ABSTRACT
Research into rare diseases is typically fragmented by data type and disease. Individual efforts often have poor interoperability and do not systematically connect data across clinical phenotype, genomic data, biomaterial availability, and research/trial data sets. Such data must be linked at both an individual-patient and whole-cohort level to enable researchers to gain a complete view of their disease and patient population of interest. Data access and authorization procedures are required to allow researchers in multiple institutions to securely compare results and gain new insights. Funded by the European Union’s Seventh Framework Programme under the International Rare Diseases Research Consortium (IRDiRC), RD-Connect is a global infrastructure project initiated in November 2012 that links genomic data with registries, biobanks, and clinical bioinformatics tools to produce a central research resource for rare diseases.
KEY WORDS: genetics, medical informatics, databases, clinical trials, Electronic health records
RD-Connect’s primary objectives are to develop:
an integrated platform to host and analyze genomic and clinical data from research projects;
clinical bioinformatics tools for analysis and integration of molecular and clinical data to discover new disease genes, pathways, and therapeutic targets;
common infrastructures and data elements for rare disease patient registries;
common standards and catalogue for rare disease biobanks; and
best ethical practices and a proposal for a regulatory framework for linking medical and personal data related to rare disease.
RD-Connect will initially incorporate data generated by two associated projects: EURenOmics, which uses multiple approaches to focus on causes, diagnostics, biomarkers, and disease models for rare kidney disorders such as steroid-resistant nephrotic syndrome and tubulopathies; and NeurOmics, which uses novel molecular approaches to improve diagnosis and treatment of rare neurodegenerative and neuromuscular disorders such as Huntington’s disease and muscular dystrophies. RD-Connect unites existing infrastructures and integrates the latest tools to create a comprehensive platform for biobanking, data analysis, and patient registry for researchers across the world.
THE RARE DISEASE ENVIRONMENT AND THE IMPACT OF NEW TECHNOLOGIES
Because 80 % of rare diseases are thought to have a genetic component, particular emphasis has been placed on the prospects offered by the rapidly expanding development of new technologies such as genomics, transcriptomics, metabolomics, and proteomics in rare disease research.1,2 These technologies offer new paths to identify novel disease and modifier genes, delineate biomarkers, and identify therapeutic targets. However, concrete achievements from these personalized and stratified medicine approaches in rare disease have been limited, particularly when it comes to translation to therapies and the clinic.3,4
The integration of the outputs of these new technologies with detailed clinical phenotype data and the combination of data across centers and across diseases is crucial for further progress. While such integrative efforts are ongoing within some medical centers, individual efforts often remain largely siloed. This is a critical problem in rare disease studies, where a given center may see only a small number of patients with a certain disease. Linking such data sets across centers and across diseases is thus an essential step. Outside the rare disease field, a number of major research infrastructures, including the International Cancer Genome Consortium and the International Human Epigenome Consortium, have shown the feasibility of robust tools for large-scale data and sample sharing across multiple research projects.5
To address this issue, major medical research funders have come together in a global effort to foster collaboration in rare disease research. The International Rare Diseases Research Consortium (IRDiRC) was launched in 2011 and now has 35 members worldwide, including the European Commission as well as key national funders, such as several institutes in the US National Institutes of Health.6 Each of these funders has pledged to spend a minimum of US$10 million on rare disease research over 5 years. The IRDiRC has set itself two headline goals to achieve by 2020: 1) to develop the means to diagnose most rare diseases and 2) to deliver 200 new therapies for rare diseases.
In this review, we provide an overview of the objectives and initial achievements of one of the first projects to be funded under the IRDiRC. Initiated in 2012, RD-Connect is a €12 million (US$19 million) IRDiRC infrastructure project funded by the European Union’s Seventh Framework Programme.7 The project brings together 27 partner institutions and works in close collaboration with two associated research projects, Neuromics (www.rd-neuromics.eu) and EURenOmics (www.eurenomics.eu) (see Table 1). Its objectives for the 6-year funding period are to develop an integrated platform connecting databases, registries, biobanks, and clinical bioinformatics for rare disease research and to contribute to the IRDiRC goals by facilitating gene discovery, diagnostics, and therapy development.
Table 1.
