Summary
Stimulation of transcriptional elongation is a key activity of leukemogenic MLL fusion proteins. Here we provide evidence that MLL-ENL also inhibits polycomb-mediated silencing as a prerequisite for efficient transformation. Biochemical studies identified ENL as scaffold that contacted the elongation machinery as well as the PRC1 (polycomb repressive complex 1) component CBX8. These interactions were mutually exclusive in vitro corresponding to an antagonistic behavior of MLL-ENL and CBX8 in vivo. CBX8 inhibited elongation in a specific reporter assay and this effect was neutralized by direct association with ENL. Correspondingly MLL-ENL defective in CBX8 binding could not fully activate gene loci necessary for transformation. Finally, we demonstrate dimerization of MLL-ENL as neomorphic activity that may augment polycomb inhibition and transformation.
Introduction
MLL fusions are highly efficient oncoproteins that transform hematopoietic progenitors and cause aggressive leukemia (Slany, 2009). These proteins are derived from chromosomal translocations that affect the MLL locus at 11q23 joining an N-terminal portion of the H3K4 histone methyltransferase MLL with a variety of different partner proteins. These replace the original methyltransferase activity contained within the MLL C-terminus and create potent transactivators that cause the inappropriate expression of target genes. Trithorax, the MLL homolog in fly acts as positive regulator of the clustered Hox-homeobox genes. In analogy also MLL fusions induce a strong overexpression of HOX, MEIS and PBX homeobox genes with the latter two coding for HOX-binding partners. Elevated levels of HOX/MEIS/PBX are sufficient to transform hematopoietic progenitor cells and deregulation of homeobox genes is mainly responsible for the oncogenic activity of MLL derivatives.
Remarkably, MLL chimeras generally do not behave as classical activators recruiting RNA Polymerase II (RNA PolII). Depending on the fusion partner they seem to affect either chromatin associated processes or more frequently they specifically stimulate transcriptional elongation. MLL partners of the ENL (ENL, AF9) and AFF (AFF1-4) families form a higher order protein complex originally named EAP (elongation assisting proteins) that was purified from nuclear extracts (Mueller et al., 2007; Mueller et al., 2009). Next to ENL/AF9 and AFF proteins (AFF1 and AFF4 are also known as AF4 and AF5q31 or short AF5) EAP also included positive transcription elongation factor b (PTEFb) and the H3K79 histone methyltransferase DOT1L. P-TEFb is a dimer of CDK9 and a cyclinT that phosphorylates RNA PolII at serine 2 within the C-terminal repeat domain. Additional substrates are proteins like NELF (negative elongation factor) and DSIF (DRB sensitivity-inducing factor) that help to keep RNA PolII stalled shortly after initiation. These modifications catalyzed by P-TEFb are a crucial prerequisite for efficient elongation of pre-initiated transcripts (Peterlin and Price, 2006). DOT1L introduces methylation of lysine79 in histone H3, a modification associated with actively transcribed chromatin. Interestingly, DOT1L was first discovered in yeast where H3K79 serves as an “anti-silencing” modification that inhibits invasion of heterochromatin into transcribed areas (Nguyen and Zhang, 2011). EAP-related complexes have been isolated by several laboratories (Bitoun et al., 2007; Monroe et al., 2011; Yokoyama et al., 2010) and some studies suggest that EAP can be separated into two subcomplexes with different functions. A super elongation complex (SEC) stimulates elongation by recruiting P-TEFb together with other elongation factors and a separate DOT1L-complex (DotCom) is responsible for chromatin modification [reviewed in (Smith et al., 2011)]. SEC may be widely involved in transcriptional control as it has also been copurified with the HIV Tat protein that is known to support viral transcription by stimulation of elongation (He et al., 2010; Sobhian et al., 2010). Although EAP is unequivocally connected to active transcription, paradoxically proteins normally associated with polycomb repressive complex 1 (PRC1) have been repeatedly demonstrated to interact and copurify with EAP components (Garcia-Cuellar et al., 2001; Hemenway et al., 2001; Monroe et al., 2011; Mueller et al., 2007).
Originally polycomb proteins were identified in Drosophila as opponents of trithorax function. The balance between trithorax-mediated activation and repression by polycomb dynamically regulates the transcriptional output of many genes, particularly those involved in self-renewal, differentiation, and developmental decisions with HOX genes as paradigmatic example. Because trithorax as well as polycomb activities involve chromatin modification the corresponding marks become heritable and constitute part of what has been termed “epigenetic memory”. In mammals this function has been conserved [reviewed in (Margueron and Reinberg, 2011)]. Major representatives of mammalian polycomb proteins can be found in two different protein complexes. Polycomb repressive complex 2 (PRC2) contains the conserved histone methyltransferase EZH2 (enhancer of zeste homolog 2) that introduces H3K27 di- and trimethylation whereas polycomb repressive complex 1 (PRC1) catalyzes histone H2A ubiquitination via the RING1/2 E3 ligases. These enzymes are accompanied by a variable set of associated factors that include PCGF (polycomb group ring finger), PHC (polyhomeotic homolog), and CBX (chromobox) proteins (Gao et al., 2012). Chromobox proteins are chromatin readers that recognize and bind to methylated H3K27. Therefore a sequential mechanism was suggested where PRC2 deposits a repressive mark that is subsequently read and interpreted by PRC1. However, also PRC2 independent recruitment of PRC1 has been demonstrated (Dietrich et al., 2012; Yu et al., 2012). Despite intensive studies it is not yet completely clear how PRC complexes actually repress. Both chromatin compaction (Eskeland et al., 2010; Gao et al., 2012) and inhibition of transcription by ubiquitinated H2A (Stock et al., 2007; Zhou et al., 2008) seem to be involved.
Here we investigated the reason for the counterintuitive copurification of PRC1 components with elongation associated proteins and demonstrate that polycomb mediated repression can be squelched by direct interaction of the ENL and CBX8 proteins. Polycomb proteins have been shown to colocalize with basal transcription factors at loci “poised” for transcription (Oguro et al., 2010; Taberlay et al., 2011) and it was speculated that PRC may block transcription after the initiation step. This would mandate neutralization of PRC function before efficient elongation can occur. We provide a likely mechanism that can achieve that effect and thus may serve as a potential therapeutic target in the context of MLL fusion proteins.
