Abstract
Background
Powdery mildew (PM) is a major fungal disease of thousands of plant species, including many cultivated Rosaceae. PM pathogenesis is associated with up-regulation of MLO genes during early stages of infection, causing down-regulation of plant defense pathways. Specific members of the MLO gene family act as PM-susceptibility genes, as their loss-of-function mutations grant durable and broad-spectrum resistance.
Results
We carried out a genome-wide characterization of the MLO gene family in apple, peach and strawberry, and we isolated apricot MLO homologs through a PCR-approach. Evolutionary relationships between MLO homologs were studied and syntenic blocks constructed. Homologs that are candidates for being PM susceptibility genes were inferred by phylogenetic relationships with functionally characterized MLO genes and, in apple, by monitoring their expression following inoculation with the PM causal pathogen Podosphaera leucotricha.
Conclusions
Genomic tools available for Rosaceae were exploited in order to characterize the MLO gene family. Candidate MLO susceptibility genes were identified. In follow-up studies it can be investigated whether silencing or a loss-of-function mutations in one or more of these candidate genes leads to PM resistance.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-618) contains supplementary material, which is available to authorized users.
Keywords: Rosaceae, MLO, Powdery mildew, Malus domestica
Background
Powdery mildew (PM) is a major fungal disease for thousands of plant species [1], including cultivated Rosaceae such as apple (Malus domestica), apricot (Prunus armeniaca), peach (Prunus persica), and strawberry (Fragaria x ananassa). Powdery mildew occurs in all major growing regions of Rosaceous crops, leading to severe losses [2] . The major PM causal agents are Podosphaera leucotricha in apple [2], Sphaerotheca pannosa var. persicae in peach [3], Podosphaera tridactyla in apricot [4] and Podosphaera aphanis (syn. Sphaerotheca macularis f. sp. fragariae) in strawberry [5]. Powdery mildew shows similar symptoms in the four species: white spots appear on young green tissues, particularly leaves in the first days after opening, whereas mature leaves show some resistance. Infected leaves crinkle, curl, and prematurely drop. Blossoms and fruits are not the primary targets of PM fungi, but infections of these tissues are possible [2, 3, 5]. In peach, apricot and apple, PM spores overwinter in buds and then in spring, with the reprise of vegetative growth, the spores start a new infection [2, 3].
Cultivars resistant to PM are fundamental in order to reduce the use of pesticides in agricultural practices. The usual strategy in breeding focuses on dominant plant resistance genes (R-genes), however these genes often originate from wild-relatives of the cultivated species, and thus interspecific crossability barriers could prevent their introgression [6]. Moreover, in case of a successful cross, several undesirable traits are incorporated with the R-gene, making extensive backcrossing necessary, which is time-consuming in woody species. Finally, the durability of R-genes is generally limited due to the appearance of virulent strains of the pathogen, which can overcome resistance in a few years [7]. Two examples are Venturia inaequalis race 6, which is able to overcome Rvi6 resistance to scab in apple [8], and P. leucotricha strains able to breakdown Pl-1 and Pl-2, two major PM R-genes of apple [9].
An alternative to the use of R-genes is based on plant susceptibility genes (S-genes), defined as genes whose loss-of-function results in recessively inherited resistance [10]. Barley mlo PM resistance, first characterized in 1942, is a remarkable example of immunity due to the absence of an S-gene, as it derives from a loss-of-function mutation in the MLO (MildewLocusO) gene, encoding for a protein with seven transmembrane domains [11, 12]. mlo resistance has long been considered as a unique form of immunity, characterized by durability, broad-spectrum effectiveness and recessive inheritance [13]. However, the characterization of the sources of resistance in other plant species, such as Arabidopsis [14], pea [15, 16] and tomato [17], has revealed that resistance resulting from loss-of-function mutations in MLO functional orthologs is more common than previously thought. Therefore, it has been suggested that the inactivation of MLO susceptibility genes could represent a valid strategy to introduce PM resistance across a broad range of cultivated species [10].
Histological characterization of mlo resistance revealed that it is based on a pre-penetration defense system, associated with the formation of cell-wall appositions [14, 18] and at least partially dependent on the actin cytoskeleton [19]. It has been suggested that functional MLO proteins negatively regulate vesicle-associated and actin-dependent defense pathways at the site of attempted PM penetration [20]. MLO proteins are therefore targeted by PM fungi as a strategy to induce pathogenesis. The early stages of PM infection are associated with an increase in transcript abundance of MLO susceptibility genes, showing a peak at six hours after inoculation. This has been shown to occur in tomato [17], barley [21], pepper [22] and grape [23, 24].
MLO susceptibility genes are members of a gene family which shows tissue specific expression patterns and are involved in a variety of physiological processes, besides the response to PM fungi: one of the 15 MLO genes of Arabidopsis, AtMLO7, is involved in pollen tube reception by the embryo sac and its mutation results in reduced fertility [25]. Two other Arabidopsis genes, named AtMLO4 and AtMLO11, are involved in the control of root architecture, as mutants with null alleles of these two genes display asymmetrical root growth and exaggerated curvature [26].
Previous phylogenetic analysis of the MLO protein family identified six clades [23]. Currently, all MLO proteins functionally related to PM susceptibility in dicot species appear in a single clade, namely Clade V [14, 17, 23, 24]. Similarly, Clade IV harbours all characterized PM susceptibility proteins from monocots [20, 27].
MLO genes have been intensively studied in many monocots and dicots, but very little has been performed in Rosaceae. In this investigation, we characterized the MLO gene family in a number of Rosaceous species, with respect to their structural, genomic and evolutionary features. Moreover, we monitored the transcript abundances of apple MLO homologs following P. leucotricha inoculation in three apple cultivars.
Results
In silicoand in vitrocharacterization of Rosaceae MLOhomologs
A database search for Rosaceae MLO homologs produced 21 significant matches in peach, 23 in strawberry and 28 in apple. Of these, six (five from M. domestica and one from F. vesca) showed a very limited alignment region with other MLO genes, whereas eight (two from M. domestica, two from P. persica and four from F. vesca) were characterized by markedly different length with respect to MLO homologs reported in the genomes of Arabidopsis and grapevine [23, 28], i.e. less than 350 amino acids (aa) or more than 700 aa. Details on genomic localization amino acid number, putative transmembrane domains and predicted exon/intron structure of the remaining homologs, together with information about the MLO homologs nomenclature chosen in this study is provided in Tables 1, 2 and 3.
Table 1.