Rare Neuromuscular, Neurodegenerative, and Renal Diseases: Initial Contributing Projects

www.rd-neuromics.eu |

www.eurenomics.eu |
|
|---|---|---|
| Coordinator | Professor Olaf Riess, MD, University of Tübingen, Germany | Professor Franz Schaefer, Heidelberg University Hospital, Germany |
| Research focus | Neuromuscular and neurodegenerative diseases: | Rare kidney diseases: |
| • Ataxia | • Steroid-resistant nephrotic syndrome | |
| • Congenital muscular dystrophy | • Membranous nephropathy | |
| • Congenital myasthenic syndrome | • Tubulopathies | |
| • Fronto-temporal lobe dementia | • Complement disorders | |
| • Hereditary motor neuropathies | • Congenital kidney malformations | |
| • Hereditary spastic paraplegias | ||
| • Huntington’s disease | ||
| • Muscular channelopathy | ||
| • Muscular dystrophy | ||
| • Spinal muscular atrophy | ||
| Project aims | • increase the number of patients with a genetic diagnosis | • utilize a wide array of high-throughput technologies to find new genes causing or predisposing to kidney diseases |
| • improve understanding of pathophysiology and identify drug targets | • utilize a wide array of high-throughput technologies to find new genes causing or predisposing to kidney diseases | |
| • develop biomarkers for clinical application | • characterize molecular signatures unique to individual disease entities | |
| • identify disease modifiers | • identify prognostic biomarkers, and screen for potential drug candidates | |
| • develop targeted therapies | ||
| • translate findings to other, related disease groups |
DEVELOPMENT OF A UNIFIED INFORMATICS PLATFORM FOR DATA SHARING AND ANALYSIS
Key data management challenges faced by rare disease research projects include the high complexity and heterogeneity of the data types involved, the variability among experimental platforms,8 the need to incorporate non-standardized sample descriptions and diverse data types, and the requirement to protect data (both pre-publication data and identifiable patient information). Furthermore, the high volume of data and the distributed nature of the sources make traditional approaches to data management impractical, and new solutions are therefore required.
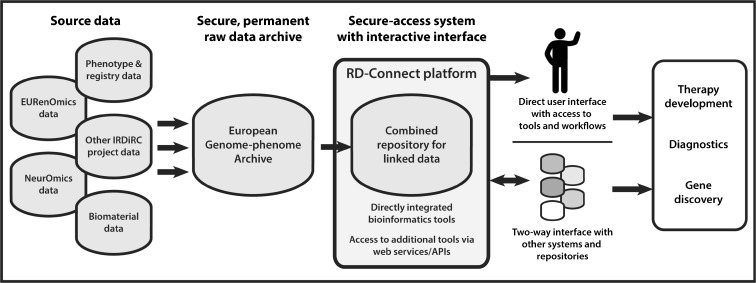
One of the primary goals of RD-Connect is to help contributing projects make their data rapidly available to the wider rare disease research community. Raw genomic data from collaborating projects is first deposited in the European Genome-phenome Archive at the European Bioinformatics Institute, a permanent resource for controlled-access archiving,9 then reprocessed in a standard manner to ensure cross-compatibility. The processed data is held in the central RD-Connect database, where it is combined with phenotypic and biomaterial information. Researchers approved by a data access committee gain secure access to an integrated data portal that enables comparison of data sets across projects; data can be explored and analyzed using a comprehensive suite of bioinformatics tools (Fig. 1).
Figure 1.
Overview of the RD-Connect integrated platform. Raw genomic data from collaborating projects, Neuromics, EURenOmics, and other IRDiRC-supported projects will be securely deposited in the European Genome-phenome Archive before being processed through a standard pipeline to ensure cross-compatibility. The processed data will be held in the central RD-Connect Data Coordination Centre, where it will be combined with other data types plus phenotypic and biomaterial information. Researchers approved by a data access committee will access data through a secure online interface that enables comparison of data sets across projects and analysis with sophisticated bioinformatics tools.
