Summary
With multiple clinical trials underway targeting autophagy against cancer, Yang et al. (1) and Karsli-Uzunbas et al. (2) address important concerns regarding autophagy inhibition in cancer patients using genetically engineered mouse models (GEMMs) that more accurately represent the tumor biology found in human pancreatic and lung cancer patients.
The consideration of autophagy as a therapeutic target in cancer has been mired by the complexities of the pathway and the seemingly dual roles it plays in tumorigenesis. On the one hand, autophagy has been described as a tumor suppressor mechanism, best exemplified by the fact that Beclin1 heterozygote mice develop spontaneous tumors (3). However, increasing evidence suggests that, especially at later stages in tumorigenesis, autophagy supports tumor growth. In addition, cytoprotective autophagy is induced by many cancer therapies as a stress response (3). Based on these findings, multiple clinical trials targeting autophagy using the antimalarial lysosomal inhibitor hydroxychloroquine (HCQ) are underway, and pharmaceutical companies are developing novel potent and specific autophagy inhibitors. More recently, concerns have arisen whether p53 status impacts the anticancer efficacy of autophagy inhibition in certain tumors as well as whether systemic autophagy inhibition will have deleterious effects on the normal tissues in cancer patients. Two articles in this issue of Cancer Discovery from Yang et al. (1) and Karsli-Uzunbas et al. (2) both provide reassurance about targeting autophagy in cancer and motivate the further testing of autophagy inhibitors in the clinic.
In the past few years, a number of investigators have moved beyond cell lines and xenograft mouse models to address the role of autophagy during cancer development and progression using tumor specific ablation of autophagy genes, such as Atg7, in immunocompetent genetically engineered mouse models (GEMMs). In BRAF-driven lung cancer, Atg7 deficiency initially promoted tumor growth. However, in lung cancers driven by either mutant KRAS or BRAF, Atg7 deletion ultimately stalled tumor growth and promoted oncocytic differentiation with and without p53 (4, 5). These results partially reconciled the dual roles of autophagy during the process of tumorigenesis, but overall supported a role for therapeutic strategies to inhibit autophagy in certain advanced cancers.
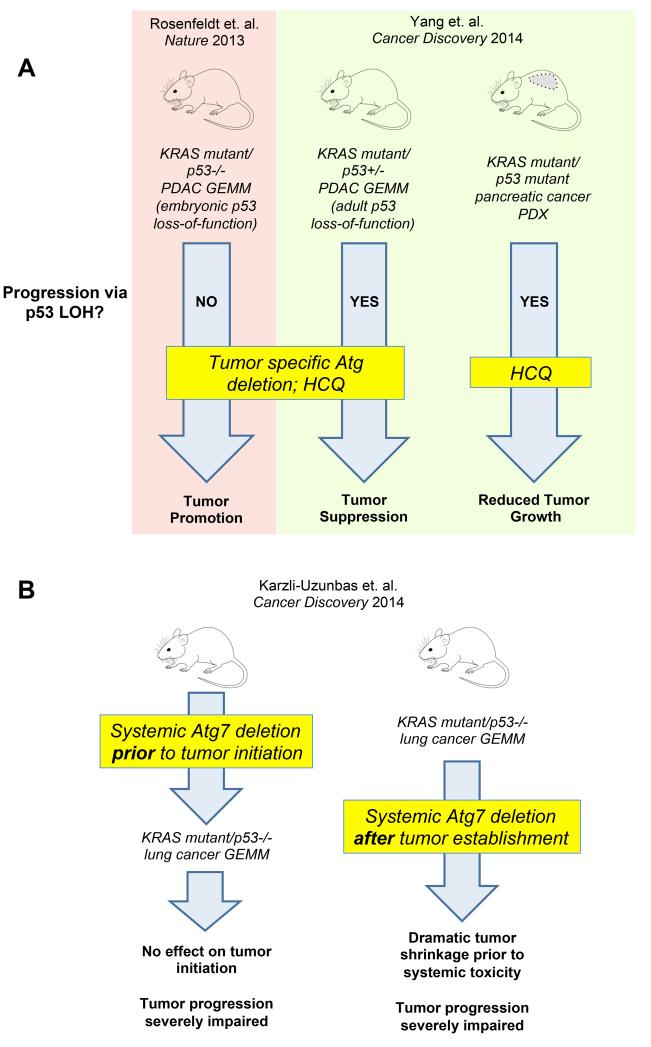
However, an important red flag was recently raised by Rosenfeldt et al. using a GEMM of pancreas cancer. In this model of pancreas-specific KRAS-mutant, p53−/− tumors, genetic ablation of atg7 or atg5 within tumors or pharmacological inhibition of autophagy with HCQ accelerated the formation of pancreatic ductal adenocarcinomas (PDACs) in mice (Figure 1A, left) (6). Although these findings illustrated the importance of molecular context in the outcome of autophagy inhibition in vivo, they also had the unfortunate consequence of motivating clinical recommendations that patients with p53 mutant pancreas cancer and possibly other p53 mutant cancers avoid clinical trials using HCQ (7). Another important concern raised in this study was that pancreas-specific deletion of ATG7 produced pancreatic atrophy and independently contributed to death of mice due to exocrine pancreas deficiency. This result was consistent with growing evidence demonstrating the importance of autophagy in maintaining organismal metabolism and tissue function (2) (3). Importantly, to date, most in vivo models of autophagy deficiency in cancer have singularly targeted the tumor cell compartment; hence, they have been unable to evaluate the collateral damage to normal tissues that systemic autophagy inhibition may produce in patients.
