Abstract
AIM: To observe the inhibition of hepatitis B virus replication and expression by transfecting vector-based small interference RNA (siRNA) pGenesil-HBV X targeting HBV X gene region into HepG2.2.15 cells.
METHODS: pGenesil-HBV X was constructed and transfected into HepG2.2.15 cells via lipofection. HBV antigen secretion was determined 24, 48, and 72 h after transfection by time-resolved immunofluorometric assays (TRFIA). HBV replication was examined by fluorescence quantitative PCR, and the expression of cytoplasmic viral proteins was determined by immunohistochemistry.
RESULTS: The secretion of HBsAg and HBeAg into the supernatant was found to be inhibited by 28.5% and 32.2% (P < 0.01), and by 38.67% (P < 0.05) and 42.86% (P < 0.01) at 48 h and 72 h after pGenesil-HBV X transfection, respectively. Immunohistochemical staining for cytoplasmic HBsAg showed a similar decline in HepG2.2.15 cells 48 h after transfection. The number of HBV genomes within culture supernatants was also significantly decreased 48 h and 72 h post-transfection as quantified by fluorescence PCR (P < 0.05).
CONCLUSION: In HepG2.2.15 cells, HBV replication and expression is inhibited by vector-based siRNA pGenesil-HBV X targeting the HBV X coding region.
Keywords: Hepatitis B virus, RNA interference, Plasmid vector, HepG2.2.15
INTRODUCTION
Hepatitis B is a severe infectious disease threatening peoples’ health all over the world. There is still no efficient therapy to control HBV persistent replication, which may lead to the development of liver cirrhosis and hepatocellualar carcinoma (HCC)[1]. RNA interference (RNAi) is a highly specific and effective mechanism of post-transcriptional gene silencing mediated by double-stranded RNA of 21-23 nt in size. Several researches have suggested that RNAi could provide a new therapeutic strategy against chronic HBV infection[2-6]. In the present study, a plasmid leading to the expression of small interfering RNA (siRNA) that targets the HBV X gene was transfected into HepG2.2.15 cells, and HBV DNA replication as well as HBV antigen expression and secretion were monitored.
MATERIALS AND METHODS
Materials
Dulbecco’s modified Eagle’s medium (DMEM) was purchased from Gibco BRL, USA. Metafectene transfection reagent was purchased from Germany Biontes. Diagnostic kits for HBsAg and HBeAg (time-resolved immunofluorometric assay) were obtained from Suzhou Xinbo Biotechnology Corporation. Mouse monoclonal antibody directed against human HBsAg and rabbit anti-mouse horseradish peroxidase (HRP)–IgG were purchased from Beijing Zhongshan Golden Bridge Biotechnology Corporation. HepG2.2.15 cells were obtained from the Institute of Infectious Disease, Beijing University Medicine School.
Plasmid construction
pGenesil, containing human U6 promoter, was used to generate a series of siRNA expression vectors by inserting annealed oligonucleotides between BamHI and HindIII sites. The oligonucleotides 5′-GAT CCG GTC TTA CAT AAG AGG ACT TTC AAG ACG AGT CCT CTT ATG TAA GAC CTT TTT TGT CGA CA-3′ (sense) and 3′-GCC AGA ATG TAT TCT CCT GAA AGT TCT GCT CAG GAG AAT ACA TTC TGG AAA AAA CAG CTG TTC GA-5′ (antisense) were used for the construction of pGenesil-HBV X targeting HBV X (N 1649 to 1667)[7]; 5′-GAT CCG CAT TG G CAA AGC GAA GCT TTC AAG ACG AGC TTC GCT TTG CCA ATG CTT TTT TGT CGA CA-3′ (sense) and 3′-GCG TAA CCG TTT CGC TTC GAA AGT TCT GCT CGA AGC GAA ACG GTT ACG AAA AAA CAG CTG TTC GA-5′ (antisense) for a control vector targeting the α-fetoprotein (AFP) gene (N 1275 to 1293); and 5′-GAT CCG ACT TCA TAA GGC GCA TGC TTC AAG ACG GCA TGC GCC TTA TGA AGT CTT TTT TGT CGA CA-3′ (sense) and 3′-GCT GAA GTA TTC CGC GTA CGA AGT TCT GCC GTA CGC GGA ATA CTT CAG AAA AAA CAG CTG TTC GA-5′ (antisense) for the control vector pGenesil-HK producing a random sequence of siRNA.
Cell culture and transfection
HepG2.2.15 cells were maintained in DMEM supplemented with 100 mL/L fetal calf serum (FCS), 100 IU/mL penicillin, 100 mg/L streptomycin and 2 mmol/L L-glutamine at 37°C in an atmosphere of 50 mL/L CO2. The cells were plated in 6-well plates which had been placed sterile cover slips (1 × 106 cells per well). Transfection was performed at about 70% confluence with pGenesil and Metafectene lipofection reagent complex at a ratio of 8 μL:2.5 µg. Under these conditions we obtained a 55% transfection efficiency (data not shown).