In addition to the technical aspects, any project involving aggregation of data has to contend with challenges relating to the willingness of researchers to share their results. Here, RD-Connect has the advantage that its two associated research projects, Neuromics and EURenOmics (Table 1), have been pilot users of the system. Thus, key policies such as implementation of staged embargo periods to protect results temporarily have been developed in close collaboration with these end users. Within the first year, this partnership has borne fruit: the first data sets have already been transferred, and time lines for release of the remainder have been approved. Additional ways of meeting the data-sharing challenge include initiatives such as microattributions10,11 and nanopublications12 to stimulate and reward data-sharing and knowledge-sharing and integration, with an explicit focus on interoperability, as opposed to massive centralization. Although it is the intention of RD-Connect to act as a central repository for IRDiRC research data, it will also be able to integrate with existing databases operating in the same field, such as the DECIPHER database of genomic variation data based at the UK Sanger Institute.13
Close integration with existing research resources such as those developed by the European Bioinformatics Institute, as well as the major European research infrastructures, including the Biobanking and Biomolecular Resources Research Infrastructure14 (BBMRI; http://bbmri.eu) and the European life-sciences infrastructure for biological Information15 (ELIXIR; http://elixir-europe.org), and important global developments in data sharing such as the Global Alliance for Genomics and Health16 (GA4GH; http://genomicsandhealth.org), are also crucial in this regard.
DATA ANALYSIS AND PROCESSING: CREATION OF NEW BIOINFORMATICS TOOLS
The multi-dimensional data compiled at the RD-Connect central hub affords an opportunity to analyze genetic etiology, functional molecular profiles, and patient-level phenotypes in relation to rare disease. Appropriate analysis tools are applied to this data with the immediate objective of generating a more complete picture of rare disease causes and mechanisms from the molecular to the physiological level, towards the eventual goal of improved diagnostics and therapy. Thus, RD-Connect will adapt, develop, and apply new bioinformatics tools to this uniquely rich data set. These tools will be integrated into the RD-Connect platform, piloted in analyses in collaboration with investigators from the Neuromics and EURenOmics projects, and be made broadly available in open-source release to the wider community for use in other projects.
A major challenge is to adapt existing sources of knowledge and methods of analysis to the scale of whole-exome and whole-genome sequencing or simultaneous characterization of thousands of transcripts and proteins. Linking data from different sources also requires the creation of novel approaches, as does the application of these results to clinical translation such as therapy selection, or to the identification of new therapeutic targets. Although the impact of pathogenic genetic variants on transcripts, proteins, and pathways can often be predicted based on genetic information, it is becoming increasingly clear that disease phenotypes are strongly influenced by additional genetic, epigenetic, and environmental factors. Therefore, large-scale data related to epigenomics, transcriptomics, proteomics, metabolomics, lipidomics, glycomics, phenomics, and secretomics need to be processed and made available to the rare disease field in a similar way to traditional genetic analysis. RD-Connect has begun developing methods to combine these data to facilitate gene and biomarker discovery.
The RD-Connect platform will enable a range of bioinformatics tools to be utilized on data held within the system—both tools that are being further developed within RD-Connect, and the related projects and external tools that can be linked in through common APIs (Application Programming Interface) and web services. These include variant interpretation and pathogenicity prediction systems, variant/phenotypic “matchmaking” tools, and integrative analysis tools using semantic web applications and frameworks, for improved data integration and access to knowledge.17–24
CROSS-LINKING DATA IN DATABASES AND BIORESOURCES
Historically there has been very limited cross-linking between biomaterial collections, registries, genomics, and trial data, with the exception of individual clinical research centers, where all the information may be held by a single investigator. Unfortunately, the more common situation is that the same patient is associated with multiple entries in different systems, with extensive phenotypic information available in a registry, biosamples available in a biobank, and genomic data in a research database—but without any possibility of linking the data sets. This is clearly suboptimal and at best can result in much duplication of effort, and at worst, missed opportunities for discovery, diagnosis, or treatment. Solving this data-linking problem is made more challenging by the need for strong data protection to ensure patient confidentiality. Furthermore, owing to the rarity of the conditions, rare disease patients are more likely than others to have data in cross-border repositories, and an international solution to this problem is therefore essential.