Results
ENL binds to PRC1 through CBX8
The PRC1 components CBX8 and RING1 have been previously copurified with ENL (Mueller et al., 2007). To confirm these data and to identify the subtype of PRC1 that interacts with ENL we purified CBX8 and the accompanying protein complex from HEK293 cells, the source of the original identification of EAP (figure 1). These cells were transduced with flag-tagged CBX8 and isolation of CBX8 interacting proteins was performed by tandem immunoprecipitation of nuclear extracts using sequential pull-down with flag- and CBX8-agarose. Non-transduced HEK293 cells served as control. Mass spectrometry identified all classical PRC1 components from eluted bands as visualized by silver staining (figure 1A, B). Consistent with a recent study classifying PRC1 subtypes (Gao et al., 2012) CBX8 occurred mainly in PRC1.2 and PRC1.4 i.e. in MEL18 and BMI1 containing PRCs. Although in substoichiometric amounts, ENL was consistently copurified with PRC1 in three independent experiments. The interaction of ENL and CBX8 could also be corroborated by standard immunoprecipitation (figure 1C). Anti-flag precipitated material from fCBX8 expressing HEK293 cells contained immunologically detectable ENL. Vice versa CBX8 could be identified by western blot in fENL precipitates.
Figure 1. Purification and molecular architecture of PRC1.
A: Silver stained gel of a typical PRC1 preparation. Nuclear extracts from HEK293 cells transduced with a flag-tagged version of CBX8 were used for tandem immunoprecipitation as schematically indicated in panel A. Non-transduced cells served as controls. The gel is a typical example of three independent experiments.
B: Mass spectrometric identification of CBX8 copurifying proteins. Not listed are heat shock proteins HSPA1/8/9 that were also present in the immunoprecipitates.
C: Co-precipitation of CBX8 and ENL. Flag-reactive material was precipitated from nuclear extracts of HEK293 expressing either flag-CBX8 (upper panel) or flag-ENL (lower panel). Presence of ENL and CBX8 was detected by western blotting. Wild-type HEK293 lysates were used as controls.
D: Typical example of a two-hybrid experiment screening for direct interactions between full-length CBX8 (bait) and PRC1 members (prey). Growth on control and selective (-histidine) plates is shown.
E: Mapping of the ENL and RING interaction domains in CBX8. CBX8 constructs as indicated were tested as in “D”. Growth on selective plates is indicated by “+”.
F: Expression of GAL4 constructs used for two-hybrid experiments. Yeast lysates were analyzed by immunoblotting with anti-GAL4 AD antibodies (upper panel) and anti-GAL4 DNA binding domain reagents (lower panels).
G: Schematic depiction of PRC1-EAP interactions.
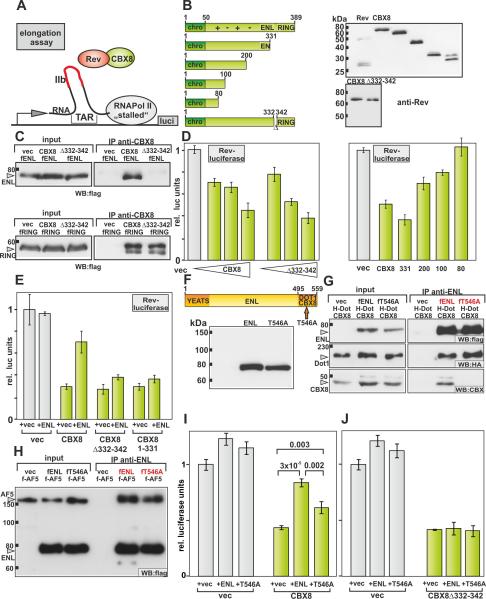
Little is known about direct protein contacts within PRC1. In order to see how ENL would fit into this interaction network we identified PRC1 components that make a direct contact to CBX8. Full length versions of the major PRC proteins were tested in a two hybrid system with CBX8 (figure 1D) or various CBX8 deletion mutants (figure 1E) as baits. Constructs were correctly expressed in yeast (figure 1F) and did not show any endogenous transactivation (not shown). Next to the known interaction with ENL, direct binding of CBX8 was observed with both RING proteins (RING1, RING2) but with none of the other PRC1 components tested. Mapping of the respective interaction domains revealed two separable regions at the CBX8 C-terminus that independently mediated binding to ENL and RING (figure 1E). A small deletion of amino acids 335 to 340 was sufficient to disrupt ENL interaction but this did not affect affinity for RING proteins. In summary, CBX8 can establish a connection between ENL and PRC (figure 1G).
CBX8 can bind ENL and RING simultaneously but ENL allows only mutually exclusive interactions
Because ENL and RING bind to CBX8 at closely neighbored sites we wanted to know if these interactions can occur at the same time. Likewise it had been shown that all direct binding partners of ENL [AF5/4 (AFF4/AFF1), DOT1L, and CBX8] contact overlapping or immediately adjacent motifs in ENL (He et al., 2011; Mueller et al., 2009). Therefore it was not clear if CBX8 could interact with ENL once it is occupied by other EAP components. To clarify these questions an elaborate set of immunoprecipitations was performed (figure 2). Various combinations of differently tagged proteins were coexpressed in 293T cells to test mutual binding in the presence of a third protein competing for the same binding site. For CBX8 a clear picture emerged indicating that the individual interactions of CBX8 (figure 2A) with ENL and RING1 can occur simultaneously. CBX8 could bridge ENL with RING suggested by coprecipitation of RING1 with ENL (figure 2A, left panels) and vice versa (figure 2A, middle panels) in the presence of CBX8. CBX8 itself contacts both proteins (figure 2A, right panels). In contrast binding of ENL to its interaction partners was only possible one at a time. Although an association of CBX8 and Dot1l with ENL could be confirmed separately (figure 2B, right panels) ENL could not “bridge” the two proteins suggesting that these contacts are not coincident (figure 2B, left and middle panels). A similar result was obtained with AF5 (figure 2C). Again CBX8 and AF5 could associate with ENL individually (figure 2C right panels) but not simultaneously (figure 2C left and middle panels). In summary these results can be best reconciled with a scaffolding function of ENL that allows only one contact at any given moment such enforcing a sequential order of events. This goes along well with previous results showing that AF5 and DOT1L also exclude each other in binding to ENL (Yokoyama et al., 2010).
