonucleases in other systems. Here we discuss the potential function of the flap endonuclease activity in T7 replication and propose a model for RNA primer removal from Okazaki fragments. In addition we discuss the possible role in DNA replication of the stimulation by gp2.5 of both the exonuclease and the flap endonuclease activities of gp6.
Flap Endonuclease Activity of gp6
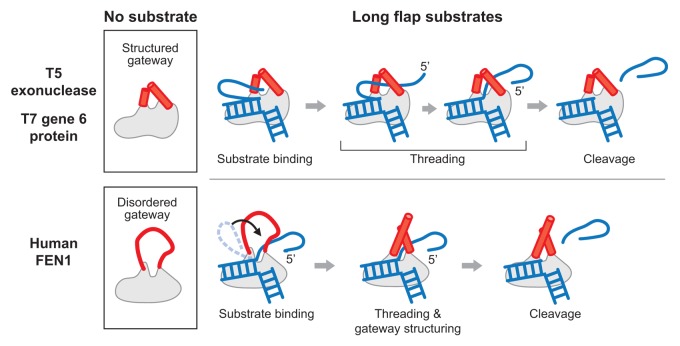
gp6 has flap endonuclease activity as well as 5′ to 3′ exonuclease activity.5 The flap endonuclease activity is considerably weaker than the exonuclease activity: the rate of hydrolysis of blunt-ended substrates ranges from 10- to 120-fold faster than that found with substrates having a 5′-overhang.5 The cleavage site for the flap endonuclease is one nucleotide into the duplex region, a site that is consistent with other flap endonuclease (FEN) homologs. The flap endonuclease activity of gp6 requires a free 5′-terminus for cleavage, as is the case for the exonuclease activity. However, the flap endonuclease activity requires the 5′-terminus to be single-stranded DNA whereas the exonuclease activity requires the 5′-terminus to be base-paired (i.e., double-stranded DNA). gp6 does not hydrolyze a flap structure if the 5′-portion of the flap is base-paired within a duplex region as is found in the gaps between Okazaki fragments. A predicted structure of gp6 generated by the program I-TASSER exhibits characteristic features shared by other FEN proteins: a helical gateway, a helical wedge, and a helix 3-turn helix motif.10-12 The gateway structure in the predicted model leads us to speculate that gp6 functions through a mechanism similar to other FEN proteins in which a 5′-flap is recognized and threaded through the gateway for cleavage. In particular, the predicted structure of gp6 is very similar to the known structure of T5 exonuclease, which also has flap endonuclease activity.13 Interestingly, the endonuclease activities of gp6 and T5 exonuclease are dependent on the length of the 5′-flap; the longer the length of the 5′-flap, the lower the flap endonuclease activity.5,14 Such dependency is not observed in eukaryote FEN proteins. Conformational changes in the gateway in these proteins may explain this dependence of 5′-flap length (Fig. 1).14-16 The eukaryote flap endonuclease homologs such as human flap endonuclease 1 (FEN1) have disordered gateway in the absence of DNA substrate, and only become structured when DNA binds to the protein (Fig. 1, lower panel). The model for the function of this class of flap endonucleases is called a disorder-thread-order mechanism.14-16 The structured gateway in flap endonucleases allows only single-stranded DNA to thread through it. The disordered state of human FEN1 has a larger aperture than those flap endonucleases that have a structured gateway, thus enabling the protein to pass long DNA flaps as a block by folding the strand or even DNA flaps containing adducts.14 As long as the size of the folded flap region exceed the size of the disordered gateway, even long flaps could pass through the gateway all at once. In contrast, the helical gateway of T5 exonuclease has a defined structure in the presence or absence of bound DNA. T5 exonuclease does not appear to undergo a conformational transition of the helical gateway, suggesting that the gateway size does not change and is just large enough to allow the passage of single-stranded DNA commencing from its 5′-terminus. Therefore, T5 exonuclease is susceptible to the length of 5′-flap compared with eukaryote homologs that have the flexible gateway (Fig. 1).14 The 5′-flap length dependency on gp6 flap endonuclease activity and its predicted structural similarity to T5 exonuclease suggest that its gateway region is similarly structured in the absence of DNA and does not change in size upon substrate binding.