Quantitative assay of HBsAg and HBeAg
The levels of HBsAg and HBeAg in culture supernatants were measured at 24, 48 and 72 h after transfection by using time-resolved immunofluorometric assay kits (TRFIA) according to the supplier’s instructions.
Assay of HBV DNA replication
HepG2.2.15 cells were harvested at 24, 48, and 72 h post-transfection. Forty µL of the supernatant were mixed with an equal volume of the DNA extractant. Samples were boiled for 10 min and then centrifuged at 10 000 × g for 5 min. Two μL of the samples were transferred into PCR reaction tubes. PCR cycling parameters consisted of denaturation at 93°C for 2 min, followed by 93°C for 45 s; 55°C for 60 s × 10 cycles and then 93°C for 30 s; 55°C for 45 s × 30 cycles.
Immunocytochemistry of HBsAg
To examine whether the effects of pGenesil-HBV X on HBsAg production were uniform with the culture media, cytoplasmic HBsAg was visualized by indirect immunocytochemistry 48 h post-transfection. pGenesil-HBV X transfected and control cells were washed with PBS, fixed in 900 mL/L ethanol for 10 min at room temperature and then washed with PBS. The fixed cells were permeabilized with 5 mL/L Triton X-100 in PBS for 15 min at 37°C and washed with PBS. To inhibit endogenous peroxidase, cells were exposed to 3 mL/L hydrogen peroxide for 10 min at 25°C. After washed with PBS, cells were incubated with mouse monoclonal anti-HBsAg antibody for 2 h at 37°C and subsequently with rabbit anti-mouse IgG conjugated horseradish-peroxidase for 30 min at 37°C. Cells were visualized with 3, 3'-diaminobenzidine tetrahydrochloride substrate and examined by light microscopy.
Statistical analysis
All statistical analysis were performed using the Microsoft SPSS 12.0 software. The graphs represented in mean ± SD and compared using unpaired t-test. P < 0.05 was regarded as a significant difference.
RESULTS
pGenesil-HBV X inhibited HBsAg and HBeAg secretion in cultured HepG2.2.15
HBsAg and HBeAg concentrations were measured in cell culture supernatants of pGenesil-HBV X treated and control cells 24, 48, and 72 h post-transfection by using TRFIA (Table 1). At 24 h in the culture media, there was no significant difference between pGenesil-HBV X treated cells and other controls (untreated control, pGenesil-AFP control , pGenesil-HK control, pGenesil alone and Metafectene alone) (P > 0.05), while HBsAg was inhibited at 48 and 72 h by 28.47% and 32.16% (P < 0.01). HBeAg was reduced at 48 and 72 h post-transfection (P < 0.05) by 38.7% and 42.9% in the media of pGenesil-HBV X treated cells compared to the controls.
Table 1.
Effect of pGenesil-siHBV X on HBsAg and HBeAg expression transfected HepG2.2.15 cells
| Group |
HBsAg (g/L) |
HBeAg (Ncu/mL) |
||||
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |
| Untreated | 7.13 ± 0.20 | 14.43 ± 0.56 | 22.50 ± 2.14 | 0.43 ± 0.02 | 0.75 ± 0.06 | 1.12 ± 0.10 |
| Metafectene | 6.80 ± 0.17 | 13.57 ± 0.88 | 21.84 ± 0.91 | 0.48 ± 0.02 | 0.69 ± 0.08 | 1.03 ± 0.06 |
| pGenesil | 6.57 ± 0.40 | 14.30 ± 0.47 | 24.15 ± 2.24 | 0.45 ± 0.06 | 0.64 ± 0.05 | 1.00 ± 0.06 |
| PGenesil-HK | 6.47 ± 0.20 | 13.63 ± 0.64 | 21.84 ± 0.51 | 0.46 ± 0.05 | 0.69 ± 0.07 | 0.97 ± 0.08 |
| PGenesil-AFP | 6.33 ± 0.35 | 13.70 ± 0.73 | 23.11 ± 1.25 | 0.47 ± 0.05 | 0.76 ± 0.08 | 1.10 ± 0.12 |
| PGenesil-HBVX | 5.97 ± 0.13 | 10.25 ± 0.32b | 15.26 ± 0.88b | 0.43 ± 0.02 | 0.46 ± 0.01c | 0.64 ± 0.04c |
P < 0.01 vs untreated control and other controls;
P < 0.05 vs untreated control and other controls.