Figure 2. Analysis of concurrent protein interactions.
A: Simultaneous binding of ENL and RING1 to CBX8. Tagged (f = flag, H = HA) and untagged versions of CBX8, ENL, and RING1 were cotransfected as indicated. Proteins targeted for precipitation are marked in red font.
B: Dot1l and CBX8 cannot bind simultaneously to ENL. No coprecipitation of Dot1l and CBX8 was observed (left and middle panels) despite the fact that ENL binds to both proteins individually (right panels). Labeling of tags and proteins are as in “A”
C: Binding of AF5 (AFF4) and CBX8 to ENL is mutually exclusive. CBX8 could not precipitate AF5 in the presence of ENL (left, middle panels) yet ENL interacts with both proteins in the same lysates (right panels). Labeling is like in “A”.
PRC1 and MLL-ENL induce opposing activities in vivo
The in vitro results suggested the possibility that CBX8/PRC1 and ENL/EAP activities may influence each other. There is no direct biological assay for ENL activity in vivo. However, in the context of an MLL fusion protein ENL function reads out as transformation capability. In order to investigate the biological consequences of CBX8/PRC1 mediated repression for cellular transformation we overexpressed CBX8 in Mll-ENL immortalized cells. Because artificially high concentrations of MLL-ENL may skew the balance and therefore affect the outcome of this experiment we chose to use a recently published (Takacova et al., 2012) Mll-ENL-ER knock-in model (Meer mice) for this test. In Meer animals an inducible Mll-ENL fusion has been created in the germ line by knock-in of ENL joined to the ligand binding domain of the estrogen receptor. Therefore Mll-ENL-ER is expressed under control of the endogenous Mll promoter (figure 3A). Isolated bone marrow progenitors from these mice can be immortalized by simple addition of tamoxifen leading to the outgrowth of permanent cell lines. To study the effect of elevated CBX8 on Mll-ENL mediated transformation, Meer cells were transduced with a pMSCV based expression construct for CBX8 leading to a large increase of CBX8 RNA and protein as compared to vector transduced controls (figure 3B). Meer/CBX8 cells were viable and could be propagated for several weeks in culture. In a quantitative assessment by CFC assays, however, they consistently showed reduced replating capability forming fewer colonies in methylcellulose (figure 3C). Phenotypically these cells displayed higher level of the differentiation marker Gr-1 on the surface indicative for a weakened transformation by Mll-ENL (figure 3D). This was mirrored at the molecular level by lower RNA concentrations of the key Mll-ENL targets Hoxa9 and Meis1 (figure 3E). In contrast to CBX8, overexpression of RING1 elicited only minor effects in Meer cells indicating that CBX8, but not RING1 is a limiting factor (supplemental figure S1).
Figure 3. Mutual inhibition of MLL-ENL and PRC1 in vivo.
A: Schematic depiction of the Mll-ENL-ER (Meer) knock-in construct. Meer bone marrow progenitors can be immortalized by simple addition of tamoxifen. pA = polyA sequence, neo = neomycin resistance.
B: Left panel: Detection of CBX8 overexpression by qRT-PCR. Meer cells were infected either with empty viruses or a viral expression construct for CBX8. Q-PCR primers were chosen that amplify mouse and human CBX8 sequence. Given are means and standard deviations of a triplicate and these data represent one out of three experiments with similar outcome.
Right panel: Immunological detection of CBX8/Cbx8 in extracts of Meer cells transduced as before. Epitope tagged human CBX8 is about 7kDa larger than the endogenous mouse protein. Ten times more total protein was loaded per lane for the vector control.
C: CBX8 overexpression reduces CFC capacity of Meer cells. Hematopoietic progenitor cells from Meer bone marrow were transduced with CBX8 or control virus. Replating assays were performed in the presence of tamoxifen. The upper panel shows a representative example of third round colonies, the bar graph charts relative colony numbers as average and standard deviation of 6 independent experiments.
D: CBX8 induces higher levels of the differentiation marker Gr-1 on the cell surface. Meer cells from the experiments shown in “C” were analyzed for Gr-1 expression by FACS analysis. The percentage of Gr-1 positive cells was calculated using the indicated region.
E: CBX8 overexpression reduces Hoxa9 and Meis1 expression in Meer cells. Q-RT PCR was performed on total RNA isolated from Meer cells transduced as indicated. Averages and standard deviations of a technical triplicate are given, representing a typical example out of three experiments in total.
F: ChIP for elongation associated chromatin modifications and marker proteins. Meer progenitor cells were transduced with vector (grey bars) or CBX8 (green bars). ChIP for modifications and factors was done as indicated. Experiments were performed in the presence of tamoxifen (dark bars, +TAM) and and 72 h after tamoxifen was removed (light bars, -TAM). Precipitation is given as % input. Unspecific IgG served as control (red hatched bars). The chart illustrates averages and standard deviations of PCR triplicates. The ChIP experiment was done on three biological replicates with essentially the same results.
G: Detection of polycomb-associated chromatin modifications. ChIP was performed as described for “F”.
H: Presence of CBX8 and Mll-ENL as detected by ChIP.
See also figure S1
To study the molecular events occurring at the respective genomic loci, ChIP experiments were done around the Hoxa9 and Meis1 transcriptional start sites (figure 3F-H). In cells with active Mll-ENL (+ tamoxifen) that overexpress CBX8 (figure 3F) a drop of four different elongation markers was observed corresponding to the reduced transcription of Hoxa9 and Meis1. H3K79me2, H3K36me2, RNA PolII serine-2 phosphorylation and ENL were moderately but consistently reduced 20% to 50% compared to vector controls. Interestingly, no significant increase of H2A ubiquitination as readout for PRC1 activity was observed in CBX8 cells as long as tamoxifen was present (figure 3G, left panel). H3K27 methylation as one potential recruiting element for CBX8 was equally present in transduced and control cells (figure 3G, right panel). The observed reduction in Hoxa9 and Meis1 transcripts in CBX8 cells was not due to decreased Mll-ENL binding as this parameter remained unchanged or even slighly increased after overexpression of CBX8 (figure 3H, left panel). Rather the reduction in transcription was correlated with a two to three-fold increase in chromatin bound CBX8 protein (figure 3H, right panel).