Figure 1. Mechanisms of cleavage of 5′-overhangs (flaps) by bacteriophage and human homologs. (Upper panel) Phage T5 exonuclease has a structured gateway domain region on the protein in the absence of DNA substrate. The predicted structure of T7 gp6 shows high similarity to the known structure of T5 exonuclease. The 5′-flap must be threaded through the gateway from the 5′-terminus until the scissile phosphodiester bond is positioned at the active site. A longer 5′-flap would require additional time to complete the threading process. (Lower panel) In human FEN1, the gateway domain region is disordered in the absence of substrate. Thus the aperture is considerably larger than that found in the structured gateway, allowing for more efficient passage of a long flap. The gateway then becomes structured after binding the substrate with the scissile phosphodiester bond in the active site. The disordered gateway is indicated as a red line and the structured gateway as two red cylinders. The DNA is shown in blue.
The flap endonuclease activity of gp6 is stimulated by the T7 single-stranded gp2.5, as is the exonuclease activity. This stimulation by gp2.5 is dependent on a single-stranded region opposite the 5′-flap. Thus, stimulation occurs when gp2.5 can bind to a region opposite the 5′-flap. It seems likely that gp2.5 physically interacts with gp6, since the gp6 is likely bound to the base of the 5′-flap where cleavage occurs. Moreover, E. coli single-strand binding protein does not stimulate gp6, suggesting that specific interactions between gp2.5 and gp6 are required for the stimulation. When gp6 is incubated with a DNA substrate that has a 5′-flap with single-stranded DNA region opposite the 5′-flap, binding can be detected by electrophoresis mobility shift assay only in the presence of gp2.5, implying that gp2.5 is improving the affinity of gp6 to these DNA substrates.
RNA Removal
One of the well-studied functions of flap endonucleases is RNA primer removal during DNA replication.17,18 Since DNA polymerases cannot initiate DNA synthesis de novo on a template strand, the synthesis of the lagging strand at a replication fork requires preformed oligonucleotides to function as primers to initiate template-dependent DNA synthesis. DNA primases synthesize oligoribonucleotides and DNA polymerases then extend these RNA primers to initiate processive DNA synthesis. The resulting Okazaki fragments are eventually covalently joined to form a continuous lagging strand. However, prior to ligation by DNA ligase the 5′-terminal RNA must first be removed.
Several models have been proposed to account for the RNA primer removal process on the lagging strand in eukaryote systems. In one mechanism, the replicative polymerase δ continues polymerization after it encounters the downstream Okazaki fragment, resulting in displacement of the RNA primer and the formation of a 5′-flap structure. FEN1 recognizes this displaced strand and cleaves it at the base to allow Ligase 1 to join the adjacent Okazaki fragments.19,20 Under some circumstance FEN1 will fail to remove the RNA primer, in which case the multifunctional Dna2 nuclease/helicase catalyzes its removal.18,21 One such example where this event will occur is when polymerase δ displaces the Okazaki fragment to such an extent that it provides a single-stranded DNA binding site for replication protein A (RPA). When the displaced strand is bound by RPA it is no longer a substrate for FEN1. RPA recruits Dna2 and stimulates its nuclease activity to cleave a large portion of the RNA-terminated flap, up to a terminal product flap ~5–6 nucleotides in length. The remaining flap is then removed by FEN1 to produce a nick for ligation. In addition Exo1, a 5′-3′ exonuclease, can specifically act as a backup for FEN1 nuclease activity. Another mechanism has been proposed in which the RNA primers are degraded by sequential action of RNase H2 and FEN1.22,23 However, genetic deletion of RNase H in Saccharomyces cerevisiae does not lead to any phenotype, suggesting that this model involving RNase H is not the primary pathway for RNA removal in those cells.24,25 Therefore, the model described above involving FEN1 and Dna2 nuclease/helicase is thought to be the dominant pathway for RNA primer removal in eukaryotes.