Members of the MdMLO gene family as predicted in M. domestica cv. Golden Delicious genome sequence
| Gene | Accession number a | Chr. | Starting position (Mb) | Clade | Introns | TM b | Amino acids | Conserved aa c |
|---|---|---|---|---|---|---|---|---|
| MdMLO1 | MDP0000177099 | 2 | 1.02 | II | 11 | 3 | 487 | 25 |
| MdMLO2 | MDP0000240125 | 2 | 11.10 | I | 11 | 3 | 571 | 20 |
| MdMLO3 | MDP0000168575 | 2 | 11.11 | I | 13 | 7 | 670 | 22 |
| MdMLO4 | MDP0000207002 | 2 | 8.79 | III | 16 | 7 | 634 | 28 |
| MdMLO5 | MDP0000163089 | 9 | 15.26 | V | 14 | 6 | 579 | 30 |
| MdMLO6 | MDP0000119433 | 3 | 33.95 | II | 0 | 7 | 504 | 30 |
| MdMLO7 | MDP0000123907 | n.d. | n.d. | V | n.d. | 6 | 561 | 28 |
| MdMLO8 | MDP0000218520 | 2 | 11.11 | I | 9 | 4 | 390 | 14 |
| MdMLO9 | MDP0000320797 | 2 | 27.20 | II | 10 | 5 | 454 | 28 |
| MdMLO10 | MDP0000196373 | 3 | 26.97 | I | 13 | 6 | 539 | 28 |
| MdMLO11 | MDP0000239643 | 4 | 9.84 | V | 12 | 8 | 575 | 28 |
| MdMLO12 | MDP0000133162 | 6 | 0.81 | III | 13 | 5 | 516 | 28 |
| MdMLO13 | MDP0000142608 | 7 | 7.48 | II | 12 | 6 | 351 | 18 |
| MdMLO14 | MDP0000191469 | 8 | 29.25 | II | 10 | 5 | 395 | 23 |
| MdMLO15 | MDP0000141595 | 9 | 7.54 | III | 15 | 6 | 647 | 24 |
| MdMLO16 | MDP0000191848 | 9 | 21.12 | VI | 14 | 6 | 606 | 29 |
| MdMLO17 | MDP0000145097 | 11 | 27.97 | I | 13 | 7 | 523 | 28 |
| MdMLO18 | MDP0000928368 | 10 | 27.97 | VII | 12 | 7 | 502 | 30 |
| MdMLO19 | MDP0000168714 | 12 | 16.23 | V | 13 | 7 | 590 | 30 |
| MdMLO20 | MDP0000134649 | 13 | 11.61 | VIII | 13 | 5 | 589 | 27 |
| MdMLO21 | MDP0000133760 | 15 | 24.99 | VI | 15 | 6 | 560 | 28 |
aAvailable at http://www.rosaceae.org/gb/gbrowse/malus_x_domestica/.
bNumber of transmembrane domains in the predicted protein, as determined by InterPro prediction software (http://www.ebi.ac.uk/interpro/).
cnumber of conserved amino acids out of the 30 identified by Elliot et al. [29].
Table 2.
Members of the PpMLO gene family as predicted in Prunus persica genome sequence
| Gene | Accession number a | Chr. | Starting position (Mb) | Clade | Introns | TM b | Amino acids | Conserved aa c |
|---|---|---|---|---|---|---|---|---|
| PpMLO1 | ppa003207m | 6 | 6.82 | V | 14 | 7 | 593 | 30 |
| PpMLO2 | ppa003435m | 7 | 18.38 | III | 14 | 8 | 574 | 30 |
| PpMLO3 | ppa003437m | 6 | 21.99 | V | 13 | 7 | 574 | 30 |
| PpMLO4 | ppa003466m | 2 | 21.03 | V | 14 | 7 | 572 | 30 |
| PpMLO5 | ppa003706m | 4 | 10.92 | I | 14 | 8 | 555 | 30 |
| PpMLO6 | ppa004012m | 7 | 22.64 | II | 14 | 6 | 535 | 29 |
| PpMLO7 | ppa004508m | 8 | 21.17 | II | 0 | 7 | 506 | 29 |
| PpMLO8 | ppa004621m | 6 | 22.01 | VI | 14 | 6 | 499 | 29 |
| PpMLO9 | ppa004687m | 4 | 2.59 | VII | 11 | 7 | 496 | 29 |
| PpMLO10 | ppa004866m | 2 | 13.73 | II | 11 | 7 | 488 | 29 |
| PpMLO11 | ppa020172m | 1 | 43.04 | I | 14 | 4 | 561 | 30 |
| PpMLO12 | ppa020311m | 5 | 0.82 | IV | 13 | 7 | 566 | 30 |
| PpMLO13 | ppa021048m | 4 | 15.57 | VIII | 12 | 5 | 510 | 24 |
| PpMLO14 | ppa022847m | 6 | 6.80 | VI | 14 | 6 | 550 | 29 |
| PpMLO15 | ppa024476m | 7 | 17.63 | I | 14 | 8 | 539 | 26 |
| PpMLO16 | ppa024488m | 5 | 0.76 | III | 14 | 6 | 504 | 30 |
| PpMLO17 | ppa024581m | 6 | 8.95 | II | 13 | 6 | 463 | 27 |
| PpMLO18 | ppa026565m | 6 | 22.00 | VI | 13 | 6 | 416 | 25 |
| PpMLO19 | ppb024523m | 1 | 42.04 | II | 13 | 5 | 446 | 23 |
aAvailable at http://www.rosaceae.org/gb/gbrowse/prunus_persica/.
bNumber of transmembrane domains in the predicted protein, as determined by InterPro prediction software (http://www.ebi.ac.uk/interpro/).
cnumber of conserved amino acids out of the 30 identified by Elliot et al. [29].
Table 3.
Members of the FvMLO gene family as predicted in Fragaria vesca genome sequence
| Gene | Accession number a | Chr. | Starting position (Mb) | Clade | Introns | TM b | Amino acids | Conserved aa c |
|---|---|---|---|---|---|---|---|---|
| FvMLO1 | mrna02774.1-v1.0-hybrid | n.d. | n.d. | V | 14 | 7 | 632 | 28 |
| FvMLO2 | mrna03210.1-v1.0-hybrid | 3 | 14.46 | II | 11 | 5 | 528 | 20 |
| FvMLO3 | mrna09651.1-v1.0-hybrid | 6 | 35.88 | III | 14 | 6 | 542 | 28 |
| FvMLO4 | mrna09653.1-v1.0-hybrid | 6 | 35.90 | V | 14 | 7 | 573 | 30 |
| FvMLO5 | mrna10166.1-v1.0-hybrid | 1 | 1.34 | II | 14 | 3 | 688 | 26 |
| FvMLO6 | mrna10346.1-v1.0-hybrid | 3 | 12.52 | II | 7 | 2 | 385 | 15 |
| FvMLO7 | mrna10363.1-v1.0-hybrid | 3 | 12.49 | II | 9 | 2 | 442 | 21 |
| FvMLO8 | mrna10558.1-v1.0-hybrid | 2 | 19.08 | II | n.d. | 6 | 514 | 28 |
| FvMLO9 | mrna11028.1-v1.0-hybrid | n.d. | n.a. | I | 10 | 4 | 434 | 18 |
| FvMLO10 | mrna13023.1-v1.0-hybrid | 1 | 7.96 | III | 13 | 6 | 557 | 27 |
| FvMLO11 | mrna14592.1-v1.0-hybrid | 1 | 8.77 | I | 13 | 7 | 548 | 28 |
| FvMLO12 | mrna23198.1-v1.0-hybrid | 7 | 15.89 | V | 14 | 7 | 507 | 29 |
| FvMLO13 | mrna26428.1-v1.0-hybrid | 7 | 17,79 | VIII | 11 | 5 | 558 | 20 |
| FvMLO14 | mrna28541.1-v1.0-hybrid | n.d. | n.a. | III | 11 | 4 | 481 | 26 |
| FvMLO15 | mrna29770.1-v1.0-hybrid | 3 | 7.36 | VII | 13 | 7 | 538 | 28 |
| FvMLO16 | mrna31264.1-v1.0-hybrid | 3 | 30.51 | I | 16 | 8 | 579 | 28 |
| FvMLO17 | mrna31498.1-v1.0-hybrid | 5 | 20.23 | IV | 11 | 5 | 531 | 27 |
| FvMLO18 | mrna29285.1-v1.0-hybrid | 5 | 19.12 | V | 6 | 4 | 357 | 18 |
aAvailable at http://www.rosaceae.org/gb/gbrowse/fragaria_vesca_v1.0-lg/ (hybrid).
bNumber of transmembrane domains in the predicted protein, as determined by InterPro prediction software (http://www.ebi.ac.uk/interpro/).
cnumber of conserved amino acids out of the 30 identified by Elliot et al. [29].