Inhibited HBV DNA replication in cultured HepG2.2.15
Levels of HBV DNA were examined by fluorescence quantitative PCR. This assay can detect HBV DNA in the range of 103 to 108 copies. The results revealed a significant decrease in DNA replication when pGenesil-HBV X treated cells were compared to untreated cells. The number of HBV DNA copies in pGenesil-HBV X treated cells was found to be reduced by 44.9% and 45.9% at 48 and 72 h after transfection, respectively (P < 0.05), while the other controls showed no significant difference to the untreated cultures at any time point (Figure 1).
Figure 1.
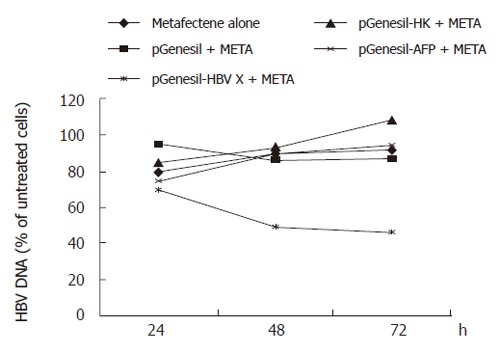
Inhibition of pGenesil-HBV X on HBV DNA in HepG2.2.15.
The effects of pGenesil-HBV X on intracellular HBsAg in HepG2.2.15
The effects of pGenesil-HBV X on intracellular HBsAg were visualized by immunocytochemistry 48 h after transfection. Intracellular HBsAg is localized in the cytoplasm of HepG2.2.15 cells at normal. In cells treated with pGenesil-HBV X, HBsAg was either decreased or non-detectable. In contrast, cells of other controls were obviously stained (Figure 2).
Figure 2.
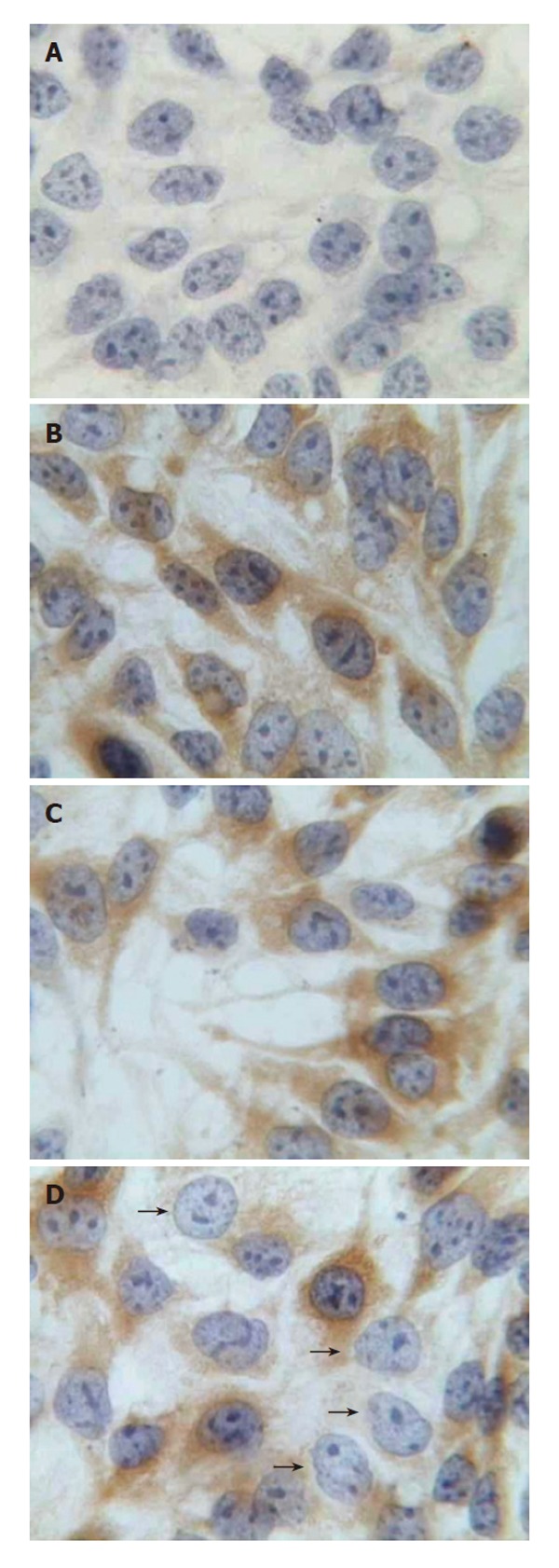
Immunocytochemistry for intracellular HBsAg in HepG2.2.15 (× 400). A: Without monoclonal antibody against HBsAg; B: pGenesil-HK treated cells; C: pGenesil-AFP treated cells; D: pGenesil-HBV X treated cells. Arrows show the cells were transfected and HBsAg expression was suppressed.
