Abstract
The QT interval, an electrocardiographic measure reflecting myocardial repolarization, is a heritable trait. QT prolongation is a risk factor for ventricular arrhythmias and sudden cardiac death (SCD) and could indicate the presence of the potentially lethal Mendelian Long QT Syndrome (LQTS). Using a genome-wide association and replication study in up to 100,000 individuals we identified 35 common variant QT interval loci, that collectively explain ∼8-10% of QT variation and highlight the importance of calcium regulation in myocardial repolarization. Rare variant analysis of 6 novel QT loci in 298 unrelated LQTS probands identified coding variants not found in controls but of uncertain causality and therefore requiring validation. Several newly identified loci encode for proteins that physically interact with other recognized repolarization proteins. Our integration of common variant association, expression and orthogonal protein-protein interaction screens provides new insights into cardiac electrophysiology and identifies novel candidate genes for ventricular arrhythmias, LQTS,and SCD.
Keywords: genome-wide association study, QT interval, Long QT Syndrome, sudden cardiac death, myocardial repolarization, arrhythmias
Introduction
Prolongation or shortening of the QT interval on the electrocardiogram are non-invasive markers of delayed or accelerated myocardial repolarization, increased risk of sudden cardiac death (SCD) and fatal arrhythmia as a side effect of medication therapy. Mendelian Long and Short QT Syndromes (LQTS, SQTS)1 stem from mutations of strong effect (QT increase or decrease per mutation > ∼20-100 msec) in genes encoding ion channels or channel-interacting proteins. In unselected community-based individuals, variation in continuous QT interval is normally distributed (ranging from 380 to 460 msec), with heritability estimates of 30-40%2. Common genetic variants that are associated individually with modest (≈1-4 msec/allele) increments in QT interval duration have been detected through candidate gene and genome-wide association studies (GWAS) in large sample sizes including the QTGEN3 and QTSCD4 consortia, as well as others5-9.
Several loci have been discovered independently in both genome-wide linkage studies of Mendelian LQTS families and GWAS of QT interval duration in unselected populations, including those harboring KCNQ1, KCNH2, SCN5A, KCNE1, and KCNJ2, highlighting the value of both approaches and the overlap of common and rare variant loci. To date, hundreds of rare mutations in 13 LQTS-susceptibility genes have been reported, with 75% of LQTS stemming from KCNQ1 (LQT1), KCNH2 (LQT2) and SCN5A (LQT3) mutations, < 5% due to LQT4-13 and ∼20% remaining genetically elusive. Identification of the causal genes underlying QT interval variation in the general population has been more challenging.
Here, the QT Interval-International GWAS Consortium (QT-IGC) performed an expanded meta-analysis of GWAS in 76,061 individuals of European ancestry with targeted genotyping in up to 33,316 additional individuals, completed mutational analysis in probands with genetically elusive LQTS, determined whether QT-associated SNPs have effects on gene expression in various tissues (eQTLs) or on other human phenotypes, and further annotated QT-associated genes using protein-protein interaction analyses.
GWAS identifies 22 novel QT loci
Clinical characteristics of 31 study cohorts of European ancestry who contributed to the stage 1 GWAS are shown in Supplementary Table 1. All studies excluded individuals with atrial fibrillation, QRS duration greater than 120 msec, bundle branch block, or intraventricular conduction delay, and when available electronic pacemaker use or QT-altering medication use. In each cohort, QT interval duration adjusted for age, sex, RR interval (inverse heart rate), and principal components of genetic ancestry was tested for association with 2.5 million directly genotyped or imputed SNPs under an additive genetic model (Supplementary Table 2 and 3). We performed inverse variance weighted meta-analysis on the GWAS results from 76,061 individuals, and observed only modest over-dispersion of the test statistics given the sample size (λGC = 1.076, Supplementary Figure 1a). Exclusion of SNPs within 500kb of the sentinel SNP at genome-wide significant loci (some identified only after incorporation of replication genotyping; see below) did not significantly attenuate the excess of low p-values, consistent with a polygenic model of QT interval variation10 (λGC = 1.069, Supplementary Figure 1b).