Peach and apricot are evolutionary very close to each other, and show a high degree of homology in DNA sequence. Phylogenetic analysis (see next paragraph) indicated peach homologs PpMLO1, PpMLO3 and PpMLO4 as candidates for being required for PM susceptibility. Therefore, we used the sequences of these genes to design primers to identify full-length apricot MLO genes. This approach resulted in the amplification and the successive characterization of three MLO sequences, which were by analogy named PaMLO1, PaMLO3, and PaMLO4 (deposited in the NCBI database with the accession numbers KF177395, KF177396, and KF177397, respectively).
Phylogenetic relations and inference of orthology
We performed a phylogenetic study on the newly identified Rosaceae MLO proteins. The dataset was completed with four homologs recently characterized in Rosa hybrida (rose) [30] (RhMLO1, RhMLO2, RhMLO3 and RhMLO4), the complete Arabidopsis thaliana AtMLO protein family [14], a series of MLO homologs which have been functionally associated with PM susceptibility, namely tomato (Solanum lycopersicum) SlMLO1 [17], pea (Pisum sativum) PsMLO1 [15, 16], pepper (Capsicum annuum) CaMLO2 [27], lotus (Lotus japonicus) LjMLO1 [15], barrel clover (Medicago truncatula) MtMLO1 [15], barley (Hordeum vulgare) HvMLO [11], rice (Oryza sativa) OsMLO2 [31], wheat (Triticum aestivum) TaMLO_B1 and TaMLO_A1b [31], and grapevine (Vitis vinifera) VvMLO14, the only dicot MLO homolog known to belong to clade IV [23]. Clustering analysis using the UPGMA algorithm resulted in a total of eight distinct clades and no divergent lineage (Figure 1). Clade numbers from I to VI were assigned based on the position of Arabidopsis AtMLO homologs and barley HvMLO, according to the previous study of Feechan et al. [23]. The two additional clades (named VII and VIII) were found to include Rosaceae MLO homologs only, both having one homolog from P. persica, one from F. vesca and one from M. domestica. Further clustering analysis with a Neighbour-Joining algorithm resulted in merging clade VII and VIII (not shown).
Figure 1.
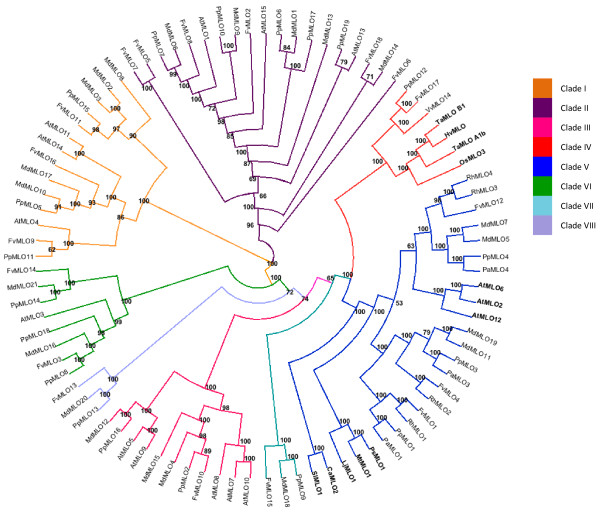
Phylogenetic tree of Rosaceae MLO. Phylogenetic relationships of predicted Rosaceae MLO amino acid sequences to MLO proteins of other plant species. The dataset includes Rosaceae MLO sequences from Rosa hybrida (RhMLO), Malus domestica (MdMLO), Prunus persica (PpMLO), Prunus armeniaca (PaMLO) and Fragaria vesca (FvMLO). The other proteins included are Solanum lycopersicum SlMLO1, Arabidopsis thaliana AtMLO, Capsicum annuum CaMLO2, Pisum sativum PsMLO1, Medicago truncatula MtMLO1, Lotus japonicus LjMLO1, Vitis vinifera VvMLO14, Hordeum vulgare HvMLO, Triticum aestivum TaMLO_B1, TaMLO_A1b and Oryza sativa OsMLO2. Proteins which have been functionally characterized as susceptibility genes are highlighted in bold. Numbers at each node represent bootstrap support values (out of 100 replicates).
Four apple MLO homologs (MdMLO5, MdMLO7, MdMLO11 and MdMLO19) and three MLO homologs from peach (PpMLO1, PpMLO3 and PpMLO4), apricot (PaMLO1, PaMLO3 and PaMLO4) and woodland strawberrry (FvMLO1, FvMLO4 and FvMLO12) were found to cluster together in the phylogenetic clade V, containing all the dicot MLO proteins experimentally shown to be required for PM susceptibility (e.g. [16, 23]. One homolog from strawberry (FvMLO17) and one from peach (PpMLO12) were found to group, together with grapevine VvMLO14, in clade IV, which contains all monocot MLO proteins acting as PM susceptibility factors (Figure 1).
We used the GBrowse-Syn tool to detect syntenic blocks encompassing P. persica, F. vesca and M. domestica MLO genes. As syntenic blocks derive from the evolution of the same chromosomal region after speciation, orthology between MLO genes could be inferred. In total, twelve orthologous relationships were predicted between P. persica and F. vesca, nine between P. persica and M. domestica and eight between F. vesca and M. domestica (Table 4, Figure 2 and Additional file 1).
Table 4.
Relations of orthology inferred between P. persica, F. vesca and M. domestica MLO homologs
| P. persicagenes | F. vescaorthologs | M. domesticaorthologs |
|---|---|---|
| PpMLO2 | FvMLO10 | MdMLO15 |
| PpMLO3 | FvMLO4 | MdMLO19 |
| PpMLO4 | FvMLO12 | - |
| PpMLO5 | FvMLO16 | MdMLO10, MdMLO17 |
| PpMLO6 | FvMLO5 | MdMLO1 |
| PpMLO7 | FvMLO8 | - |
| PpMLO8 | FvMLO3 | - |
| PpMLO9 | FvMLO15 | MdMLO18 |
| PpMLO10 | FvMLO2 | MdMLO9 |
| PpMLO14 | FvMLO14 | MdMLO21 |
| PpMLO15 | FvMLO11 | - |
| PpMLO16 | - | MdMLO12 |
| PpMLO18 | FvMLO3 | - |
Relations of orthology between PpMLO1, PpMLO3, PpMLO4 and apricot PaMLO1, PaMLO3, PaMLO4 were clearly suggested by the high percentage of sequence identity between these homolog genes, which was 97,3%, 98,8% and 96,7%, respectively.
Figure 2.
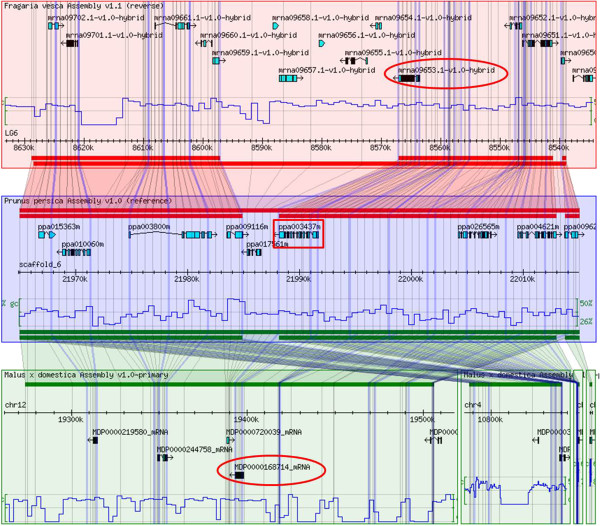
Synteny between apple, peach and strawberry. Results of search for F. vesca and M. domestica chromosomal regions syntenic to a P. persica 50 kb stretch including the MLO homolog PpMLO3 (corresponding to ppa003437m in the genomic database of Rosaceae), boxed. Shaded polygons indicate aligned regions between genomes. Grid lines are meant to indicate insertions/deletions between the genomes of F. vesca and M. domestica with respect to the P. persica reference sequence. Strawberry FvMLO4 and apple MdMLO19 (named in the figure as mrna09653.1-v1.0-hybrid and MDP0000168714, according to the nomenclature provided in this paper), predicted to be PpMLO3 orthologs, are indicated with circles.