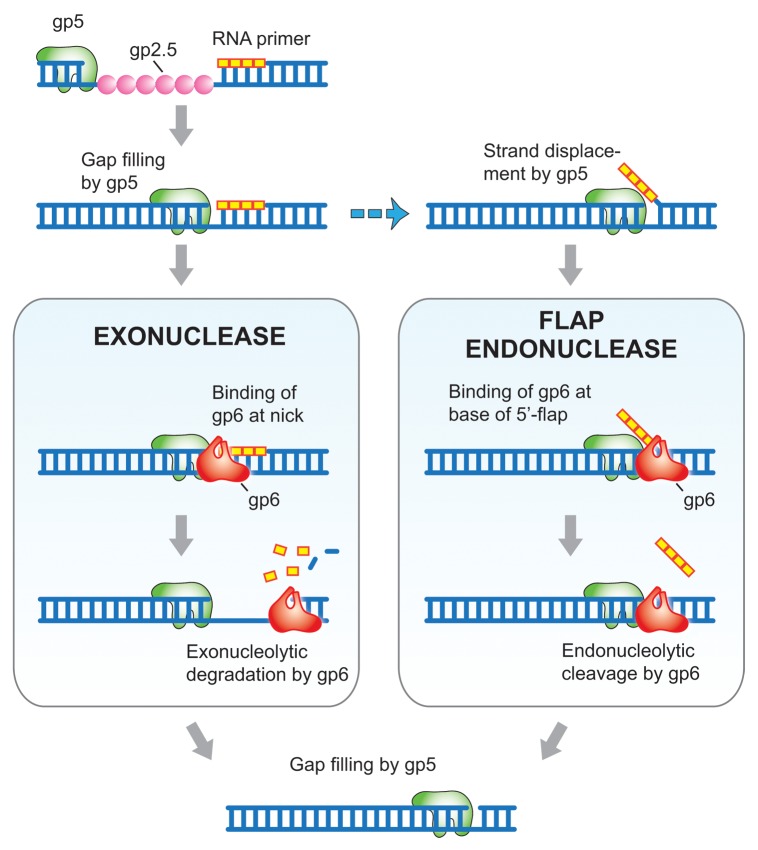
In T7 replication, gene 5 DNA polymerase (gp5) is the replicative DNA polymerase for both the leading and lagging strands. gp5 normally does not catalyze strand displacement synthesis.26 When gp5 encounters downstream duplex regions, it halts synthesis and idles as its 3′-5′-proofreading exonuclease activity removes nucleotides and they are replaced by the polymerization activity. Consequently the RNA primer on an Okazaki fragment should not normally be displaced but remain annealed to the template strand. Thus, gp6 can remove the RNA primers exonucleolytically without using its flap endonuclease activity (Fig. 2).27 Is there any possibility for a flap to occur at the 5′-terminus of an Okazaki fragment? gp5 deficient in the 3′-5′-proofreading exonuclease activity catalyzes strand displacement synthesis for several hundred nucleotides. gp5 is subject to an interesting reaction wherein reactive oxygen species selectively modify residues within the exonuclease domain provided that Fe2+ ions are bound in the Mg2+ site of the exonuclease active site.28 Whether or not this form of DNA polymerase exists in phage-infected cells is not known but the possibility does exist. Moreover, gp2.5 enables gp5 to catalyze limited strand displacement synthesis at a nick in duplex.29,30 On the lagging strand, a region within the lagging strand replication loop upstream of the preceding Okazaki fragment is single-stranded, awaiting copying by the lagging strand DNA polymerase as it completes an Okazaki fragment.31 Clearly, both gp5 and gp2.5 are in close proximity as the synthesis of the Okazaki fragment is completed. In this situation, strand-displacement synthesis by gp5 could occur. Moreover, E. coli single-strand binding protein, an abundant protein in T7 phage-infected cells, also has the ability to promote strand-displacement synthesis by gp5.29 In these situations, the RNA-primer region would be displaced by gp5 as observed in eukaryote systems and subject to the flap endonuclease activity of gp6. However, it remains likely that the processing of most Okazaki fragments occurs via the exonuclease activity of gp6, resulting in removal of the RNA primer (Fig. 2). The flap endonuclease activity of gp6 would be a backup pathway during replication: flap structures resulting from failure of proper polymerization and RNA removal are processed to maintain T7 genome integrity.
Figure 2. Model for the role of gp6 in RNA primer removal. T7 DNA polymerase (gp5 in green) fills the gap between Okazaki fragments, in the process displacing T7 single-stranded DNA binding protein (gp2.5 in pink) that is coating the single-stranded DNA template. When gp5 encounters the preceding Okazaki fragment two distinct pathways can occur: (i) The exonuclease activity of gp6 (red) degrades the RNA primer and continued hydrolysis results in the removal of some deoxyribonucleotides (left panel). (ii) Strand displacement synthesis by gp5, an event that can occur with exonuclease deficient gp5 or with exonuclease proficient gp5 in the presence of gp2.5, creates a flap containing the RNA primer (right panel). The flap endonuclease activity of gp6 cleaves the flap one nucleotide into the duplex, releasing the RNA primer. gp5 fills the gap resulting from removal of the flap and the resulting nick is sealed by T7 DNA ligase. DNA is indicated in blue and ribonucleotides are represented in yellow.