At an interim meta-analysis of GWAS results, SNPs were selected for two forms of replication. First, a set of the top 35 independent SNPs (one per locus) were selected for targeted replication genotyping in as many as 31,962 individuals of European ancestry (Supplementary Table 1) on a variety of platforms (Supplementary Table 2). Second, a set of ∼5,000 LD-pruned (r2 > 0.2) SNPs with nominal evidence of association with QT interval (P ≤ 0.015) were included in a custom genotyping array (Metabochip) and genotyped in 1,354 individuals (Supplementary Note, Supplementary Tables 1, 4, 5)11. Meta-analysis of all GWAS and replication genotyping results in up to 103,331 individuals (Supplementary Note) identified a total of 35 genome-wide significant (P < 5×10-8) loci, of which 22 were novel and 13 have been reported previously3-5,9 (Table 1, Figure 1, Supplementary Table 6). Some SNPs initially selected for replication genotyping were not ultimately the most significant SNP at a locus (Supplementary Table 7). Many loci had evidence of multiple independent signals of association based on having low LD (r2 < 0.05) to other genome-wide significant SNPs, with a total of 68 independent SNPs at 35 loci (Supplementary Note, Supplementary Table 8a, Supplementary Figure 2).
Table 1.
Common genetic variants at loci associated with QT interval (P < 5×10-8) on meta-analysis of GWAS+replication results (Supplementary Table 6). N is the effective number of samples contributing to the signal. For a given SNP, the effective sample size is the sum of the product of the cohort-specific sample size and imputation quality (ranging from 0 to 1). Function (Fxn) shown for coding variants with r2 = 1 to sentinel SNP or proxy with 1.0 > r2 > 0.8 (-p) to the sentinel SNP. Expression quantitative trait loci (eQTL) transcripts are shown if associated at P < 5×10-8 with sentinel SNPs or their close proxies (r2 > 0.8, Supplementary Table 12, bolded if eQTL found in left ventricle for the sentinel SNP). Protein-protein interactor (PPI) relationships for nearby genes to genes in loci previously established to influence myocardial repolarization are provided (Supplementary Table 17). Interactors from immunoprecipitation (IP) experiments are shown from murine cardiac tissue using 5 baits (K1=KCNQ1, K2=KCNH2, CV=CAV3, CA=CACNA1C, S1=SNTA1) with protein identified in parentheses if different from the nearest gene listed.Loci at which a SNP (index or secondary) or a close proxy (r2>0.8) falls in a left ventricular enhancer are marked with I or S. Brackets indicate annotations for secondary signals of association (Supplementary Table 8a).