Transcription of putative apple MLOgenes in response to Podosphaera leucotrichainoculation
To identify MLO genes that respond to the PM fungus P. leucotricha, we measured the transcript abundance of 19 out of 21 apple MLO genes in leaves 4, 6, 8 and 24 hours after artificial inoculation with the pathogen, and compared these data with the ones of non-inoculated leaves. Three cultivars, Golden Delicious, Braeburn and Gala, were analysed in order to investigate whether up-regulation was comparable among them and results could therefore be generalized for all apple cultivars. Three genes, namely MdMLO11, MdMLO18 and MdMLO19, were found to be significantly up-regulated after inoculation with the pathogen (Figure 3 and Additional file 2). Up-regulation of these genes was about 2-fold compared to non-inoculated plants, with peaks of 4-fold up-regulation at very early time points (‘Braeburn’- MdMLO11 - 6 hpi; ‘Gala’- MdMLO18 - 4 hpi; ‘Golden Delicious’- MdMLO19 - 6hpi). MdMLO11 and MdMLO18 were up-regulated in all cultivars, while MdMLO19 was only up-regulated in ‘Braeburn’ and ‘Golden Delicious’.
Figure 3.
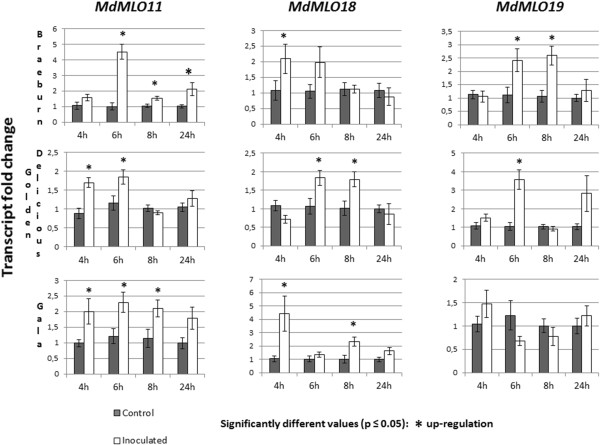
Transcriptional variation of three apple MLO genes following inoculation with P. leucotricha. Transcript abundances of three MLO genes in leaves of the apple cultivars ‘Braeburn’, ‘Golden Delicious’ and ‘Gala’, following powdery mildew (PM) inoculation. Here we show only MLO genes that were more than one time significantly up or down regulated following PM inoculation at one of the four time points examined (4, 6, 8 and 24 hpi). The set of results of all investigated genes is shown in Additional file 1: Figure S1. Each bar shows the average of four to eight biological replicates. The Ct values have been normalized for three reference genes: actin, ubiquitin and elongation factor 1. Statistical significance was determined with a t-test for each individual pair of inoculated and non-inoculated samples at each time point. The error bars show standard errors of the means. Significant differences between inoculated samples and control samples are indicated with an asterisk (P < 0.05).
Two of the genes, MdMLO11 and MdMLO19 belong to Clade V, while MdMLO18 belongs to the newly identified Clade VII (Figure 1).
Discussion
Genomic organization and phylogenetic relations between Rosaceae MLOhomologs
We report here the identification, through an in silico approach, of 19 MLO homologs in the genome of peach and 18 in the genome of strawberry. This is consistent with the results of previous genome-wide studies carried out on dicotyledonous species, indicating the presence of 15 MLO homologs in Arabidopsis, 17 in grapevine and 16 in tomato [9, 13]; Appiano et al., unpublished results; [24]. Conversely, the number of MLO homologs detected in apple (21) is lower than expected, considering that a relatively recent genome-wide duplication event has occurred in the Pyreae tribe [32].
Most PpMLO, FvMLO and MdMLO homologs appeared to be widely distributed within the respective genomes (Tables 1, 2 and 3), indicating segmental duplication as the prevailing evolutionary mechanism for the Rosaceae MLO gene family. However, we also found cases of clusters of adjacent homologs (PpMLO3, PpMLO8 and PpMLO18, PpMLO12 and PpMLO16, PpMLO1 and PpMLO14, FvMLO3 and FvMLO4, FvMLO6 and FvMLO7, MdMLO2, MdMLO3 and MdMLO8), which are likely the result of tandem duplication events.
Inference of phylogenetic relationships between MLO proteins revealed the presence of apple, strawberry, peach and apricot homologs in the clade V, containing all dicot MLO homologs shown so far to be involved in PM susceptibility, thus making them candidates to act as susceptibility factors. Although the simple clustering in clade V is not enough to recognize a gene as a susceptibility factor, it does provide the first evidence for functionality and allows for the reduction in the number of candidates for further functional analysis. Clade IV, that contains functional MLO susceptibility homologs from monocots, was found to include one homolog from F. vesca (FvMLO17) and one from P. persica (PpMLO12). In accordance with this finding, a MLO homolog from the dicot species V. vinifera also clusters in clade IV [23, 24]. Figure 1). Interestingly, phylogenetic analyses carried out in this study also revealed the presence of one or two additional clades, depending on the type of phylogenetic reconstruction (UPGMA or Neighbour-Joining), which were not reported to occur in earlier investigations. Moreover, they appear to be characteristic of Rosaceae, since they contain only homologs from this family. Clearly, the exclusivity for Rosaceae of these clade(s) needs to be confirmed by further studies containing a larger dataset of MLO proteins. Additional studies could be also addressed to the functional characterization of Rosaceae MLO homologs grouped in clade VII. Indeed, this appears to be basal to both clade IV and clade V (Figure 1), and thus might have contained ancestral proteins which later on evolved into PM susceptibility factors.
Synteny between apple, peach and woodland strawberry MLOgenes
We took advantage of recent developments in Rosaceae genomics in order to detect synteny between P. persica, F. vesca and M. domestica chromosomal regions containing MLO homologs. This permitted the inference of ortholgous relationships between MLO genes in these species. Notably, all predicted MLO orthologs from different Rosaceae species, fell in the same phylogenetic clade (Tables 1, 2 and 3; Figure 1 and Additional file 1). This is to be expected, since orthologs generally share the same function and thus are characterized by a high level of sequence conservation. It is noteworthy that the chromosomal localization of predicted MLO orthologs between P. persica, M. domestica and F. vesca is in accordance with the results of the synteny study performed after the release of the three genomes [33, 34]. In particular, genes situated on peach scaffold 2, 7 and 8 were predicted to have orthologs on strawberry chromosome 7, 1 and 2, respectively, whereas genes on peach scaffold 4 were predicted to have orthologs on strawberry chromosomes 2 or 3 (Table 4). FvMLO3 was predicted to be orthologous to two peach MLO genes, PpMLO8 and PpMLO18, which are localised in close proximity to each other on peach scaffold 6 and grouped together in clade VI. In this case, we hypothesize a relation of co-orthology due to the occurrence of a recent tandem duplication event in the peach genome. Similarly, PpMLO5 and FvMLO16 were predicted to be orthologs of two apple MLO genes, MdMLO10 and MdMLO17, located on chromosomes 3 and 11. This is consistent with indications of duplications of large segments of these two chromosomes during the evolution of the apple genome [32].