To investigate this phenomenon we repeated the ChIP experiments 72h after inactivation of Mll-ENL by removal of tamoxifen. As it has been shown before (Milne et al., 2005) this leads to cellular differentiation and in parallel to an exit of Mll-ENL from its target loci. As a consequence elongation markers were almost completely lost with exception of H3K36 dimethylation that remained detectable at this time point (figure 3F). Remarkably, inactivation of Mll-ENL lead to a large increase of PRC1 and also PRC2 activity exclusively in CBX8 cells whereas this effect was almost negligible in controls (figure 3G). This was not correlated with the levels of chromatin bound CBX8 (figure 3H, right panel) as additional overexpression of CBX8 in differentiating Mll-ENL cells (-TAM) even led to a reduction of ChIP-detectable CBX8 bound to the locus. In contrast, there was an inverse association with remaining Mll-ENL (figure 3H, left panel). that exited earlier from the Hox/Meis loci in CBX8 cells compared to vector controls and corresponding to the more advanced state of differentiation in CBX8 cells. This suggests that Mll-ENL has to fall below a certain threshold before PRC1 can become active supporting a suppressive role of Mll-ENL on CBX8 and polycomb function.
CBX8 bound by ENL loses repressor activity in an elongation reporter assay
Because ENL and MLL-ENL mainly stimulate transcriptional elongation, we wanted to assess the consequences of the ENL/CBX8 interaction with respect to this parameter. Elongation can be specifically measured with a specialized reporter assay (Rev-assay) that makes use of the fact that the HIV LTR is known to be controlled mostly after initiation has occurred (Gold and Rice, 1998). A luciferase-based reporter system (figure 4A) driven by a modified HIV LTR contains an engineered binding motif (SLIIb loop) for the RNA-binding protein Rev. This loop is located within the short RNA produced after RNA PolII initiates transcription. In that way any protein of interest can be recruited to the paused RNA polymerase allowing readout of either stimulating or repressive activity. A series of Rev-CBX8 mutants including deletions of the C-terminal RING binding domain and a CBX8 derivative without the ENL interaction motif (CBX8Δ332-342) were constructed and tested for correct expression (figure 4B). Coimmunoprecipitations confirmed that the CBX8Δ332-342 mutant has lost the capacity to bind ENL whereas the interaction with RING was untouched (figure 4C). Both CBX8 and CBX8Δ332-342 demonstrated a clear, concentration dependent repressor activity in Rev-assays (figure 4 D, left panel). This indicates a CBX8 intrinsic inhibitory function that does not require binding to ENL. Corroborating a previous study (Grau et al., 2011) the CBX8 encoded repressor function was not contingent on RING binding as a loss of the RING binding domain in the CBX8_1-331 construct did not affect its inhibitory activity (figure 4D, right panel). Rather the repressor function relied on an extended, highly charged region within the central portion of the protein. ShRNA experiments were also consistent with a RING independent repressor activity for CBX8 (supplemental figure S2).
Figure 4. In vitro inhibition of transcriptional elongation by CBX8 is blocked by direct contact with ENL.
A: Schematic description of the Rev elongation reporter system. A modified HIV LTR drives a luciferase reporter gene. The Tat-interacting TAR mRNA loop is modified to contain a SLIIb recognition site for the RNA binding protein Rev. Rev-fusion proteins are directed to the RNA bound, “stalled” RNA Polymerase II. In this way any influence on elongation can be specifically read out by alterations of luciferase activity.
B: CBX8 mutants tested in Rev-assays. Left panel: Representation of various C-terminal CBX8 deletions. The N-terminal chromobox (chro) and the central charged region (+-), as well as the ENL and RING binding domains are labeled. Right panel: Anti-Rev western blot of Rev-CBX8 derivatives.
C: An 11 amino acid deletion in CBX8 selectively abrogates ENL binding. CBX8 and a CBX8 mutant missing amino acids 332 to 342 were tested in co-immunoprecipitation for their interaction with ENL (upper panel) and RING1 (lower panel).
D: Repression of transcriptional elongation by CBX8 is independent of ENL and RING and relies on a charged region. Increasing amounts of Rev-CBX8 and Rev-CBX8Δ332-342 constructs were cotransfected together with elongation reporter into 293T cells (left panel). To determine the repressor domain in CBX8 a series of C-terminal deletion mutants was tested in the same assay (right panel).
E: Direct interaction with ENL neutralizes the CBX8-induced repression of elongation. CBX8, CBX8Δ332-342 (ENL binding site deletion) and a truncated CBX8_1-331 deleting both ENL and RING1 interaction domains were cotransfected together with reporter and an expression construct for ENL or a vector control as before.
F: A point mutation that had been shown to disable interaction of the ENL homolog AF9 with CBX8 was introduced at the corresponding residue at the C-terminus of ENL where binding sites for Dot1l and CBX8 overlap.
G: The ENLT546A mutant abrogates CBX8 binding but leaves interaction with Dot1l intact. ENL, ENLT546A and a vector only control were expressed together with Dot1l and CBX8. Precipitation of ENL indicated that the interaction with Dot1l is unharmed whereas CBX8 association is abrogated in ENLT546A.
H: Coimmunoprecipitation of WT-ENL and ENLT546A with AF5. Similar affinities of WT-ENL and the CBX8 binding defective point-mutant ENLT546A were observed in these experiments.
I: ENLT546A does not efficiently rescue CBX8 mediated repression. ENL, ENLT546A or a vector control were cotransfected with CBX8 and the elongation reporter. Whereas some activity remains for ENLT546A, this mutant is significantly weaker than WT ENL in rescuing CBX8 mediated repression of elongation (p values were calculated by a Student's T-test, n=3).
J: Combining ENLT546A with the CBX8Δ332-342 mutant completely abolishes any effect of ENL on CBX8 induced repression.