Other projects facing similar issues have evaluated solutions involving the generation of a “globally unique identifier” (GUID) from a set of personally identifiable information associated with the research participant. This allows data on a single individual to be accumulated across projects over time, regardless of where and when the data was collected, and enables researchers to define a study population using data collected in different centers. In the United States, the National Institutes of Health (NIH)-funded National Database for Autism Research (NDAR)25 has developed an identification system that is now being extended under the auspices of the NIH Office of Rare Diseases Research for use in linking patient clinical information with biospecimens. Similarly, a unique identifier for Huntington’s Disease patients taking part in international research studies has been developed.26 RD-Connect intends to implement such a system prospectively on a voluntary basis across participating registries and biobanks, to allow data sets to be cross-linked in full compliance with current data-protection policies. However, to ensure that no data from contributing projects has to be excluded, the GUID will not be a prerequisite for entry of data into the system.
GENERATION OF A COMPREHENSIVE, SEARCHABLE, ONLINE CATALOGUE FOR HUMAN RARE DISEASE BIOMATERIALS
Making biological materials and associated data from rare disease patients accessible and available to the scientific community in an internationally coordinated manner is a core aim of the RD-Connect platform. This initiative is based on collaboration between the two major relevant biobanking infrastructures in Europe: BBMRI, which historically has focused primarily on population biobanks, and EuroBioBank.27 EuroBioBank has historically been composed principally of neuromuscular biobanks, but will extend to incorporate rare disease biobanks of all types.
Within the first year, RD Connect investigators have begun a mapping exercise to ensure outreach to all biobanks holding biomaterials related to rare diseases. Progress has also been made towards the development of a new online interface for a rare disease biomaterial catalogue, including primary cells, tissue, DNA, serum, RNA, and human-induced pluripotent stem cell lines. This will also include information related to biobanking standards, including an overview of major existing standards and guidelines for biobanking and “Minimal Information Standards” for sample collections.
The design of existing databases, registries, and biobanks does not often allow the sharing of information in a computer-accessible fashion. For this reason, participating resources are being offered the opportunity to make use of linked data and semantic web approaches: computational standards and methods that enable them to make their data more accessible and interoperable.28
USE AND FURTHER DEVELOPMENT OF PHENOTYPE ONTOLOGIES
It is increasingly recognized that advances in sequencing technology do not replace the need for detailed clinical assessment of patients with rare disease. On the contrary, deep phenotyping is more important than ever in order to interpret whole-exome and whole-genome sequencing results. However, where clinical notes are on paper systems in hospitals, or where clinicians enter free text in electronic systems, the power of computation cannot be leveraged to support analysis. Ontologies are structured representations of knowledge using a standardized, controlled vocabulary for data integration, organization, searching, and analysis. To ensure the searchability of the data in the central system, RD-Connect makes use of ontologies of both phenotypic features (signs and symptoms of diseases) and diseases and disease groups (disease classifications or nosologies).
With over 10,000 classes (terms) describing human phenotypic abnormalities and over 13,000 subclass relations between the classes, together with extensive annotation and cross-referencing with other ontologies, the Human Phenotype Ontology (HPO)29 is a leading example of a phenotypic ontology. Phenotypic data contributed by the associated research projects, Neuromics and EURenOmics, is being captured using the HPO, enabling standardized cross-cohort comparisons and filtering, as well as implementation of algorithms for automated “matchmaking” to help find cases with clinically similar presentations and variants in the same gene. Collaboration between RD-Connect, Neuromics, and the developers of the PhenoTips30 software has enabled development of user-friendly online forms for clinicians to enter disease-specific phenotypic information for neuromuscular and neurodegenerative diseases using the HPO. As part of the collaboration with HPO developers, terminology workshops with expert clinicians are planned to augment the HPO (still under active expansion) with further phenotypic classes. For rare disease classification, RD-Connect will also use the Orphanet Rare Disease Ontology,31 a nosology system cross-referenced with ICD10, HPO, and other systems.