Transcription of apple putative MLOgenes in response to P. leucotrichainoculation
In barley, pea and tomato, only one of the clade V MLO homologs seems to be involved in powdery mildew susceptibility, whereas in A. thaliana three MLO genes in Clade V are required to be inactivated in order to achieve a fully resistant phenotype [16, 27]. This implies that, within Clade V MLO genes, a further selection might be required to identify PM susceptibility genes. Accumulating evidence indicates that MLO susceptibility genes are up-regulated upon challenge with powdery mildew fungi [17]. Therefore, we analysed the expression level of apple MLO genes identified in this study in response to the interaction with P. leucotricha. Three pathogen-dependent gene up-regulations were detected. Two up-regulated MLO homologs, MdMLO11 and MdMLO19, encode for proteins falling in clade V, thus making them likely candidates to act as PM susceptibility genes in apple. MdMLO11 and MdMLO19 are located on chromosomes 4 and 12 respectively, and are therefore both generated from a duplication event in the 9-chromosome ancestor of apple [32]. A third pathogen-dependent up-regulated gene, MdMLO18, was found, which encodes a protein grouping in the newly identified Clade VII (Figure 1). The presence of a PM upregulated gene outside clade V is consistent with transcriptome analyses recently performed in tomato (Appiano et al., unpublished results). Apple clade V also contains two genes, MdMLO5 and MdMLO7, which show no significant changes in expression following inoculation. Accordingly, the lack of up-regulation of some clade V MLO genes has been observed in grapevine and tomato [23, 24]; Appiano et al., unpublished results). The possible role of these genes as susceptibility factors has not yet been highlighted.
PpMLO3, PaMLO3 and FvMLO4 are likely to represent true orthologs of MdMLO19 (Table 4). Since orthologs often maintain the same function during evolution, we conjecture that the expression of these genes might also be responsive to PM fungi attacking corresponding species. Moreover, FvMLO15 and PpMLO9 are likely orthologs of MdMLO18, so they should also be considered as putative transcriptionally responsive genes to PM fungi attack. Further studies aimed at the functional characterization of these genes (e.g. through the application of reverse genetic approaches of targeted mutagenesis or gene silencing), in apple but also in peach and strawberry, might lead to the identification of resistant phenotypes, which could be used for the development of PM resistant cultivars. Particularly, studies on MdMLO18 could lead to the characterization of a possible role for clade VII in the interaction with PM fungi.
Conclusions
Our work led to the identification of 19 MLO homologs in peach, 17 in strawberry and 21 in apple. Three, three and four homologs, respectively, belong to clade V and therefore are candidates for being S-genes. Due to the high similarity between peach and apricot, we were able to amplify and characterize three Clade V apricot MLO genes.
The phylogenetic analysis revealed two new Rosaceae specific clades for the MLO family, although this needs to be confirmed by the use of a larger MLO proteins dataset.
Through inoculation of apple with P. leucotrica, we identified three up-regulated genes, i.e. MdMLO11, MdMLO18 and MdMLO19. MdMLO11 and MdMLO19, that belong to Clade V, are positioned in duplicated regions and have high sequence identity, therefore they are likely to be recent paralogs. MdMLO18 belongs to the newly identified Clade VII.
Methods
In silicoidentification and comparison of MLO predicted proteins in peach, woodland strawberry and apple
Predicted peptides from the peach genome (v. 1.0) and the strawberry genome (v.1.0) gene model databases, available at the website of the Genomic Database for Rosaceae (http://www.rosaceae.org) [35], were queried for the presence of MLO homologs protein sequences. First, a BLAST search, using the tomato SlMLO1 amino acid sequence as query was carried out. A further search was performed with the HMMER programme, which uses a method for homolog searches based on the profile hidden Markov probabilistic model [36]. The sequences obtained with the previously mentioned BLAST search, were used together with other known MLO sequences from dicot and monocot species, namely: four RhMLOs from Rosa hybrida, 15 AtMLOs from Arabidopsis thaliana, SlMLO1 from Solanum lycopersicum, CaMLO2 from Capsicum annuum, PsMLO1 from Pisum sativum, MtMLO1 from Medicago truncatula, LjMLO1 from Lotus japonicus, VvMLO14 from V. Vinifera, HvMLO from Hordeum vulgare, TaMLO1_A1b and TaMLO_B1 from Triticum aestivum and OsMLO2 from Oryza sativa. MLO protein sequences from apple (Malus domestica cv. Golden Delicious) were identified by searching for the MLO domain profile (IPR004326) in the apple genome available at FEM-IASMA computational biology web resources (http://genomics.research.iasma.it). The resulting list was integrated with a BLAST search, carried out with the amino acid sequences previously listed for the HMMER search in peach and strawberry.
Chromosomal localization and predicted introns/exons structure of each MLO gene of apple, peach and strawberry was deduced based on the available genomic information at the GDR database. The presence and number of membrane spanning helices was predicted using the online software InterPro (http://www.ebi.ac.uk/interpro). Alignments for conserved amino-acids analysis were performed with the CLC Sequence Viewer v. 6.9 software (http://clcbio.com).
A total of 90 MLO protein sequences, including three apricot MLO sequences isolated in vitro (see next paragraph), were used for Clustal alignment (http://www.ebi.ac.uk/Tools/msa/clustalw2/). UPGMA-based and Neighbour-Joining-based phylogenetic trees were obtained with the CLC sequence viewer software. The UPGMA clustering algorithm was further used as input for the Dendroscope software, suitable for the visualization of large phylogenetic trees [37].
Relationships of orthology between MLO candidate genes from peach, woodland strawberry and apple were inferred by running the GBrowse-Syn tool available at GDR (http://www.rosaceae.org/gb/gbrowse_syn/peach_apple_strawberry) [35, 38]. This displays syntenic regions among the three available genomes of Rosaceae, as detected by the Mercator programme [35, 39]. For 50 Kb chromosomal stretches flanking each P. persica PpMLO homolog, syntenic regions from F. vesca and M. domestica were searched. Orthology was called upon the identification of F. vesca or M. domestica MLO homologs within syntenic blocks.
In vitroisolation of apricot MLOhomologs
RNA from apricot leaves (cultivar Orange Red) was extracted by using the SV Total RNA Isolation System Kit (Promega), and corresponding cDNA was synthesized by using the QuantiTect Reverse Transcription Kit (Qiagen) with oligo(dT) primers. Sequences of the peach MLO homologs PpMLO1, PpMLO3 and PpMLO4, are phylogenetically close to MLO homologs functionally associated to PM susceptibility, and were therefore used to design the primer pairs 5 ′ -ATGGCAGCCGCAACCTCAGGAAGA-3 ′ /5 ′ -TTATATACTTTGCCTATTGTCAAAC-3 ′ , 5 ′ -ATGGCAGGGGGAAAAGAAGGACG-3 ′ /5 ′ -TCAACTCCTTTCTGATTTCTCAA-3 ′ and 5 ′ -ATGGCCGAACTAAGTAAAGA-3 ′ /5 ′ TCAACTTCTTGATTTTCCTTTGC-3 ′ , respectively. These were employed to amplify full-length cDNA sequences of apricot putative orthologs, by using the AccuPrime Taq polymerase (Invitrogen). Amplicons were purified by using the NucleoSpin Extract II kit (Macherey-Nagel) and ligated (molar ratio 1:1) into the pGEM-T Easy vector (Promega). Recombinant plasmids were cloned in E. coli DH10β chemically competent cells and recovered by using the Qiaprep spin miniprep kit (Qiagen). Sequencing reactions were performed twice, by using universal T7 and SP6 primers (Eurofins MWG Operon).