| Nearest gene |
SNP | chr | position hg18 |
coded/ noncoded allele |
coded allele freq |
N | effect msec (SE) |
P | LQTS gene locus |
Fxn | eQTL transcript |
PPI interactor known QT loci |
IP interactor |
LV enhancer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Previously discovered loci | ||||||||||||||
|
| ||||||||||||||
| RNF207 | rs846111 | 1p36 | 6,201,957 | C/G | 0.28 | 47,041 | 1.73 (0.13) | 7×10-40 | G603A | |||||
| NOS1AP | rs12143842 | 1q23 | 160,300,514 | T/C | 0.24 | 75,053 | 3.50 (0.11) | 1×10-213 | S | |||||
| ATP1B1 | rs10919070 | 1q24 | 167,365,661 | C/A | 0.13 | 75,707 | -1.68 (0.14) | 1×10-31 | intron | ATP1B1,NME7 | ATP1B1-K1, ATP1B1-K2, ATP1B1-CA, ATP1B1-CV | I,S | ||
| SLC8A1 | rs12997023 | 2p22 | 40,606,486 | C/T | 0.05 | 70,311 | -1.69 (0.22) | 5×10-14 | ANK2,CAV3 | I,S | ||||
| SCN5A-SCN10A | rs6793245 | 3p22 | 38,574,041 | A/G | 0.32 | 73,697 | -1.12 (0.10) | 4×10-27 | LQT3 | intron | SCN5A-SNTA1 | S | ||
| SLC35F1-PLN | rs11153730 | 6q22 | 118,774,215 | T/C | 0.50 | 74,932 | -1.65 (0.10) | 2×10-67 | PLN-CV, PLN-CA | S | ||||
| KCNH2 | rs2072413 | 7q36 | 150,278,902 | T/C | 0.27 | 65,331 | -1.68 (0.11) | 1×10-49 | LQT2, SQT1 | intron | KCNE1 | S | ||
| KCNQ1 | rs7122937 | 11p15 | 2,443,126 | T/C | 0.19 | 72,978 | 1.93 (0.12) | 1×10-54 | LQT1, SQT2 | intron | C11ORF21,PHEMX,TSPAN32 | KCNE1,KCNH2 | I | |
| LITAF | rs735951 | 16p13 | 11,601,037 | A/G | 0.46 | 62,994 | -1.15 (0.10) | 2×10-28 | LITAF | S | ||||
| CNOT1 | rs246196 | 16q21 | 57,131,754 | C/T | 0.26 | 76,513 | -1.73 (0.11) | 2×10-57 | intron | NDRG4,CNOT1 | GOT2-CV, GOT2-K1 | I,S | ||
| LIG3 | rs1052536 | 17q12 | 30,355,688 | C/T | 0.53 | 75,961 | 0.98 (0.10) | 6×10-25 | 3′UTR | LIG3,CCT6B | UNC45B-K1, UNC45B-CV | I | ||
| KCNJ2 | rs1396515 | 17q24 | 65,942,588 | C/G | 0.52 | 77,058 | -0.98 (0.09) | 2×10-25 | LQT7, SQT3 | S | ||||
| KCNE1 | rs1805128 | 21q22 | 34,743,550 | T/C | 0.01 | 20,061 | 7.42 (0.85) | 2×10-18 | LQT5 | D85N | KCNQ1,KCNH2 | |||
| Novel loci | ||||||||||||||
| TCEA3 | rs2298632 | 1p36 | 23,583,062 | T/C | 0.50 | 83,031 | 0.70 (0.09) | 1×10-14 | intron | TCEA3 | ||||
| SP3 | rs938291 | 2q31.1 | 174,450,854 | G/C | 0.39 | 101,902 | 0.53 (0.09) | 6×10-10 | ||||||
| TTN-CCDC141 | rs7561149 | 2q31.2 | 179,398,101 | C/T | 0.42 | 85,299 | -0.52 (0.09) | 7×10-9 | CCDC141-CV,TTN-K2, TTN-CV | I | ||||
| SPATS2L | rs295140 | 2q33 | 200,868,944 | T/C | 0.42 | 103,331 | 0.57 (0.09) | 2×10-11 | SPATS2L | SGOL2-SCN5A | I | |||
| C3ORF75 | rs17784882 | 3p21 | 47,519,007 | A/C | 0.40 | 76,184 | -0.54 (0.10) | 3×10-8 | intron | KLHL18,PTPN23, SCAP,SETD2 | MYL3-CA | I | ||