See also figures S2 and S3
Interestingly, CBX8-mediated repression was largely neutralized by coexpression of ENL (figure 4E). This phenomenon was absolutely reliant on a direct ENL/CBX8 interaction as ENL-binding defective CBX8 mutants remained repressors even in the presence of ENL. In an attempt to confirm these results in a reciprocal fashion an ENL mutant was constructed that has lost CBX8 affinity but keeps all other interactions intact. Because DOT1L and CBX8 bind to coinciding regions in ENL we chose to test a point mutant that has been found to specifically abrogate CBX8 binding in the homologous AF9 protein (Tan et al., 2011). The corresponding T546A exchange was introduced into ENL and correct protein expression was confirmed by western blotting (figure 4F). Interactions between DOT1L and ENLT546A were unaffected but affinity for CBX8 was no longer detectable in Co-IP experiments under stringent washing conditions (figure 4G). Interaction with AF5 also remained intact (figure 4H). In contrast to WT-ENL, ENLT546A was significantly weaker in “rescuing” transcriptional elongation from CBX8-mediated repression (figure 4I). The remaining activity of ENLT546A was likely due to residual binding to CBX8 that was not completely abrogated by introduction of the single point mutation. A complete ablation of the ENL/CBX8 interaction by using an ENL-binding defective CBX8 mutant (CBX8Δ332-342) fully eliminated the ENLT546A induced effect on CBX8 induced repression (figure 4J). Because ENLT546A retained some affinity for CBX8 we tried to introduce an additional amino acid exchange (T534A) that has been demonstrated to be important for CBX8 binding in AF9. The introduction of the second mutation, however, also reduced affinity for Dot1l (supplemental figure S3) and therefore this construct was not tested further.
MLL-ENL interaction with CBX8 is required for efficient transformation
To assess the in vivo consequences of interfering with CBX8 binding, the ENLT546A mutation was introduced into an MLL-ENL context (figure 5A). The single amino acid change did not affect expression of the protein, yet this mutant blunted the transforming capacity of the respective MLL-ENL fusion. Primary hematopoietic progenitors transduced with MLL-ENLT546A formed about 70% fewer colonies than those transformed by the WT counterpart (figure 5B). In addition these cells could not be propagated in liquid culture for more than 2 weeks with terminal differentiation and proliferation arrest eventually prevailing (not shown). Concomitant with the weaker replating efficiency endogenous concentrations of Hoxa9 and Meis1 transcripts were significantly lower in MLL-ENLT546A cells compared to controls (figure 5C). H3K79 dimethylation was reduced but nevertheless present at Hoxa9 and Meis1 loci confirming that DOT1L still can interact with the MLL-ENLT546A mutant. H2A ubiquitination was slightly to moderately increased in MLL-ENLT546A cells and H3K27 methylation was barely detectable in this retroviral overexpression model (figure 5D). These data can be best reconciled with an inability of MLL-ENLT546A to make the target gene loci sufficiently permissive for transcription. This occurs despite the fact that retroviral transduction leads to overexpression of the MLL fusion.
Figure 5. MLL-ENL needs interaction with CBX8 for efficient transformation.
A: Graphical representation and expression of the MLL-ENL construct containing the T546A mutation in the ENL portion. Wild-type and MLL-ENLT546A were expressed at equal levels in phoenix packaging cells. *= unspecific band.
B: Reduced CFC capability of MLL-ENLT546A transformed cells. Upper panel: Representative example of third round colonies formed by progenitors transduced by MLL-ENL or MLL-ENLT546A viruses as indicated. Lower panel: Aggregated results (average and standard deviation) of 3 biological replicates.
C: Expression of Hoxa9 and Meis1 in MLL-ENLT546A transduced cells is reduced. QRT-PCR was done on RNA isolated from cells after two rounds of replating.
D: Histone patterns are shifted towards repression in MLL-ENLT546A cells. Chromatin was isolated from cells transduced either with MLL-ENL or MLL-ENLT546A after two rounds of replating and subject to ChIP as indicated. Enrichment is given as percent of input. Bars depict the average and standard deviation of a PCR triplicate and this experiment was done twice with essentially the same result.
Global knock-down of Cbx8 blunts transformation by MLL-ENL
Because MLL-ENL must overcome a CBX8 (PRC1) induced barrier to immortalize cells, we speculated that global reduction of this protein might aid in transformation. To test this assumption, Meer cells were transduced with a shRNA construct targeting Cbx8 or with a control vector. After antibiotic selection the cells were screened for surface marker expression and for gene expression at two different time points. Concordant with our hypothesis, knock-down of Cbx8 initially (11 days after transduction) caused a shift towards cells with lower Gr-1 levels indicating a stronger block in differentiation. However, this phenomenon was short-lived and the knock-down effect was lost after further culture (figure 6A). This transient phenotype was correlated with a rebound of Cbx8 from approximately 50% suppression at day 11 to nearly normal levels at day 19 (figure 6B) indicating outgrowth of cells that have lost shRNA expression. A potential reason for this observation became apparent when we checked the transcripts of the Cdkn2 (Ink4) tumor suppressor family (Cdkn2a, b, c, d) that are a known PRC1 targets. In line with previous findings (Dietrich et al., 2007) knock-down of Cbx8 was inversely correlated to the concentrations of Cdkn2a and Cdkn2b, two powerful inhibitors of cell cycle progression that are transcribed from the same locus. Concordant and in parallel with the surge in Cdkn2a/b transcription a pronounced but transient G1 arrest was observed in Cbx8 knock-down cells (figure 6C). This phenomenon was correlated to a reduced plating efficiency of the respective cells in methylcellulose (figure 6D). Thus de-repression of the Cdkn2a/b genes is a likely explanation for the growth disadvantage of cells with globally reduced Cbx8 concentrations.
Figure 6. Global knock-down of Cbx8 is incompatible with transformation by MLL-ENL.
A: Surface Gr-1 marker expression on Meer cells transduced with a control vector or a shRNA plasmid targeting Cbx8. Overlay FACS plots are shown of cell populations after 11 days and 19 days of selective culture and the percentage of Gr-1 positive cells is given.
B: Knock-down of Cbx8 induces tumor suppressor genes. RNA was harvested from control and Cbx8 knock-down cells at 11 days and 19 days of culture as indicated. Expression of Cbx8 and the Cdkn2 family was determined by qRT-PCR. The insert shows a Cbx8 specific immunoblot.
C: Cell cycle analysis of Cbx8 knockdown cells. Meer cells containing a Cbx8 shRNA (blue line) or vector only cells (black line) were analyzed for the cell cycle phase by propidiumiodide (PI) staining 11 days and 19 days after transduction.
D: Numerical evaluation of CFC assays performed at early (11 days) and late passage (19 days). Cells were plated in triplicates into cytokine supplemented methocel at the indicated time points and colonies were enumerated 4 to 5 days later.