Glasshouse test with apple cultivars
A total of 192 apple plants from three cultivars (Braeburn, Golden Delicious and Gala) were used to measure transcript abundance of MLO genes. Budwoods from these cultivars were grafted on M9 rootstocks in January 2012. The grafts were kept at −1°C for 2 months, and potted at the beginning of March in greenhouse. The plants grew for 6 weeks in the greenhouse at 20°C during the day, 17°C during the night, relative humidity of 70% and natural day/night cycle.
P. leucothrica was collected from apple trees in an unsprayed test orchard and used to infect greenhouse grown apple seedlings from ‘Gala Galaxy’ seeds. Four weeks after inoculation, conidia were used for the inoculation experiment, or transferred to new seedlings, to keep them viable. We inoculated by touching the plants with heavily infected apple seedlings. Control plants were not inoculated and kept separated in the same greenhouse of the inoculated plants. Inoculated and control plants were grown in the greenhouse at the growing conditions previously mentioned. The leaf samples were collected 4, 6, 8 and 24 hours post-inoculation (hpi).
Eight experimental repeats were performed and each sample contained three or four young leaves collected from each single plant. Every plant was used for sampling only once, to avoid any possible effect of wounding on the expression of MLO genes. The smallest statistical unit was a plant. The leaves were flash-frozen and ground in liquid nitrogen, and stored at −80°C until RNA extraction.
qPCR analysis of transcript levels
RNA extraction was carried out with the MagMAX-96 Total RNA isolation kit (Applied Biosystem) that includes DNAse treatment. The kit yielded between 50 and 200 ng/ul, of good quality RNA per sample.
Primers for gene expression analysis were designed with NCBI Primer Designing Tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Four serial dilutions of cDNA (1/5 - 1/25 – 1/125 – 1/625) were used to calculate the efficiency of each primer pair with iCycler software (Biorad). In case of efficiency lower than 1.80 or greater than 2.20, the primer pair was discarded and a new one tested, with the exception of MdMLO9, for which was not possible to design a primer pair with better efficiency. It was only possible to analyse 19 MLO genes because for MdMLO12 and MdMLO16 was not possible to design specific and efficient primer pairs, despite numerous attempts. Presence of a specific final dissociation curve was determined after each qPCR amplification reaction with progressive increment of temperature from 65°C to 95°C (0.5°C each step, 5 sec) and the size of the product was confirmed by agarose gel electrophoresis.
Quantitative Real Time-PCR (qPCR) was performed with SYBR greenER mix (Invitrogen) in a 15-μL reaction volume, using a Bio-Rad iCycler iQ detection system, run by the Bio-Rad iCycler iQ multicolor 3.1 software. The software applies comparative quantification with an adaptive baseline. Samples were run in two technical replicates with the following thermal cycling parameters: 95°C 3 min – 95°C 15 sec, 60°C 1 min (repeated 40 times) – 95°C 10 sec.
Reference genes β-actin (NCBI accession number DT002474; Plaza accession number MD00G171330 - http://bioinformatics.psb.ugent.be/plaza/), ubiquitin (Plaza accession number MD05G001920) and elongation factor 1 (Plaza accession number MD09G014760) were used as reference genes (Table 5). All these three genes were used in previous works [40–42]. For additional control, we assessed the stability of our genes with the software geNorm (medgen.ugent.be/~jvdesomp/genorm/). An M-value lower than 1.5 is generally considered as stable enough [43–45] and all three reference genes in all three cultivars considered are within this threshold. We saw differences in stability between cultivars: ‘Golden Delicious’ was the most stable cultivar (actin: 0.824 – ubiquitin: 0,852 – elongation factor 1: 0,926), whereas ‘Braeburn’ was the less stable (actin: 1.246 – ubiquitin: 1,293 – elongation factor 1: 1,369) and ‘Gala’ showed intermediate stability (actin: 1.039 – ubiquitin: 1,152 – elongation factor 1: 1,078).
Table 5.
Gene-specific primers and amplicon sizes in qRT-PCR detection of 19 MdMLO -like genes based on Malus domestica cv. Golden Delicious genome sequence
| Gene | Forward primer (5 ′ – 3 ′ ) | Reverse primer (5 ′ – 3 ′ ) | Size (bp) | Efficiency |
|---|---|---|---|---|
| MdMLO1 | GTGGGCTCGGTCGGCCAAAA | CCAGCACCAGCACCAGAACCA | 81 | 2.06 |
| MdMLO2 | CGTTGGATCAACCACTGCGCCT | TGAGCTGCAGCCAGTGGGATCT | 87 | 1.83 |
| MdMLO3 | CCACTGCGCCTCTCTGAAGCA | CCACCAAAACGGCTCTCCAGGT | 93 | 2.12 |
| MdMLO4 | TGTTGCAGACACTATGCTGCCATGT | GGCAGCAGCTAAAGATCTGCGT | 109 | 1.87 |
| MdMLO5 | TCGTCAGGCTCTCATTCGGGGT | GTGCTGCTGCCACTCCCTC | 132 | 1.80 |
| MdMLO6 | TTCGCGGAGGAGGGGTCGTT | TTCGAGCGACAGCAACGGCA | 72 | 2.15 |
| MdMLO7 | TGGAGCAAGTCACCAGTCTCCAT | CGCTTCCTGGTGCCAAATGTGC | 127 | 2.12 |
| MdMLO8 | GTCAAGCTAATCTTACCACGCGCT | GGCTGGAAGGAAGGACAGCCA | 85 | 1.95 |
| MdMLO9 | GCTGCAACACGTAATCACCC | AGAACGCCATTTCGAAAGCA | 173 | 2.30 |
| MdMLO10 | GCGATCGTTGGCCTTGACTCC | TTCCGCGCTCGACAAGCAGA | 86 | 1.92 |
| MdMLO11 | CCGTTCCATCACCAAGACGA | ATTGCTCTCCGAGTTACGCC | 102 | 1.90 |
| MdMLO13 | ACATTGTCCCCAGGCTTGTT | GCCCAACCAATAAGTCCCGA | 151 | 2.00 |
| MdMLO14 | TGCACTTGTCAGCCAGATGGG | GCATCTCCCACCCACGAACCG | 81 | 2.15 |
| MdMLO15 | GCGCCTTTCTCTCTGCTGGGT | CGCGTGCGAGGTGGTCTCTT | 90 | 2.01 |
| MdMLO17 | TTGCCACTGTATGCCTTGGT | TGCTTGCTTCTGTGCGAATG | 163 | 2.15 |
| MdMLO18 | AAGGAAGGCTCTCATTCAGGCTCT | TGCAATTGGCTTTTGACCAACGGT | 100 | 2.22 |
| MdMLO19 | CAGAGTGGCGACTGCACTTA | GGGACATGGAGTGCAAAGGA | 110 | 1.97 |
| MdMLO20 | AAAAAGCTCCACCAACCCCA | TTTCTCTCCCATGACGCTCG | 165 | 2.11 |
| MdMLO21 | CCTTGTTCGAGGCCGTAGAG | ACCAAGTGCTTTGGTGGTTT | 176 | 1.95 |
| β-actin | CTATGTTCCCTGGTATTGCAGACC | GCCACAACCTTGATTTTCATGC | 82 | 1.90 |
| Ubiquitin | CATCCCCCCAGACCAGCAGA | ACCACGGAGACGAAGCACCAA | 349 | 1.91 |
| Elongation Factor 1 | TACTGGAACATCACAGGCTGAC | TGGACCTCTCAATCATGTTGTC | 308 | 2.07 |
Each of the biological replicates was analysed in duplicate and the average of these two replicates was used for further analysis. In case of excessive difference between the two replicates (one Ct or more), the run was repeated. Considering the high number of samples and genes of interest, we opted for this approach in order to reduce the number of total runs. Data analysis was performed according to Hellemans et al. [46], using the statistical package SPSS (IBM). This analysis method takes into account the efficiency value of each primer pair. For some genes it was necessary to apply a natural log transformation to the data, in order to obtain normal distribution of residues. To investigate the differences between control and inoculated samples, we used T-test (p ≤ 0.05).