| SLC4A4 | rs2363719 | 4q13 | 72,357,080 | A/G | 0.11 | 70,821 | 0.97 (0.16) | 8×10-10 | intron | |||||
| SMARCAD1 | rs3857067 | 4q22 | 95,245,457 | A/T | 0.46 | 101,382 | -0.51 (0.08) | 1×10-9 | ||||||
| GFRA3 | rs10040989 | 5q31 | 137,601,624 | A/G | 0.13 | 87,942 | -0.85 (0.13) | 5×10-11 | FAM13B | ETF1-RPL22 | ||||
| GMPR | rs7765828 | 6p22 | 16,402,701 | G/C | 0.40 | 93,262 | 0.55 (0.09) | 3×10-10 | intron (F256I-p) | ATXN1 | ATXN1-ACOT7,ATXN1-KCNAB2 | I | ||
| CAV1 | rs9920 | 7q31 | 115,987,328 | C/T | 0.09 | 102,060 | 0.79 (0.14) | 3×10-8 | 3′ UTR | CAV-ATP1B1, CAV2-ATP1B1 | CAV1-CA, CAV1-S1, CAV1-CV, CAV2-CV | |||
| NCOA2 | rs16936870 | 8q13 | 71,351,896 | A/T | 0.10 | 74,196 | 0.99 (0.16) | 1×10-9 | intron | I | ||||
| LAPTM4B | rs11779860 | 8q22.1 | 98,919,506 | C/T | 0.47 | 73,404 | -0.61 (0.10) | 2×10-10 | intron | I | ||||
| AZIN1 | rs1961102 | 8q22.3 | 104,002,021 | T/C | 0.33 | 82,677 | 0.57 (0.10) | 3×10-9 | ||||||
| GBF1 | rs2485376 | 10q24 | 104,039,996 | A/G | 0.39 | 70,552 | -0.56 (0.10) | 3×10-8 | intron | ACTR1A-CV | I | |||
| FEN1-FADS2 | rs174583 | 11q12 | 61,366,326 | T/C | 0.34 | 100,900 | -0.57 (0.09) | 8×10-11 | intron | FADS1, FADS2, FADS3 | I | |||
| ATP2A2 | rs3026445 | 12q24 | 109,207,586 | C/T | 0.36 | 95,768 | 0.62 (0.09) | 3×10-12 | intron | VPS29, GPN3, ARPC3, C12ORF24 | PLN | ATP2A2-CV, ATP2A2-CA | I | |
| KLF12 | rs728926 | 13q22 | 73,411,123 | T/C | 0.36 | 69,219 | 0.57 (0.10) | 2×10-8 | intron | KLF12 | I | |||
| ANKRD9 | rs2273905 | 14q32 | 102,044,752 | T/C | 0.35 | 83,532 | 0.61 (0.09) | 4×10-11 | 5′ UTR | ANKRD9 | I | |||
| USP50-TRPM7 | rs3105593 | 15q21 | 48,632,310 | T/C | 0.45 | 77,240 | 0.66 (0.10) | 3×10-12 | ||||||
| CREBBP | rs1296720 | 16p13.3 | 3,813,643 | C/A | 0.20 | 59,812 | 0.83 (0.13) | 4×10-10 | intron | CV-TRAP1 | ||||
| MKL2 | rs246185 | 16p13.12 | 14,302,933 | C/T | 0.34 | 77,411 | 0.72 (0.10) | 3×10-13 | ||||||
| PRKCA | rs9892651 | 17q24 | 61,734,255 | C/T | 0.43 | 74,683 | -0.74 (0.10) | 3×10-14 | intron | PRKCA | CACNA1C,KCNE1 | I | ||
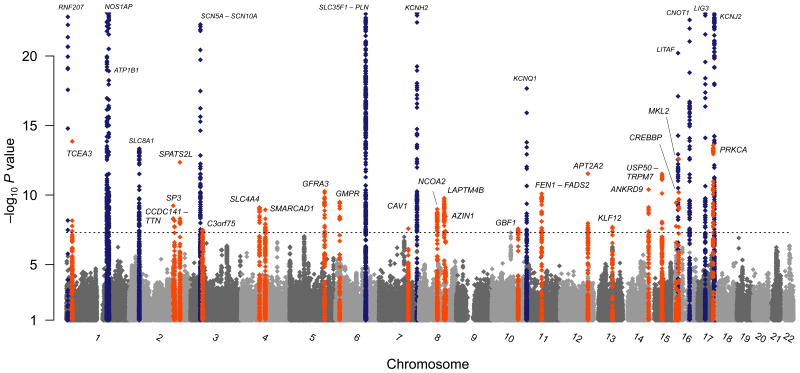
Figure 1.
Genome-wide association results for GWAS meta, annotated with gene names. Shown are association results from meta-analysis of QT interval GWAS in 76,198 individuals of European ancestry across 22 autosomes. Loci meeting P < 5×10-8 upon meta-analysis with replication data are annotated for novel (large font) and previously reported (small font) loci. Nearest genes are used for annotation but the causal gene at any given locus is unknown.