MLL-ENL can dimerize to allow simultaneous occurrence of normally separated processes
The coimmunoprecipitation results and the mutational analysis suggested that under normal circumstances binding of CBX8 and DOT1L/P-TEFb to ENL cannot occur simultaneously. This presumably allows for regulation and may control the extent of transcriptional stimulation. Nevertheless, genes activated by MLL fusion proteins are at the same time hypermethylated at H3K79 (Krivtsov et al., 2008), highly transcribed and not blocked by PRC1. This suggests that ENL may circumvent normal control mechanisms in the context of the fusion. To investigate the molecular basis of this effect we probed for dimerization of MLL-ENL because this would allow for recruitment of elongation factors despite the presence of CBX8 (PRC1). In addition it is known that fusions of MLL with strong dimerization domains are weakly transforming (Martin et al., 2003; Xia et al., 2003). To check for self-association of MLL-ENL, flag- and HA-tagged versions of this protein were coexpressed. Precipitation with a flag-specific antibody (figure 7A, left panels) clearly brought down also HA-tagged MLL-ENL. This was not due to an unspecific association with DNA as all extracts were extensively digested with benzonase to remove interfering nucleic acids. Unexpectedly, dimerization was not contingent on the ENL fusion partner. The amino-terminal MLL moiety up to the fusion point at aa 1444 could replace full length MLL-ENL as precipitating agent without loss of efficiency (figure 7A, right panels). A deletion analysis identified two regions within MLL that were responsible for dimer formation. The first self-association domain coincided with the AT-hook motif at the very MLL N-terminus (figure 7B) and the second comprised the CxxC domain further C-terminal (figure 7C). Remarkably both domains could also heterodimerize (figure 7D). Homo- and heterodimerization capabilities could be separated. N-terminal portions were necessary for homodimerization of the AT-hook and the CxxC motif whereas C-terminal parts of the respective peptides were sufficient for heterologous interaction (supplemental figure S4). Homo- and heterodimer formation could also be demonstrated by GST-pulldown (supplemental figure S4). Purified GST-fusions of the AT-hook and CxxC peptides efficiently interacted with their tagged counterparts in nuclear extracts. Dimerization of MLL-ENL with WTMLL was not possible under indentical conditions (figure 7E). WT-MLL is postranslationally processed and forms a dimer of the respective MLLN and MLLC moieties. In cells coexpressing untagged WT-MLL (=MLLN and MLLC after processing within the cell) and flag-tagged MLL-ENL immunoprecipitation with an antibody that binds to MLLN brought down the MLLN-interacting MLLC (figure 7E lower panel). In contrast the same procedure done with anti-flag precipitated only flagMLL-ENL (figure 7E, middle panel) but not MLLC (figure 7E, upper panel) indicating that the fusion protein does not interact with the MLLN/MLLC dimer.
Figure 7. Dimerization of MLL-ENL.
A: MLL-ENL dimerizes through the N-terminal MLL moiety. Flag-tagged and HA-tagged MLL-ENL as depicted were coexpressed and nuclear extracts were precipitated with anti-flag agarose (left panels). Dimerization was not dependent on the ENL fusion partner as an N-terminal MLL moiety (aa 1-1444) was sufficient to induce self-association (right panels). ME = MLL-ENL, f = flag, numbers in subscript indicate the last amino acid of C-terminal deletion mutants. The protein/peptide targeted for precipitation is labeled in red font.
B: The AT-hook motif dimerizes. Co-immunoprecipitations were done as in “A” with two differently tagged, N-terminal subregions of MLL comprising amino acids 1 to 331. Note that IgG heavy chain is detected at around 50kDa.
C: The CxxC domain contains a second dimerization domain. Amino acids 1146 to 1337 of MLL containing the CxxC core and flanking regions were differentially tagged and coprecipitation was done as above.
D: The AT-hooks and CxxC domain heterodimerize. Co-immunoprecipitations were done with AT-hooks and either a CxxC peptide as in “C” or a fragment thereof (aa1146 to 1252) containing only the core CxxC motif and a downstream basic region.
E: MLL-ENL does not dimerize with WT-MLL. Flag-tagged MLL-ENL was co-expressed together with WT-MLL or empty vector as control. Full length MLL is subject to natural post-translational processing yielding a stable dimer of MLLN (300kDa) and MLLC (180kDa) fragments. The MLLC product can be detected with specific antibodies. Anti-flag antibodies brought down substantial amounts of MLL-ENL but no MLLC (upper and middle panel) whereas antibodies against MLLN that recognize WT-MLL as well as MLL-ENL could successfully coprecipitate MLLC under the same conditions.
F: Dimerization of MLL-ENL allows for simultaneous binding of CBX8 and Dot1l. CBX8, Dot1l, and either ENL or MLL-ENL were coexpressed and precipitation was done with a CBX8 specific antibody.
G: MLL-ENL bridges CBX8 to AF5. Experiment was done as in “F” probing for Co-IP of CBX8 and AF5 in the presence of ENL or MLL-ENL.
See also figure S4
In contrast to monomeric ENL, the MLL-ENL fusion was able to connect CBX8 with Dot1l as well as CBX8 with AF5 in precipitation assays. This is in line with a capability for di/multimer formation that is unique to the MLL fusion protein and that is absent in ENL (figure 7F,G).
Discussion
The balance between trithorax/MLL and polycomb activity is an important factor determining the output of a transcriptional unit. Here we reveal another aspect of this “ying-yang” relationship providing evidence that stimulation of transcription by the EAP activator complex and polycomb mediated repression are rivaling activities with ENL and CBX8 as key regulators. Inhibition of PRC1 adds yet another function to the repertoire of MLL fusion proteins contributing to their potent transforming potential and it explains the consistent copurification of repressor proteins with ENL.
Because polycomb complexes encode enzymatic activities that may be potentially “druggable,” the role of PRCs in hematological malignancies, and cancer in general, is of great interest. With respect to MLL fusions, two recent studies have shown that global reduction of PRC2 function by conditional knock-out of essential components impairs MLL-AF9 induced leukemogenesis in vivo (Neff et al., 2012; Tanaka et al., 2012). Loss of PRC2 lead to a widespread de-repression of genes involved in cell cycle control and differentiation. This included known tumor suppressors like Cdkn2a as well as genes inducing maturation as for example Egr1. In addition, a transformation associated “Myc-gene expression module” was suppressed in PRC2 knock-out cells. These findings are consistent with our results after general knock-down of Cbx8 and similar observations have been made also in solid tumors. In normal cells CBX8 bypasses senescence by direct binding to the INK4A-ARF region (Dietrich et al., 2007). In general de-repression of tumor suppressors is the rationale for clinical attempts to use EZH2 inhibitors as therapeutics. However, it has been known for a long time that also the oncogenic HOX loci are under polycomb control and here we show that inhibition of polycomb-mediated repression contributes towards transformation in MLL-fusion induced leukemia.