Figure 1. Effects of autophagy inhibition in mouse models of pancreatic and lung cancer.
(A) In genetically engineered mouse models (GEMM) and patient-derived xenografts (PDX) of KRAS mutant pancreatic ductal adenocarcinoma (PDAC), both genetic autophagy ablation and hydroxychloroquine (HCQ) treatment elicited contrasting effects on tumorigenesis. The phenotypes observed upon autophagy inhibition in each model are likely the result of the developmental stage at which p53 tumor suppression function is lost (embryonic or during tumor progression), and whether PDAC progression involves p53 loss of heterozygosity (LOH). (B) In a KRAS mutant, p53−/− lung cancer GEMM, the effects of systemic autophagy inhibition were analyzed following inducible genetic Atg7 deletion in the entire adult mouse. Systemically inhibiting autophagy prior to tumor onset did not affect early initiation; rather, it severely impaired the progression to advanced cancers (left). In mice with established lung tumors, acute autophagy deletion led to significant tumor regression, which occurred prior to the onset of any systemic toxicity or normal tissue degeneration secondary to the complete loss of autophagy in the whole animal (right).
A major aspect of the KRAS mutant, p53−/− PDAC model in Rosenfeldt et al. is that it utilizes embryonic pancreas-specific homozygous deletion of p53 in the context of KRAS mutation, resulting in advanced carcinoma during early development (Figure 1A, left) (6). In contrast, p53 is most commonly found as missense point mutations in KRAS mutant pancreatic cancers. The heterozygous expression of mutant p53 in the setting of oncogenic KRAS is proposed to facilitate the formation of precancerous lesions called pancreatic intraepithelial neoplasias (PanINs); subsequent loss of heterozygosity (LOH) of the wild type p53 allele drives the progression from PanIN to PDAC (1). Thus, the very rapid and aggressive disease observed in this mouse PDAC model employing early developmental deletion of p53 does not fully recapitulate the typical stepwise progression of pancreas cancer found in humans.
To address this salient issue, Yang et al. (1) employed a pancreas specific KRAS mutant p53+/− mouse model that exhibits LOH of the wild type p53 allele during PDAC progression, thereby reproducing the stepwise human development of pancreas cancer more faithfully than the KRAS mutant, p53−/− model. When Atg5 was genetically ablated in the KRAS mutant, p53+/− model, the number of PanIN lesions was increased, but the progression of PanIN to PDAC was significantly prevented, and mice with autophagy-deficient tumors survived longer. Importantly, the investigators confirmed that p53 LOH was responsible for the conversion of PanIN to PDAC in the context of ATG5 deficiency (Figure 1A, right). These beneficial effects of autophagy inhibition were also observed upon treating KRAS mutant PDAC tumor cells, possessing either deleted or mutant p53, with chloroquine (CQ). In addition, this paper interrogated a large collection of genetically characterized patient-derived xenografts (PDX) treated with HCQ in vivo (Figure 1A, right). Impressive tumor growth reduction was observed uniformly in 100% of KRAS mutant, p53 mutant pancreatic PDX lines. Altogether, this focus on a key aspect of p53 genetics in mouse models convincingly ameliorates concerns about developing autophagy inhibitors for p53 mutant pancreas cancers and possibly other p53 mutant tumors. It also teaches us a major lesson that extrapolation of mouse model data to help make clinical decisions for therapeutic strategies needs to be done with careful consideration, and that the details of the model really do matter. While it now clearly appears premature to turn patients with p53 mutant tumors away from clinical trials of autophagy inhibitors, the overall results from these two seminal studies in pancreatic cancer highlight the importance of obtaining p53 genotypes in patients enrolled on autophagy inhibitor trials (1) (6).