GENERATION OF COMMON DATA ELEMENTS AND STANDARD OPERATING PROCEDURES FOR PATIENT REGISTRIES
To enable standardized aggregation of patient information, in addition to the use of standardized ontologies, it is helpful to establish common data elements for data collection. Work in this area builds on existing protocols and best-practice recommendations produced by the leading database and registry initiatives represented within RD-Connect, including the disease-specific networks established for neuromuscular diseases,32 cystic fibrosis,33 and Huntington’s disease,34 the recommendations of the European Platform for Rare Disease Registries project, and the rare disease common data elements developed and published by the NIH’s Office of Rare Diseases Research,35 all of which will be leveraged to provide an initial framework for participating registries. Best practices for information collection by biomaterial collections have been developed by established biobanks such as EuroBioBank,36 the Telethon Network of Genetic Biobanks,37 and BBMRI, and their further development will also be promoted by connection with the BioMedBridges initiative, whose mission statement includes “Building data bridges and services between biological and medical infrastructures in Europe”, and which comprises the ten biological and medical sciences research infrastructures selected by the European Strategic Forum for Research Infrastructures.
DEVELOPING SOLUTIONS TO ETHICAL AND REGULATORY ISSUES
Ethical issues surrounding sharing of sensitive personal data have to be dealt with robustly in a project of this nature. Data-protection and ethics-approval mechanisms must be taken seriously, but solutions must not excessively hinder research. In rare diseases, this is a particular issue when differences in national procedures can pose significant barriers. For example, in a recent trial for juvenile dermatomyositis, the participation of 103 clinical centers was needed to recruit 130 patients, and the ethics approval process took 2 years.
Working closely with the regulators at a European and global level, RD-Connect is developing recommendations to overcome such hurdles, recommending a risk-based approach to ethical review38 that simplifies the process for information-based research. The risks associated with research that uses genetic information, stored biospecimens, or information from databases, medical registries, patient records, and questionnaires are not physical, but informational, e.g., related to unauthorized release of information. In such cases, a more expedient review process is suggested, paving the way for a simplified procedure for data-sharing research. On a practical level, a rare disease data-sharing charter and standardized templates for informed consent procedures are being developed. Patient issues and stakeholder inclusion are also recognized as central, and a patient-centered approach ensures patient views are taken into account across all aspects of the work. This is coordinated by a 16-member Patient Advisory Council made up of patient representatives from associated projects and patient organizations.
CHALLENGES AND ISSUES TO BE ADDRESSED
A number of challenges facing the RD-Connect project are summarized in Table 2. While some of the technical and scientific challenges are specific to this project, others such as economic, ethical, societal, regulatory, and political issues apply to rare disease research in general.
Table 2.
Challenges and Issues to Overcome while Developing the RD-Connect Infrastructure
| Area | Challenge | Strategies to address challenge |
|---|---|---|
| General | Re-inventing the wheel and failing to capitalize on previously invested resources | Global collaboration with partners with existing high-quality databases, registries, biobanks, and programs. |
| Registries and biobanks | Fragmentation of patient data | Collaborate with other organizations and databases/registries without duplicating efforts; for prospective data and sample collection the use of de-identified identifiers will allow the connection of data from different sources and organizations. |
| Registries and biobanks | Lack of patient participation in registries and poor endorsement by patients and patient organizations for the donation of biological materials | Feedback to patients by the provision of information on the research resulting from data and samples. Engagement of patients and patient organizations through open consultation, data protection, and ethical conduct and by invitations of key representatives in relevant meetings. |