Therefore the situation in malignant disease is more complex and artifical interference with repressor activities may have unwanted side-effects even exacerbating oncogene activity. Reflecting this dualism it seems logical that EZH2 mutations in hematological malignancies may either enhance or destroy the catalytic activity depending if tumor suppressors or oncogenes play the major role in the transformation process (Hock, 2012). With respect to novel treatments for MLL-induced leukemia it would seem more advisable to target the interaction of MLL-fusions with CBX8 directly, rather than intend to inhibit global PRC-activity. In particular patient cells that have lost the CDKN2 tumor suppressor locus, an event occurring frequently in leukemia (Sulong et al., 2009), may respond unexpectedly to such a generalized “epigenetic” therapy.
Interestingly MLL fusion proteins have evolved different strategies to overcome polycomb-induced repression. A recent report (Tan et al., 2011) showed that MLL-AF9 evokes a “moonlighting” function in CBX8 by using it as an intermediate to recruit the histone acetylase TIP60. In this way MLL-AF9 induced local histone acetylation that was associated with HOX expression and transformation. Our results suggest MLL-ENL works differently as we could not find any evidence for an interaction of ENL or CBX8 with TIP60 either by biochemical purification or in direct Co-IP attempts (not shown). This is not without precedent as ENL and AF9 are homologs but they are not identical. For example ENL copurifies with CDK9/CYCT2 (Mueller et al., 2007), but AF9 associates with the alternative CDK9/CYCT1 conformation of P-TEFb (Monroe et al., 2011).
Unfortunately, not much is known about the molecular details of polycomb-mediated repression. It would be tempting to speculate that part of the repressor activity of CBX8 may be due to interference with ENL-function by physically displacing other ENL-bound factors. The structural basis for mutually exclusive binding was explored in a recent publication for AF9 (Leach et al., 2012). Interaction of AF9 with various proteins is mediated by an intrinsically disordered domain, that “adapts” different conformations depending on the respective binding partner and thus restricting interaction to a single protein at any time. However competition cannot be the sole reason for repression by CBX8 because CBX8 without an ENL binding domain still acts as repressor. Therefore alternative mechanism(s) must exist. An attractive possibility described previously (Grau et al., 2011) is chromatin compaction that can occur in vitro and in vivo. This process has been demonstrated to be a prerequisite for silencing at HOX loci (Eskeland et al., 2010). Compaction is independent of histone modification and can be induced by various polycomb group proteins that carry highly charged regions like those present within the central portion of CBX8. In this respect it is interesting to note that a feature of CBX8, a 16-fold repeat of a dipeptide with alternating charge (DR/ER repeat), is present also in negative elongation factor E (NELF-E). NELF-E is responsible for stalling RNA PolII by binding to nascent RNA and this block is released by phosphorylation through P-TEFb (Gilchrist et al., 2012).
Finally we provide yet another example for a potential role of dimerization in transformation, a feature that has been observed for many oncoproteins (So and Cleary, 2004). ENL complexes control transcription also in normal cells and the sequential order of events that is enforced by mutually exclusive binding of the ENL interaction partners would be a perfect opportunity for regulation of this process. The high transcriptional output seen at MLL-ENL controlled loci in leukemic cells suggests a bypass of this mechanism. This could be achieved by dimerization that allows multiple simultaneous interactions. The ability to dimerize seems to be restricted to the fusion proteins as MLLENL did not interact with WT-MLL. Unfortunately, at present the exact importance of dimerization for MLL-ENL mediated transformation cannot be experimentally tested because the regions in the CxxC domain that mediate self-association are also responsible for binding to the PAF (Polymerase associated factor) complex, a necessary cofactor for all MLL fusion proteins (Milne et al., 2010; Muntean et al., 2010). In this regard it is important to mention that the add-on of a strong dimerization motif alone is sufficient to convert the MLL N-terminus to a weakly transforming protein. This was shown experimentally (Martin et al., 2003; Xia et al., 2003) and it is as well suggested by the numerous rare translocation partners that encode for cytoplasmatic proteins with dimerization domains. Dimerizing MLL fusions would circumvent normal control circuits and recruit more PAF complex. PAF, in turn, interacts with SEC and therefore ENL (He et al., 2011). It is tempting to speculate that fusion partners either increase intrinsic dimerization to offset the indirect ENL interaction or they provide direct access to ENL/SEC which makes endogenous dimerization suffice.
Experimental procedures
Plasmids, cell culture, animals, antibodies
CDNAs used for cloning are listed in supplemental methods. Retroviral packaging was done in the Phoenix-E packaging line (Swift et al., 2001). Protein expression and precipitation were performed in 293T cells. Primary hematopoietic progenitors were isolated from Meer mice that carry a knock-in of an inducible ENL-ER fusion that is joined to genomic Mll sequences reconstituting an Mll-ENL protein analogous to human leukemia derived samples. For a complete description of the Meer model see (Takacova et al., 2012). Culture conditions for Meer cells are given in the supplement.
Purification of PRC1 complex
PRC1 was purified by tandem affinity precipitation of tagged CBX8 essentially as described before for EAP (Mueller et al., 2007). In short HEK293 cells were stably transduced with a flag-CBX8 construct. Nuclear extracts from these cells were precipitated with immobilized flag-agarose, bound material was eluted by addition of flag-peptide and a second precipitation was done with anti-CBX8 agarose. Final precipitates were eluted by acid treatment (100mM glycin, pH 2.9) and analyzed by gel-chromatography, silver staining as well as mass spectrometry. Similarly treated extracts from non-transduced HEK293 cells served as controls. Purification was done on 3 independent biological samples.
Interaction studies: two-hybrid, co-immunoprecipitation, GST-pulldown
Two-hybrid analysis was performed according to standard procedures exactly as described (Garcia-Cuellar et al., 2009).