Association of QT SNPs in individuals of African Ancestry
We examined the association of 67 of these SNPs in 13,105 individuals of African ancestry in the CARe-COGENT consortium12 (one SNP was poorly imputed due to low minor allele frequency, MAF). Despite the limited power due to smaller sample size, 10 SNPs at 9 loci were significantly associated with QT interval (P < 0.0007 = 0.05/67) in the same direction as in QT-IGC (Supplementary Table 9). The SNP direction of effect was concordant between European- and African-derived samples for 51/67 SNPs (binomial P = 5×10-5) and effects were highly correlated (r = 0.60, P = 9×10-10, Supplementary Table 9). These findings are consistent with the hypothesis that a majority of common variants are associated with QT interval in both ancestral populations.
Variants with additional non-QT effects
Because heart rate (HR) is a strong determinant of unadjusted QT interval (r2 ∼0.5-0.8), we examined the 68 independent SNPs at 35 QT loci in a HR GWAS in 92,355 individuals of European or Indian Asian ancestry13. Among the 35 loci examined, we found significant association with HR of 5 SNPs at the 4 loci including PLN, FEN1—FADS2, ATP2A2, and SCN5A-SCN10A (p < 0.0007 = 0.05/68, Supplementary Table 10). Arguing against inadequate heart rate adjustment as the source of association of QT variants with HR was the modest correlation of QT effects (in models adjusting for RR interval, inverse heart rate) and HR effects for QT associated SNPs (r2 = 0.16). Only 38 of 68 of the SNP effects on HR showed the inverse relationship that has been well established between QT interval duration and HR (binomial p = 0.20).In ARIC (n = 8,524), we found no evidence that QT-SNP associations were altered with additional adjustment for RR2, RR3, RR1/2 or RR1/3. In total, these findings favor independent pleiotropic effects of the SNPs on heart rate and QT interval.
QRS prolongation due to bundle branch block can result in delayed myocardial repolarization and QT prolongation, hence our exclusion of individuals with prolonged QRS duration/bundle branch block (Supplementary Note). We examined QT interval SNPs in a published GWAS of QRS duration (N = 40,407), which reflects electrical impulse propagation in the cardiac ventricles14. Among the 68 QT-associated SNPs, 15 were significantly associated with QRS duration (p < 0.0007) at 8 loci (Supplementary Table 10)7,14.Because QRS duration is a subinterval of QT interval on the ECG, it is perhaps not surprising that some QT-prolonging variants are also positively associated with QRS duration. However, the significant genetic effects show concordant (N = 6) as well as discordant effects (N = 9).Across all SNPs there was no significant excess of concordant vs discordant effects (37 vs 31, binomial p = 0.27) or significant correlation of effect sizes (r2 = 0.03, p = 0.18, Supplementary Table 10). Collectively, these findings suggest that while the fundamental electrophysiologic mechanisms underlying the SNP-QT relationships for some SNPs may be shared, many involve cell-type-specific effects and that a consistent general relationship between SNP effects on QT interval and QRS duration does not hold.
Examination of the NHGRI GWAS database (Supplementary Note) revealed additional associations of our QT associated SNPs (or their close proxies with r2 > 0.8) at SCN5A-SCN10A with PR interval15, at MKL2 with age of menarche16 and at FEN1-FADS2 with high-density lipoprotein cholesterol, triglycerides17, n-3 fatty acids18, fasting plasma glucose and HOMA-B19 and alkaline phosphatase20 (Supplementary Table 11), which may point to novel repolarization mechanisms or simply reflect independent (pleiotropic) effects of the same genetic variation in different tissues.
Functional annotation of associated variants
Because common variants that code for changes in protein structure have an increased potential to be causal, we investigated the presence of coding variants among QT-associated loci using 1000 Genomes Project data (CEU).Among 68 total genome-wide significant SNPs at 35 loci, there were 5 loci in which the index or a highly correlated SNP was nonsynonymous.(Supplementary Table 8b).While most loci have multiple genes in associated intervals (Supplementary Figure 2), the genes that harbor genome-wide significant missense SNPs highly correlated with the top SNP are high-priority candidates to underlie the QT interval association at those loci.