| Registries and biobanks | Possible resistance of databases/registries/biobanks to adapt to new common data elements and the Rare Disease-Identification (RD-ID) | Provide different stages of implementation of the common data element, starting form the most common element. |
| Registries and biobanks | Difficulties in protecting patient identity in de-identified databases/registries/biobanks, especially those suffering from ultra-rare diseases | Develop standard operating procedures for data protection and appropriate informed consent forms to provide patients with full information. |
| Platform development/Ethical | Implementation of a unique identifier (RD-ID) creates ethical and data-protection difficulties | Build on existing highly robust systems for data sharing with privacy protection that are already in use in projects of a similar complexity and implement an appropriate level of data encryption and access control. Develop and publish guidelines on data protection within the project. Gather appropriate informed consents at all stages. |
| Platform development | Highly variable data types are to be integrated into the platform | High level of flexibility built in; agreement on data standards and operating procedures. |
| Ethical/social | Lack of willingness of participating researchers and organizations to share data | Implement mechanisms for protecting unpublished data and for “incentivization” of data sharing already developed by partners (e.g., microattributions, database journals). Develop a strategy in collaboration with IRDiRC funders to ensure that data sharing becomes a condition of getting the grant. |
| Impact | Delay in implementation of “ -omics” knowledge into new diagnostic and therapeutic advances for rare disease patients | Communication through website, publications, annual conference and training activities. |
CONCLUSION
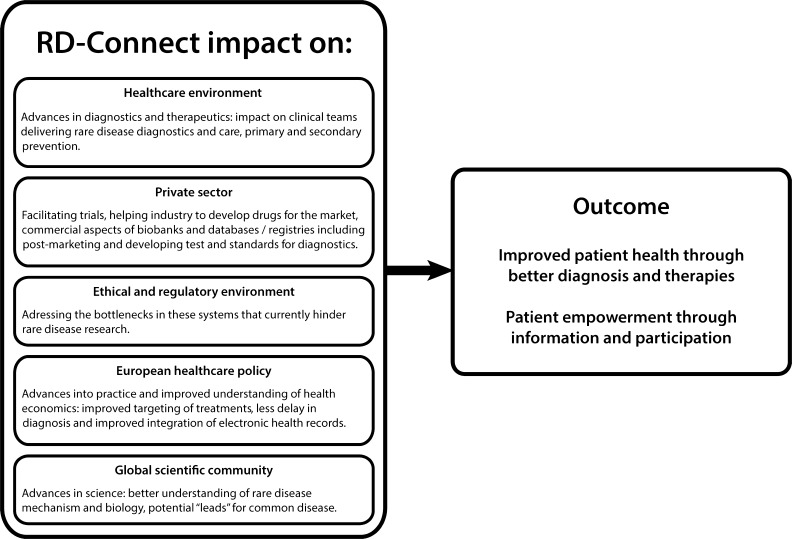
Patient registries,39 biobanks,40 and bioinformatics support are key infrastructure tools required for genomic research in rare disease; data sharing and linking of patients, samples, and analysis is also essential. The infrastructure developed by RD-Connect supports research in rare disease to find new genes, biomarkers, and therapeutic targets more quickly and efficiently. Its ultimate goal will be to improve outcomes for rare disease patients via major improvements in diagnostics and therapeutics (Fig. 2). The therapeutics market in rare disease has strong growth potential due to the high (and unmet) medical need for most rare diseases. Genomic research and development will thus be highly relevant for many markets, including genetic testing, biomarkers, and therapeutics. The 2011 Orphanet report on rare disease research41 noted that networking initiatives resulting in easy and secure access to resources (databases and registries, biobanks, reference data sets, and analysis tools) and a close working relationship with patient groups are clear predictors of success for translational efforts in rare disease. As the field evolves and embraces the opportunities of the new technologies, there will be further challenges relating to access to sufficient patient numbers, plus sufficient high-quality biological samples annotated with harmonized ontologies and associated with detailed molecular data, analyzed by standardized analysis pipelines. RD-Connect will enable the elucidation of pathways relevant across rare diseases and identify shared therapeutic targets for groups of rare and common disorders. Ultimately, it will enable cross-linking and efficient distribution of quality-controlled data to the rare disease research community in a secure ethical and legal framework, which is crucial for achieving the IRDiRC goals.
Figure 2.
Impact of RD-Connect
Acknowledgements
The authors would like to acknowledge the contribution of the RD-Connect scientific advisory board, chaired by Bartha Maria Knoppers, as well as all partners and associated partners in RD-Connect. Further acknowledgement is due to Olaf Riess, Neuromics coordinator; Franz Schaefer, EURenOmics coordinator; and all Neuromics and EURenOmics partners contributing data to RD-Connect. We thank the PhenoTips developers for collaboration on standardized phenotypic data entry forms. The efforts of the IRDiRC Executive and Scientific Committees in policy and guideline development are appreciated. The work leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement No. 305,444 (RD-Connect), 305,121 (Neuromics) and 305,608 (EURenOmics). TREAT-NMD was funded under EU FP6 contract number 036,825 (2007 to 2011) and TREAT-NMD Operating Grant - Second Public Health Programme (contract number 2012 3,307).