Availability of supporting data
The following files are available on: mynotebook.labarchives.com.Figure 1 – Phylogenetic tree of Rosaceae MLO.
DOI: 10.6070/H4Z60M0N.
Additional file 1 - Synteny between apple, peach and strawberry.
DOI: 10.6070/H4TD9V8C.
Electronic supplementary material
Additional file 1: Synteny between apple, peach and strawberry. Results of search for F. vesca and M. domestica regions syntenic to 50 kb P. persica chromosomal stretches containing the PpMLO homologs identified in this study. Shaded polygons indicate aligned regions between genomes. Grid lines are drawn to indicate insertions/deletions between the genomes of F. vesca and M. domestica with respect to the P. persica reference sequence. P. persica, F. vesca and M. domestica MLO homologs, named according to the nomenclature of the Genomic Database of Rosaceae, are boxed. (PDF 817 KB)
Additional file 2: Transcriptional variation of 19 apple MLO genes in three cultivars following inoculation with P. leucotricha. Transcription abundances of 19 MLO-like genes following powdery mildew (PM) inoculation in ‘Golden Delicious’ (1a), ‘Gala’ (1b) and ‘Braeburn (1c) leaf samples. The graphs show expression values of inoculated samples relative to control samples, averaged from four to eight biological replicate, normalized, that are in turn the average of two experimental replicates. The Ct values have been normalized with three reference genes: actin, ubiquitin and elongation factor 1. Statistical significance was determined with a t-test for each individual pair of inoculated and control samples at each time point (4, 6, 8 and 24 hpi). The error bars show standard errors of the means. Significant differences between inoculated samples and control samples are indicated with a *(P < 0.05). (PDF 1 MB)
Acknowledgment
The authors would like to thank Remmelt Groenwold (WUR) for helping with apple inoculation with P. leucotricha and Lorenza Dalla Costa (FEM) for all the valuable advices about qPCR data analysis.
This work has been founded by Fondazione Edmund Mach and GMPF PhD programme.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
SPe carried out the inoculation and gene expression analysis, identified MLO homologs in apple and wrote the major part of the manuscript. SPa contributed to design the experiment, identified MLO homologs in peach, apricot and strawberry, performed the analysis of synteny and contributed to write the manuscript. DC contributed to design the experiment, performed the phylogenetic analysis and revised the manuscript. AG contributed to design the experiment and revised the manuscript. YB contributed to the design of the experiments. RGFV contributed to the design of the experiments and revised the manuscript. MM contributed to the design of the experiments, contributed to the identification of MLO homologs in apple and revised the manuscript. HS contributed to the design of the experiments and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Stefano Pessina, Email: stefano.pessina@fmach.it.
Stefano Pavan, Email: stefano.pavan@uniba.it.
Domenico Catalano, Email: domenico.catalano@igv.cnr.it.
Alessandra Gallotta, Email: alessandra.gallotta@uniba.it.
Richard GF Visser, Email: richard.visser@wur.nl.
Yuling Bai, Email: bai.yuling@wur.nl.
Mickael Malnoy, Email: mickael.malnoy@fmach.it.
Henk J Schouten, Email: henk.schouten@wur.nl.
References
- 1.Glawe DA. The powdery mildews: a review of the world most familiar (yet poorly known) plant pathogens. Annu Rev Phytopathol. 2008;46:27–51. doi: 10.1146/annurev.phyto.46.081407.104740. [DOI] [PubMed] [Google Scholar]
- 2.Turechek WW, Carroll JE, Rosenberger DA. Powdery Mildew of Apple. New York State Integrated Pest Management Program: Cornell Universit; 2004. [Google Scholar]
- 3.Foulongne M, Pascal T, Pfeiffer F, Kervella J. QTLs for powdery mildew resistance in peach × Prunus davidiana crosses: consistency across generations and environments. Mol Breed. 2003;12:33–50. doi: 10.1023/A:1025417507358. [DOI] [Google Scholar]
- 4.Boesewinkel HJ. Differences between the conidial states of Podosphaera tridactyla and Sphaerotheca pannosa. Annales de Phytopathologie. 1979;11:525–527. [Google Scholar]
- 5.Xiao CL, Chandler CK, Price JF, Duval JR, Mertely JC, Legard DE. Comparison of epidemics of botrytis fruit rot and powdery mildew of strawberry in large plastic tunnel and field production systems. Plant Dis. 2001;85:901–909. doi: 10.1094/PDIS.2001.85.8.901. [DOI] [PubMed] [Google Scholar]
- 6.Fu XL, Lu Y, Liu XD, Li JQ. Crossability barriers in the interspecific hybridization betweenOryza sativaandO.Meyeriana. J Integr Plant Biol. 2009;51:21–28. doi: 10.1111/j.1744-7909.2008.00728.x. [DOI] [PubMed] [Google Scholar]
- 7.Parlevliet JE. What is durable resistance, a general outline. In: Jacobs TH, Parlevliet JE, editors. Durability of Disease Resistance. Dordrecht: Kluwer; 1993. pp. 23–29. [Google Scholar]
- 8.Parisi L, Lespinasse Y, Guillaumes SJ, Kruger J. A new race of Venturia inequalis virulent to apples with resistance due to the Vf gene. Phytopathology. 1993;85:533–537. doi: 10.1094/Phyto-83-533. [DOI] [Google Scholar]
- 9.Krieghoff O. Entwicklung einer In-vitro-selektionsmethode auf Resistenz von Malus-Genotypen gegenüber Podosphaera leucotricha (Ell. Et Ev.) Salm. und In-vitron-Differenzierung von Virulenzunterschieden des Erregers. Berlin: PhD dissertation, Humboldt-Universität; 1995. [Google Scholar]
- 10.Pavan S, Jacobsen E, Visser RGF, Bai Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed. 2010;25:1–12. doi: 10.1007/s11032-009-9323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88(5):695–705. doi: 10.1016/S0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 12.Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, Von Heijne G, Schulze-Lefert P. Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem. 1999;274:34993–35004. doi: 10.1074/jbc.274.49.34993. [DOI] [PubMed] [Google Scholar]
- 13.Jørgensen JH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63:141–152. doi: 10.1007/BF00023919. [DOI] [Google Scholar]
- 14.Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, Somerville SC, Panstruga R. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38(6):716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 15.Humphry M, Reinstädler A, Ivanov S, Bisseling T, Panstruga R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol. 2011;12(9):866–878. doi: 10.1111/j.1364-3703.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavan S, Schiavulli A, Appiano M, Marcotrigiano AR, Cillo F, Visser RGF, Bai Y, Lotti C, Ricciardi L. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor Appl Genet. 2011;123:1425–1431. doi: 10.1007/s00122-011-1677-6. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y, Pavan S, Zheng Z, Zappel N, Reinstädler A, Lotti C, De Giovanni C, Ricciardi L, Lindhout P, Visser RGF, Theres K, Panstruga R. Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of MLO function. Mol Plant Microbe Interact. 2008;21:30–39. doi: 10.1094/MPMI-21-1-0030. [DOI] [PubMed] [Google Scholar]
- 18.Aist JR, Bushnell WR. Invasion of plants by powdery mildew fungi, and cellular mechanisms of resistance. In: Cole GT, Hoch HC, editors. The Fungal Spore and Disease Initiation in Plants and Animals. New YorK: plenum press; 1991. pp. 321–345. [Google Scholar]