For co-immunoprecipitation studies tagged and native versions of the proteins were expressed in 293T cells. EAP as well as PRC1 are endogenously present in these cells therefore normal interactions should be able to form. In case subregions of proteins without an endogenous nuclear localization sequence were used a bona fide NLS was fused to the N-terminus. The detailed procedures for IP and GST pulldown can be found in the supplementary section.
Chromatin immunoprecipitation, qPCR
Chromatin immunoprecipitation (ChIP) was performed with formaldehyde crosslinking according to a standard protocol (Milne et al., 2009) with the modification that magnetic proteinG beads (Diagenode, Liege, Belgium) were used instead of agarose coupled proteinG. Evaluation of ChIP was done by qPCR in technical triplicates on at least two to three different biological samples. The primers used for qPCR of ChIP precipitates were designed to amplify a region immediately downstream of the respective transcription start sites and they are listed in supplemental materials together with antibody sources.
Rev-elongation assays
Elongation was quantified by a special reporter system as developed by Gold and Rice (Gold and Rice, 1998). In summary, this reporter uses a modified HIV LTR that has been engineered to contain the sequence of the SLIIb stem-loop Rev binding structure. Proteins of interest can be recruited through a fusion with Rev to RNA and therefore they are brought into the vicinity of a RNA polymerase stalling at the known LTR pause point. Influence on elongation will readout as luciferase activity. Because the elongation machinery is universally expressed, experiments were done in 293T cells.
Supplementary Material
Highlights.
- The MLL fusion partner ENL copurifies with polycomb repressive complex 1
- A direct interaction of ENL with CBX8 blocks polycomb repressive activity
- Inhibition of polycomb is necessary for MLL-ENL induced transformation
- Dimerization of MLL-ENL can combine polycomb-inhibition with stimulation of elongation
Acknowledgments
We thank Renate Zimmermann for technical assistance. This work was supported by research funding from DFG (Deutsche Forschungsgemeinschaft) grant SL27/7-1 and co-financed by the Bavarian Ministry of Sciences, Research and the Arts in the framework of the Bavarian Molecular Biosystems Research Network to RKS. JLH is supported by a Specialized Center of Research Grant from the Leukemia and Lymphoma Society of America. VD was supported by the Czech Ministry of Education (NPV2B06077, MSM6198959205), and in part by the Palacky University Institutional Funding (LF_2012_16). EM, MPGC, CB and RKS performed and analyzed experiments, ST and VD contributed the Meer animals, JH provided access to mass-spectrometry and helped to interpret data. RKS designed research and wrote the paper.
Footnotes
The authors have no conflict of interest to declare.
References
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007;26:1637–1648. doi: 10.1038/sj.emboj.7601632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N, Rappsilber J, Sodersten E, Hansen K. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012;8:e1002494. doi: 10.1371/journal.pgen.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cuellar MP, Mederer D, Slany RK. Identification of protein interaction partners by the yeast two-hybrid system. Methods Mol Biol. 2009;538:347–367. doi: 10.1007/978-1-59745-418-6_18. [DOI] [PubMed] [Google Scholar]
- Garcia-Cuellar MP, Zilles O, Schreiner SA, Birke M, Winkler TH, Slany RK. The ENL moiety of the childhood leukemia-associated MLL-ENL oncoprotein recruits human Polycomb 3. Oncogene. 2001;20:411–419. doi: 10.1038/sj.onc.1204108. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MO, Rice AP. Targeting of CDK8 to a promoter-proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res. 1998;26:3784–3788. doi: 10.1093/nar/26.16.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Chan CK, Sobhian B, Chou S, Xue Y, Liu M, Alber T, Benkirane M, Zhou Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc Natl Acad Sci U S A. 2011;108:E636–645. doi: 10.1073/pnas.1107107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway CS, de Erkenez AC, Gould GC. The polycomb protein MPc3 interacts with AF9, an MLL fusion partner in t(9;11)(p22;q23) acute leukemias. Oncogene. 2001;20:3798–3805. doi: 10.1038/sj.onc.1204478. [DOI] [PubMed] [Google Scholar]
- Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26:751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach BI, Kuntimaddi A, Schmidt CR, Cierpicki T, Johnson SA, Bushweller JH. Leukemia Fusion Target AF9 Is an Intrinsically Disordered Transcriptional Regulator that Recruits Multiple Partners via Coupled Folding and Binding. Structure. 2012 doi: 10.1016/j.str.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, Gibbs D, Brock HW, Slany R, Hess JL. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, Allis CD. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38:853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Milne TA, Zhao K, Hess JL. Chromatin immunoprecipitation (ChIP) for analysis of histone modifications and chromatin-associated proteins. Methods Mol Biol. 2009;538:409–423. doi: 10.1007/978-1-59745-418-6_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe SC, Jo SY, Sanders DS, Basrur V, Elenitoba-Johnson KS, Slany RK, Hess JL. MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia. Exp Hematol. 2011;39:77–86. e71–75. doi: 10.1016/j.exphem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, Xie H, Orkin SH, Armstrong SA. Polycomb repressive complex 2 is required for MLLAF9 leukemia. Proc Natl Acad Sci U S A. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H, Nakauchi H, Iwama A. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell. 2010;6:279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Cleary ML. Dimerization: a versatile switch for oncogenesis. Blood. 2004;104:919–922. doi: 10.1182/blood-2004-03-0992. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Sulong S, Moorman AV, Irving JA, Strefford JC, Konn ZJ, Case MC, Minto L, Barber KE, Parker H, Wright SL, et al. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100–107. doi: 10.1182/blood-2008-07-166801. [DOI] [PubMed] [Google Scholar]
- Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1017cs31. Chapter 10, Unit 10 17C. [DOI] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacova S, Slany R, Bartkova J, Stranecky V, Dolezel P, Luzna P, Bartek J, Divoky V. DNA damage response and inflammatory signaling limit the MLLENL-induced leukemogenesis in vivo. Cancer Cell. 2012;21:517–531. doi: 10.1016/j.ccr.2012.01.021. [DOI] [PubMed] [Google Scholar]
- Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Miyagi S, Sashida G, Chiba T, Yuan J, Mochizuki-Kashio M, Suzuki Y, Sugano S, Nakaseko C, Yokote K, et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012;120:1107–1117. doi: 10.1182/blood-2011-11-394932. [DOI] [PubMed] [Google Scholar]
- Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.