Conflict of interest
The authors declare that they do not have a conflict of interest.
REFERENCES
- 1.Gut IG. New sequencing technologies. Clin Transl Oncol. 2013;15(11):879–81. doi: 10.1007/s12094-013-1073-6. [DOI] [PubMed] [Google Scholar]
- 2.Aymé S., Rodwell C., eds., 2013 Report on the State of the Art of Rare Disease Activities in Europe, July 2013.
- 3.Bushby K, Lochmüller H, Lynn S & Straub V. Interventions for muscular dystrophy: molecular medicines entering the clinic. Lancet 374, 2009;28:1849–56. [DOI] [PubMed]
- 4.Tremblay JP, Xiao X, Aartsma-Rus A, et al. Translating the genomics revolution: the need for an international gene therapy consortium for monogenic diseases. Mol Ther. 2013;21(2):266–8. doi: 10.1038/mt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The International Cancer Genome Consortium. International network of cancer genome projects. Nature 464, 2010;15:993–98. [DOI] [PMC free article] [PubMed]
- 6.International Rare Disease Research Consortium (IRDiRC) Policies and Guidelines [pdf]. Available at: http://www.irdirc.org/wp-content/uploads/2013/06/IRDiRC_Policies_Longversion_24May2013.pdf. Accessed April 1, 2014.
- 7.Commission E. Rare diseases—How Europe is meeting the challenges. Luxembourg: Publications Office of the European Union; 2013. [Google Scholar]
- 8.’t Hoen PA, Friedländer MR, Almlöf J. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31(11):1015–22. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 9.Church DM, Lappalainen I, Sneddon TP, et al. Public data archives for genomic structural variation. Nat Genet. 2010;42(10):813–4. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groth P, Gibson A, Velterop J. The anatomy of a nanopublication. Information Services and Use. 2010;30:51–6. [Google Scholar]
- 11.Patrinos GP, Cooper DN, van Mulligen E, Gkantouna V, Tzimas G, Tatum Z, Schultes E, Roos M, Mons B. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Hum Mutat. 2012;33(11):1503–12. doi: 10.1002/humu.22144. [DOI] [PubMed] [Google Scholar]
- 12.Giardine B, Borg J, Higgs DR, et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat Genet. 2011;43(4):295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84(4):524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuille M, van Ommen GJ, Bréchot C, et al. Biobanking for Europe. Brief Bioinform. 2008;9(1):14–24. doi: 10.1093/bib/bbm050. [DOI] [PubMed] [Google Scholar]
- 15.Crosswell LC, Thornton JM. ELIXIR: a distributed infrastructure for European biological data. Trends Biotechnol. 2012;30(5):241–2. doi: 10.1016/j.tibtech.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Editorial. Open to interpretation. Nat Biotechnol. 2013;31 (8):661. [DOI] [PubMed]
- 17.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v. 2.0: the next generation in gene variant databases. Hum Mutat. 2011;32(5):557–63. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 18.Tuffery-Giraud S, Béroud C, Leturcq F, et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30(6):934–45. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 19.Frédéric MY, Lalande M, Boileau C, et al. UMD-predictor, a new prediction tool for nucleotide substitution pathogenicity – application to four genes: FBN1, FBN2, TGFBR1, and TGFBR2. Hum Mutat. 2009;30(6):952–9. doi: 10.1002/humu.20970. [DOI] [PubMed] [Google Scholar]
- 20.Robinson P, Köhler S, Oellrich A et al. Improved exome prioritization of disease genes through cross species phenotype comparison. Genome Res. 2013 Oct 25. [E-pub ahead of print]. [DOI] [PMC free article] [PubMed]
- 21.Brudno M, Gîrdea M, Buske O et al. PhenomeCentral: An Integrated Portal for Sharing and Searching Patient Phenotype Data for Rare Genetic Disorders. Figshare. http://dx.doi.org/10.6084/m9.figshare.939458