- 19.Miklis M, Consonni C, Bhat RA, Lipka V, Schulze-Lefert P, Panstruga R. Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol. 2007;144:1132–1143. doi: 10.1104/pp.107.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panstruga R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem Soc Transact. 2005;33(Pt 2):389–392. doi: 10.1042/BST0330389. [DOI] [PubMed] [Google Scholar]
- 21.Piffanelli P, Zhou FS, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129:1076–1085. doi: 10.1104/pp.010954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Z, Nonomura T, Bóka K, Matsuda Y, Visser RGF, Toyoda H, Kiss L, Bai Y. Detection and quantification of Leveillula taurica growth in pepper leaves. Phytopathology. 2013;103:6. doi: 10.1094/PHYTO-08-12-0198-R. [DOI] [PubMed] [Google Scholar]
- 23.Feechan A, Jermakow AM, Torregrosa L, Panstruga R, Dry IB. Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct Plant Biol. 2008;35:1255–1266. doi: 10.1071/FP08173. [DOI] [PubMed] [Google Scholar]
- 24.Winterhagen P, Howard SF, Qiu W, Kovács LG. Transcriptional up-regulation of grapevine MLO genesin response to powdery mildew infection. Am J Enol Vit. 2008;59:2. [Google Scholar]
- 25.Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330:968. doi: 10.1126/science.1195211. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Noir S, Kwaaitaal M, Hartmann A, Wu MJ, Mudgil Y, Sukumar P, Muday G, Panstruga R, Jones AM. Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell. 2009;21:1972–1991. doi: 10.1105/tpc.108.062653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinstädler A, Müller J, Czembor JH, Piffanelli P, Panstruga R. Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant Biol. 2010;10:31. doi: 10.1186/1471-2229-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devoto A, Hartmann HA, Piffanelli P, Elliott C, Simmons C, Taramino G, Goh CS, Cohen FE, Emerson BC, Schulze-Lefert P, Panstruga R. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J Mol Evol. 2003;56(1):77–88. doi: 10.1007/s00239-002-2382-5. [DOI] [PubMed] [Google Scholar]
- 29.Elliott C, Muller J, Miklis M, Bhat RA, Schulze-Lefert P, Panstruga R. Conserved extracellular cysteine residues and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem l J. 2005;385(Pt 1):243–254. doi: 10.1042/BJ20040993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufmann H, Qiu X, Wehmeyer J, Debener T. Isolation, molecular characterization, and mapping of four rose MLO orthologs. Front Plant Sci. 2012;3:244. doi: 10.3389/fpls.2012.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elliott C, Zhou F, Spielmeyer W, Panstruga R, Schulze-Lefert P. Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol Plant Microbe Interact. 2002;15(10):1069–1077. doi: 10.1094/MPMI.2002.15.10.1069. [DOI] [PubMed] [Google Scholar]
- 32.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, et al. The genome of the domesticated apple (Malus × domestica Borkh) Nat Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 33.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, Burns P, Davis TM, Slovin JP, Bassil N, Hellens RP, Evans C, Harkins T, Kodira C, Desany B, Crasta OR, Jensen RV, Allan AC, Michael TP, Setubal JC, Celton J, Rees DJG, Williams KP, Holt SH, Ruiz-Rojas JJ, Chatterjee M, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung S, Cestaro A, Troggio M, Main D, Zheng P, Cho I, Folta K, Sosinski B, Abbot B, Celton JM, Arùs P, Shulaev V, Verde I, Morgante M, Rokhsar D, Velasco R, Sargent DJ. Whole genome comparisons ofFragaria,Prunus and Malus reveal different modes of evolution between Rosaceous subfamilies. BMC Genomics. 2012;13:129. doi: 10.1186/1471-2164-13-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung S, Ficklin SP, Lee T, Cheng C, Blenda A, Zheng P, Yu J, Bombarely A, Cho I, Ru S, Evans KM, Peace C, Abbott A, Mueller L, Olmstead M, Main D. The genome database for rosaceae (GDR): year 10 update. Nucleic Acid Res. 2014;42:D1237–D1244. doi: 10.1093/nar/gkt1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huson D, Richter D, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKay SJ, Vergara IA, Stajich JE. Using the generic synteny browser (GBrowse_syn) Curr Protoc Bioinformatics. 2010;Chapter 9(Unit 9):12. doi: 10.1002/0471250953.bi0912s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dewey CN. Aligning multiple whole genomes with Mercator and MAVID. Methods Mol Biol. 2007;395:221–236. doi: 10.1007/978-1-59745-514-5_14. [DOI] [PubMed] [Google Scholar]
- 40.Kürkcüoglu S, Degenhardt J, Lensing J, Al-Masri AN, Gaul AE. Identification of differentially expressed genes in Malus domestica after application of the non-pathogenic bacterium Pseudomonas fluorescensBk3 to the phyllosphere. J Exp Bot. 2007;58(3):733–741. doi: 10.1093/jxb/erl249. [DOI] [PubMed] [Google Scholar]
- 41.Giorno F, Guerriero G, Baric S, Mariani C. Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics. 2012;13:639. doi: 10.1186/1471-2164-13-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dal Cin V, Danesin M, Boschetti A, Dorigoni A, Ramina A. Ethylene biosynthesis and perception in apple fruitlet abscission (Malus domestica L. Borck) J Exp Bot. 2005;56(421):2995–3005. doi: 10.1093/jxb/eri296. [DOI] [PubMed] [Google Scholar]
- 43.Ling D, Salvaterra PM. Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One. 2011;6:3. doi: 10.1371/journal.pone.0017762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Hiel MB, Van Wielendaele P, Temmerman L, Van Soest S, Vuerinckx K, Huybrechts R, Broeck JV, Simonet G. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. BMC Mol Biol. 2009;10:56. doi: 10.1186/1471-2199-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strube C, Buschbaum S, Wolken S, Schnieder T. Evaluation of reference genes for quantitative real-time PCR to investigate protein disulfide isomerase transcription pattern in the bovine lungworm Dictyocaulus viviparus. Gene. 2008;425:36–43. doi: 10.1016/j.gene.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Synteny between apple, peach and strawberry. Results of search for F. vesca and M. domestica regions syntenic to 50 kb P. persica chromosomal stretches containing the PpMLO homologs identified in this study. Shaded polygons indicate aligned regions between genomes. Grid lines are drawn to indicate insertions/deletions between the genomes of F. vesca and M. domestica with respect to the P. persica reference sequence. P. persica, F. vesca and M. domestica MLO homologs, named according to the nomenclature of the Genomic Database of Rosaceae, are boxed. (PDF 817 KB)
Additional file 2: Transcriptional variation of 19 apple MLO genes in three cultivars following inoculation with P. leucotricha. Transcription abundances of 19 MLO-like genes following powdery mildew (PM) inoculation in ‘Golden Delicious’ (1a), ‘Gala’ (1b) and ‘Braeburn (1c) leaf samples. The graphs show expression values of inoculated samples relative to control samples, averaged from four to eight biological replicate, normalized, that are in turn the average of two experimental replicates. The Ct values have been normalized with three reference genes: actin, ubiquitin and elongation factor 1. Statistical significance was determined with a t-test for each individual pair of inoculated and control samples at each time point (4, 6, 8 and 24 hpi). The error bars show standard errors of the means. Significant differences between inoculated samples and control samples are indicated with a *(P < 0.05). (PDF 1 MB)