Degradation of Host DNA
The degradation of host DNA to nucleoside 5′-monophosphates following infection with T7 phage provides the precursors for deoxynucleoside 5′-triphosphates, the substrates for DNA polymerase. More than 80% of the nucleotides found in newly synthesizing T7 DNA are derived from the breakdown of the host DNA.32 Gene 3 endonuclease (gp3) carries out endonucleolytic breakdown of the host chromosome into DNA fragments.6,33 gp6 then hydrolyzes these fragments to yield nucleoside 5′-monophosphates. Inactivation of either gene 3 or 6 results in a lack of degradation the E. coli DNA after T7 infection and little DNA synthesis is observed, indicating that both proteins are required for degradation of host DNA during infection.4,6
In vitro, gp6 degrades duplex DNA non-processively from 5′-termini liberating nucleoside 5′-monophophates.1,2 gp3 hydrolyzes duplex DNA endonucleolytically and hence its nucleolytic products are predominately duplex. However, the identity of the termini of the relatively high molecular weight DNA (1 × 106) products resulting from the breakdown of E. coli DNA is not known. It seems likely that these fragments would contain both 5′ and 3′-single-stranded DNA overhangs since they originate from nicks in close proximity to one another. The flap endonuclease activity of gp6 would be indispensible for degradation of duplex DNA containing the 5′-overhangs.
After gp6 removes several nucleotides from the 5′-terminus of the host DNA, the 3′-terminus of the strand opposite of the degraded strand is exposed. We have found that such a 3′-single-stranded DNA extension inhibits gp6 nuclease activity by ~50% for the flap endonuclease activity and by ~20% for the exonuclease activity.5 Hence, the formation of 3′-extensions decrease the efficiency of degradation of these endonucleolytic products of gp3. However, gp2.5 can overcome this inhibition since it stimulates both activities of gp6. When gp6 is incubated with a 500-bp duplex DNA, the exonuclease activity is 3.5-fold more active in the presence of gp2.5 than in its absence.5 This stimulation does not occur when E. coli single-strand binding protein is substituted for gp2.5. In addition, gp2.5 lacking its C-terminal tail does not stimulate gp6 activity, implying a specific interaction occurs between these two proteins through the acidic C-terminal tail of gp2.5 (data not shown). This C-terminal tail of gp2.5 is known to interact with both T7 gp5 and T7 helicase.34
Concluding Remarks
The flap endonuclease activity of gp6 is likely to be involved in several aspects of DNA metabolism in T7-infected cells. We postulate that the flap endonuclease activity functions to remove RNA primers from Okazaki fragments when they are present in 5′-single-stranded overhangs created by strand-displacement synthesis by T7 DNA polymerase. In addition, during recombination, 5′-single-stranded regions may arise as a result from annealing of homologous strands; these structures must be removed by the flap endonuclease activity of gp6 prior to repair of the resulting gap by T7 DNA polymerase and subsequent ligation by the T7 DNA ligase. Finally, during the degradation of host DNA, the flap endonuclease activity of gp6 is necessary to prepare fragments created by gp3 endonuclease for subsequent hydrolysis of the double-stranded DNA by the exonuclease activity of gp6. Several intriguing questions remain. For example, by what mechanism does gp2.5 stimulate gp6 nuclease activity and what is the nature of their physical interaction? It is particularly interesting to examine a possible interaction of gp6 with gp5 since in many instances these two proteins function together. It is reasonable to propose that gp6, through interactions with one or more of the other T7 replication proteins, is an inherent and essential component of the T7 replisome.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Steven Moskowitz for help with preparation of figures.
References
- 1.Kerr C, Sadowski PD. Gene 6 exonuclease of bacteriophage T7. I. Purification and properties of the enzyme. J Biol Chem. 1972;247:305–10. [PubMed] [Google Scholar]
- 2.Kerr C, Sadowski PD. Gene 6 exonuclease of bacteriophage T7. II. Mechanism of the reaction. J Biol Chem. 1972;247:311–8. [PubMed] [Google Scholar]
- 3.Studier FW. The genetics and physiology of bacteriophage T7. Virology. 1969;39:562–74. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- 4.Center MS, Studier FW, Richardson CC. The structural gene for a T7 endonuclease essential for phage DNA synthesis. Proc Natl Acad Sci U S A. 1970;65:242–8. doi: 10.1073/pnas.65.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsunobu H, Zhu B, Lee SJ, Tabor S, Richardson CC. Flap endonuclease activity of gene 6 exonuclease of bacteriophage t7. J Biol Chem. 2014;289:5860–75. doi: 10.1074/jbc.M113.538611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadowski PD, Kerr C. Degradation of Escherichia coli B deoxyribonucleic acid after infection with deoxyribonucleic acid-defective amber mutants of bacteriophage T7. J Virol. 1970;6:149–55. doi: 10.1128/jvi.6.2.149-155.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinozaki K, Okazaki T. RNA-linked nascent DNA pieces in T7 phage-infected Escherichia coli cells. I. Role of gene 6 exonuclease in removal of the linked RNA. Mol Gen Genet. 1977;154:263–7. doi: 10.1007/BF00571281. [DOI] [PubMed] [Google Scholar]