The development of a mouse model that better recapitulates the human response to systemic therapy allowed another group of investigators to tackle the concern that autophagy inhibition may be too toxic to pursue in the clinic due to the protective roles of autophagy in normal cells. Karsli-Uzunbas et al. (2) developed an inducible model of systemic ATG7 ablation that allowed the study of systemic autophagy inhibition in adult mice, as well as the effects of acute autophagy ablation on the initiation, progression and maintenance of KRAS-driven lung cancers. The systemic loss of ATG7 in adult mice surprisingly showed little toxicity for 5 weeks, although over two to three months, these autophagy-null mice did develop liver and muscle injury, depletion of white adipose tissue, and lethal neurodegeneration. Moreover, these mice did not tolerate fasting, and rigorous metabolic analysis uncovered a critical role for ATG7 in this context for the maintenance of glucose homeostasis in starved animals. Next, the investigators systemically deleted Atg7 and then initiated lung cancer by activating KRAS and deleting p53 (Figure 1B). Although early tumor initiation was independent of ATG7 status, the lung tumors that did arise in the setting of systemic ATG7 deficiency failed to progress to aggressive cancers and displayed the histological features of benign oncocytomas.
Finally, when KRAS mutant, p53−/− lung cancers were allowed to grow in adult mice, upon which Atg7 was systemically ablated in the context of established tumors, there was substantial antitumor activity, with tumor growth arrest, increased apoptosis, and loss of RAS-driven oncogenic signaling (Figure 1B). Tumor clusters that were examined by histology consisted largely of dead cells and cells with oncocytic features. Importantly, the anti-tumor effects of systemic autophagy inhibition occurred rapidly and prior to the onset of any normal tissue degeneration attributable to systemic Atg7 deletion. Notably, this last experiment most closely recapitulates the clinical scenario of treating advanced KRAS mutant lung cancer patients with pharmacological autophagy inhibitors, including anti-malarials like HCQ. Although the multi-organ toxicity associated with long-term Atg7 deletion continues to illustrate that certain toxicities should be considered when applying autophagy inhibitors to humans, it is very important to recognize that pharmacological autophagy inhibitors, both present and future, are unlikely to achieve the complete and irreversible autophagy null phenotype obtained in this model. In addition, due to disruption of protein-protein interactions, the complete loss of the ATG7 protein may have unique implications in comparison to either pharmacological inhibition of ATG7 enzymatic activity or lysosomal inhibition using HCQ or other anti-malarial agents.
The important findings of these two mouse models arrive at an opportune time as the first series of HCQ clinical trials in cancer patients have been recently published; three examples include (8-10). In general, the non-hematological toxicity profile across these clinical trials was mild and manageable, despite achieving very high doses of HCQ in certain instances. Remarkably, there was no evidence of extensive metabolic problems, liver injury, or neurological impairment. Taken together with the GEMM data, it now appears that ample evidence supports moving forward with autophagy inhibition in cancer patients, either with anti-malarials or the next generation of more specific upstream autophagy inhibitors.
Footnotes
Conflicts of interest: The authors have no conflicts of interest to disclose.
References
- 1.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karsli-Uzunbas G, Guo JY, Price SM, Teng X, Laddha SV, Khor S, et al. Autophagy is Required for Glucose Homeostasis and Lung Tumor Maintenance. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes & development. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes & development. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer discovery. 2013;3:1272–85. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 7.Iacobuzio-Donahue CA, Herman JM. Autophagy, p53, and pancreatic cancer. The New England journal of medicine. 2014;370:1352–3. doi: 10.1056/NEJMcibr1400189. [DOI] [PubMed] [Google Scholar]
- 8.Rangwala R, Leone R, Chang C, Fecher L, Schucter L, Kramer A, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10 doi: 10.4161/auto.29118. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10 doi: 10.4161/auto.28984. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogl DT, Stadtmauer EA, See Tan K, Heitjan DF, Davis LE, Pontiggia L, et al. Combined autophagy and proteasome inhibition: A phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10 doi: 10.4161/auto.29264. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]