DISCUSSION
RNAi is a mechanism of post-transcriptional gene silencing mediated by double-stranded RNA of 21-23 nt in length. RNAi has been applied as a highly specific and efficient tool to interfere with viral replication as it has been shown for HIV[8-12], hepatitis C virus[13-16], myetitis virus[17], gamma herpesvirus[18] or influenza virus[19]. HBV DNA replication requires reverse transcription to form a pregenomic RNA that is similar to reverse-transcription virus. 3.2kb pregenomic mRNA is not only translated into HBV proteins including HBeAg, HBcAg and HBV DNAP, but it is also a template for the synthesis of viral DNA to continue replication. Furthermore, all four transcripts include the HBV X protein coding region. Selecting a conserved sequence in the X gene region as a target, we expect to inhibit the expression of HBV antigens and the replication of HBV DNA. To set up an in vitro model which is more stable and more similar to natural condition of viral infection is a base of experiment and is a key prerequisite to evaluate the efficiency of anti-virus therapy. Previous studies were carried out by co-transfecting siRNA or siRNA expression vector and a plasmid cloned with full-length HBV DNA or HBV target region into cells[7,20,21]. It is obvious that cells transfected with siRNA or siRNA expression vectors must get HBV expression plasmid at same time, which is better to observe and evaluate the specificity and efficiency by siRNA. But only selecting out the cells co-transtected successfully for research does not coincide with natural conditions of viral infection and will not reflect the exact effects of siRNA on HBV target gene.
The HepG2.2.15 cell line, a derivative of the human HepG2 hepatoma cell line that has been stably trans-formed with a head-to-tail dimer of HBV DNA[22], was chosen as a model because it produces HBV infectious particles constitutively and expresses HBV antigens stably. To transfected siRNA expression vector into HepG2.2.15 cells, and culturing transfected cells and untransfected cells under one system, we could simulate the nature condition that virus still replicate and express constitutively in untransfected cells. As a result, we found that HBsAg and HBeAg in the supernatant were inhibited by 28.5% and 38.7% at 48 h, and decreased by 32.2% and 42.9% 72 h post-tranfection with pGenesil-HBV X against HBV X. Levels of HBV DNA were also found to be reduced. Moreover, immunocytochemistry revealed that the amount of intracellular HBsAg parallels the decline in HBV serum markers in cultures treated with pGenesil-HBV X. Controls (including untreated cells and treated with Metafectent reagent alone, pGenesil plasmid alone, pGenesil-AFP or pGenesil-HK expressing random siRNA) failed to reduce HBV expression and replication. At the premises of targeting same sequence in HBV X region, the efficiency we got was lower than that of Shlomai[7] who used a co-transfection approach to Huh-7 cells. At present, there isn’t any reagent that achieves 100% transfection efficiency neither to cells nor to animals. Selecting HepG2.2.15 cell line as a model is better for us to evaluate the effects of siRNA in consideration of the efficiency of transfection, and thus it will be valuable for us to evaluate the effects of siRNA on clinical application research in the future.
By measuring the levels of AFP which is constitutively secreted by HepG2.2.15 cells no change was found between pGenesil-HBV X treated cells and other controls except pGenesil-AFP treated cells. This result shows that pGenesil-HBV X specifically inhibits HBV gene expression.
ACKNOWLEDGMENTS
Jiang-Nan Feng and Wuhan Genesil Biotechnology Corporation for kindly helping us construct pGenesil-HBV X.
Footnotes
S- Editor Pan BR L- Editor Mihm S E- Editor Ma WH
References
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 4.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- 8.Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 10.Park WS, Miyano-Kurosaki N, Hayafune M, Nakajima E, Matsuzaki T, Shimada F, Takaku H. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interference. Nucleic Acids Res. 2002;30:4830–4835. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coburn GA, Cullen BR. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capodici J, Karikó K, Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- 13.Seo MY, Abrignani S, Houghton M, Han JH. Small interfering RNA-mediated inhibition of hepatitis C virus replication in the human hepatoma cell line Huh-7. J Virol. 2003;77:810–812. doi: 10.1128/JVI.77.1.810-812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci U S A. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 18.Jia Q, Sun R. Inhibition of gammaherpesvirus replication by RNA interference. J Virol. 2003;77:3301–3306. doi: 10.1128/JVI.77.5.3301-3306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, Chen J. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XN, Xiong W, Wang JD, Hu YW, Xiang L, Yuan ZH. siRNA-mediated inhibition of HBV replication and expression. World J Gastroenterol. 2004;10:2967–2971. doi: 10.3748/wjg.v10.i20.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang N, Huang AL, Zhang BQ, Yan G, He TC. [Potent and specific inhibition of hepatitis B virus antigen expression by RNA interference] Zhonghua Yixue Zazhi. 2003;83:1309–1312. [PubMed] [Google Scholar]
- 22.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]