- 8.Powling A, Knippers R. Some functions involved in bacteriophage T7 genetic recombination. Mol Gen Genet. 1974;134:173–80. doi: 10.1007/BF00268418. [DOI] [PubMed] [Google Scholar]
- 9.Lee M, Miller RC., Jr. T7 exonuclease (gene 6) is necessary for molecular recombination of bacteriophage T7. J Virol. 1974;14:1040–8. doi: 10.1128/jvi.14.5.1040-1048.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy A, Yang J, Zhang Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 2012;40:W471–7. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceska TA, Sayers JR, Stier G, Suck D. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature. 1996;382:90–3. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 14.Patel N, Atack JM, Finger LD, Exell JC, Thompson P, Tsutakawa S, Tainer JA, Williams DM, Grasby JA. Flap endonucleases pass 5′-flaps through a flexible arch using a disorder-thread-order mechanism to confer specificity for free 5′-ends. Nucleic Acids Res. 2012;40:4507–19. doi: 10.1093/nar/gks051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA. Unpairing and gating: sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem Sci. 2012;37:74–84. doi: 10.1016/j.tibs.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobhy MA, Joudeh LI, Huang X, Takahashi M, Hamdan SM. Sequential and multistep substrate interrogation provides the scaffold for specificity in human flap endonuclease 1. Cell Rep. 2013;3:1785–94. doi: 10.1016/j.celrep.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Balakrishnan L, Bambara RA. Flap endonuclease 1. Annu Rev Biochem. 2013;82:119–38. doi: 10.1146/annurev-biochem-072511-122603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balakrishnan L, Bambara RA. Okazaki fragment metabolism. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gloor JW, Balakrishnan L, Bambara RA. Flap endonuclease 1 mechanism analysis indicates flap base binding prior to threading. J Biol Chem. 2010;285:34922–31. doi: 10.1074/jbc.M110.165902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budd ME, Campbell JL. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci U S A. 1995;92:7642–6. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turchi JJ, Huang L, Murante RS, Kim Y, Bambara RA. Enzymatic completion of mammalian lagging-strand DNA replication. Proc Natl Acad Sci U S A. 1994;91:9803–7. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murante RS, Henricksen LA, Bambara RA. Junction ribonuclease: an activity in Okazaki fragment processing. Proc Natl Acad Sci U S A. 1998;95:2244–9. doi: 10.1073/pnas.95.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank G, Qiu J, Somsouk M, Weng Y, Somsouk L, Nolan JP, Shen B. Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J Biol Chem. 1998;273:33064–72. doi: 10.1074/jbc.273.49.33064. [DOI] [PubMed] [Google Scholar]
- 25.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 1999;19:8361–71. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lechner RL, Engler MJ, Richardson CC. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J Biol Chem. 1983;258:11174–84. [PubMed] [Google Scholar]
- 27.Engler MJ, Richardson CC. Bacteriophage T7 DNA replication. Synthesis of lagging strands in a reconstituted system using purified proteins. J Biol Chem. 1983;258:11197–205. [PubMed] [Google Scholar]
- 28.Tabor S, Richardson CC. Selective oxidation of the exonuclease domain of bacteriophage T7 DNA polymerase. J Biol Chem. 1987;262:15330–3. [PubMed] [Google Scholar]
- 29.Nakai H, Richardson CC. The effect of the T7 and Escherichia coli DNA-binding proteins at the replication fork of bacteriophage T7. J Biol Chem. 1988;263:9831–9. [PubMed] [Google Scholar]
- 30.Ghosh S, Marintcheva B, Takahashi M, Richardson CC. C-terminal phenylalanine of bacteriophage T7 single-stranded DNA-binding protein is essential for strand displacement synthesis by T7 DNA polymerase at a nick in DNA. J Biol Chem. 2009;284:30339–49. doi: 10.1074/jbc.M109.024059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Chastain PD, 2nd, Griffith JD, Richardson CC. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage t7 replication proteins. J Mol Biol. 2002;316:19–34. doi: 10.1006/jmbi.2001.5325. [DOI] [PubMed] [Google Scholar]
- 32.Calendar R. The Bacteriophages. 2nd ed. Oxford; New York: Oxford University Press, 2006 [Google Scholar]
- 33.Sadowski PD. Bacteriophage T7 endonuclease. I. Properties of the enzyme purified from T7 phage-infected Escherichia coli B. J Biol Chem. 1971;246:209–16. [PubMed] [Google Scholar]
- 34.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–43. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]