- 22.Hunter AA, Macgregor AB, Szabo TO, Wellington CA, Bellgard MI. Yabi: An online research environment for grid, high performance and cloud computing. Source Code Biol Med. 2012;7(1):1. doi: 10.1186/1751-0473-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopes P, Oliveira JL. COEUS: “semantic web in a box” for biomedical applications. J Biomed Semantics. 2012;3(1):11. doi: 10.1186/2041-1480-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopes P & Oliveira JL. An innovative portal for rare genetic diseases research: The semantic Disease card. J Biomed Inform. 2013; 21. [epub ahead of print] [DOI] [PubMed]
- 25.Hall D, Huerta MF, McAuliffe MJ, Farber GK. Sharing heterogeneous data: the national database for autism research. Neuroinformatics. 2012;10(4):331–9. doi: 10.1007/s12021-012-9151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orth M, Handley OJ, Schwenke C, Dunnett SB, Craufurd D, Ho AK, Wild E, Tabrizi SJ, Landwehrmeyer GB, the European Huntington’s Disease Network Tio. Observing Huntington’s Disease: the European Huntington’s Disease Network’s REGISTRY. PLOS Currents Huntington Disease. 2010 Sep 28. Edition 1 [DOI] [PMC free article] [PubMed]
- 27.Lochmüller H, Aymé S, Pampinella F, et al. The Role of Biobanking in Rare Diseases: European Consensus Expert Group Report. Biopreservation and Biobanking. 2009;7(3):155–56. doi: 10.1089/bio.2010.7302. [DOI] [PubMed] [Google Scholar]
- 28.Roos M, Marshall MS, Gibson AP et al. Structuring and extracting knowledge for the support of hypothesis generation in molecular biology. BMC Bioinformatics. 2009;1;10 Suppl 10:S9 [DOI] [PMC free article] [PubMed]
- 29.Köhler S, Doelken SC, Mungall CJ. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucl. Acids Res. 2013 doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girdea M, Dumitriu S, Fiume M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat. 2013;34(8):1057–65. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 31.Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33(5):803–8. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 32.Bushby K, Lynn S, Straub V. Collaborating to bring new therapies to the patient—the TREAT-NMD model. Acta Myol. 2009;28(1):12–15. [PMC free article] [PubMed] [Google Scholar]
- 33.McCormick J, Mehta G, Olesen HV, Viviani L, Macek M, Jr, Mehta A. European Registry Working Group. Comparative demographics of the European cystic fibrosis population: a cross-sectional database analysis. Lancet. 2010;375(9719):1007–13. doi: 10.1016/S0140-6736(09)62161-9. [DOI] [PubMed] [Google Scholar]
- 34.Tabrizi SJ, Scahill RI, Owen G, et al. Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington’s disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013;12(7):637–49. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 35.Global Rare Diseases Patient Registry and Data Repository. CDE Overview. Available at: https://grdr.ncats.nih.gov/index.php?option = com_content&view = article&id = 3&Itemid = 5 Accessed 15 April 2014
- 36.Lochmüller H, Schneiderat P. Biobanking in rare disorders. Adv Exp Med Biol. 2010;686:105–13. doi: 10.1007/978-90-481-9485-8_7. [DOI] [PubMed] [Google Scholar]
- 37.Filocamo M, Baldo C, Goldwurm S, et al. Telethon Network of Genetic Biobanks: a key service for diagnosis and research on rare diseases. Orphanet J Rare Dis. 2013;8(1):129. doi: 10.1186/1750-1172-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansson MG, van Ommen GJ, Chadwick R, Dillner J. Patients would benefit from simplified ethical review and consent procedure. Lancet Oncol. 2013;14(6):451–3. doi: 10.1016/S1470-2045(13)70129-3. [DOI] [PubMed] [Google Scholar]
- 39.Bellgard M, Beroud C, Parkinson K, et al. Dispelling myths about rare disease registry system development. Source Code Biol Med. 2013;8(1):21. doi: 10.1186/1751-0473-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wichmann HE, Kuhn KA, Waldenberger M, et al. Comprehensive catalog of European biobanks. Nat Biotechnol. 2011;29(9):795–7. doi: 10.1038/nbt.1958. [DOI] [PubMed] [Google Scholar]
- 41.Orphanet Report Series: Disease Registries in Europe. January 2013. Available at. http://www.orpha.net/orphacom/cahiers/docs/GB/Registries.pdf . Accessed 15 April 2014.