Since non-coding variants may influence gene expression, we examined the index SNPs or proxies (r2 > 0.8) at the 68 SNPs at 35 loci in publicly available eQTL datasets from diverse tissues. Twelve QT interval loci are associated with variable expression of at least one gene in one or more tissues with high correlation (r2 > 0.8) between the top QT SNP and the top eQTL SNP (Supplementary Note, Table 1, Supplementary Table 12). QT-associated SNPs are associated with expression of the nearest gene at loci including ANKRD9, ATP1B1, CNOT1, FADS1, LIG3, and TCEA3. The eQTL data help point to specific genes at these loci as a potential source of the repolarization association signal, presumably through regulatory variation. However, some loci are associated with expression of multiple genes. We did not observe a significant signature of eQTLs among the QT loci that implicate a specific tissue or cell type in an atlas of human and mouse expression (P>0.01, Supplementary Note)21.
Because genetic variants that influence gene expression may do so in a cell-type specific manner, we examined the association of QT-associated SNPs with gene expression in a collection of 313 left ventricular biopsy samples in the MAGNet consortium. This collection included samples from the hearts of individuals transplanted for heart failure (n=177) or healthy hearts from potential donors (n=136) that were ultimately not used (Supplementary Note). We examined 63 of the 68 QT-associated SNPs that were well imputed from genome-wide genotyping in relation to cis-expression of all genes within 1Mb of each SNP.
After adjusting for age, sex, study site and presence of heart failure, 9 SNPs at 8 loci were significantly associated with one of 9 transcripts (one SNP was associated with 2 transcripts), correcting for multiple testing (P < 4.4 × 10-5 = 0.05/1,146 SNP-transcript associations, Supplementary Note, Table 2). After adjustment for the best eSNP, the QT SNP association became non-significant (all P ≥ 0.01) for 8 of the 9 SNP-transcript associations, consistent with a potentially causal effect for these 8 eQTLs. Inclusion of interaction terms for heart failure status did not alter the results (data not shown). In sum, these findings highlight several genes that are plausibly modulated by the QT SNP (or a correlated variant) and thus high priority targets for further experimental work.
Table 2. Association of QT SNPs with gene expression in human left ventricle.
Shown are QT SNPs associated with expression of a transcript within 1Mb at experiment-wide significance (Supplementary Note). For each transcript, the best eSNP for that transcript is shown. The QT SNP association with transcript is shown with and without adjustment for the best eSNP for that transcript. QT SNPs and transcripts are bolded if the QT SNP and best eSNP are highly correlated (r2 > 0.8), show attenuation of association in conditional models and show comparable strength of association with QT interval for both the QT SNP and best eSNP.
| QT SNP | chr | position | transcript | best eSNP for transcript | r2 between QT SNP & eSNP | direction of eSNP effect for QT increasing allele | Transcript association of QT SNP (P) | Transcript association of QT SNP with adjustment for best eSNP (P) | attenuated significance | QT association of QT SNP (P) | QT association of best eSNP (P) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17457880 | 1 | 160,434,778 | FCGR2B | rs17457880 | same | ↑ | 1×10-5 | 0.99 | YES | 3×10-10 | same |
| rs17457880 | 1 | 160,434,778 | FCGR3A | rs9727076 | 0.00 | ↑ | 1×10-7 | 9x10-9 | NO | 3×10-10 | NA |
| rs295140 | 2 | 200,868,944 | SPATS2L | rs295113 | 0.53 | ↓ | 8×10-7 | 0.74 | YES | 4×10-13 | 0.76 |
| rs174583 | 11 | 61,366,326 | FADS2 | rs174548 | 0.80 | ↓ | 6×10-8 | 0.94 | YES | 1×10-10 | 8×10-8 |
| rs3026445 | 12 | 109,207,586 | VPS29 | rs6606686 | 0.86 | ↑ | 1×10-6 | 0.84 | YES | 1×10-8 | 2×10-7 |
| rs728926 | 13 | 73,411,123 | KLF12 | rs1886512 | 0.93 | ↑ | 4×10-5 | 0.25 | YES | 2×10-8 | 4×10-8 |
| rs735951 | 16 | 11,601,037 | LITAF | rs7187498 | 0.93 | ↑ | 4×10-13 | 0.62 | YES | 2×10-28 | NA |
| rs246196 | 16 | 57,131,754 | SETD6 | rs42945 | 0.30 | ↑ | 4×10-7 | 0.20 | YES | 2×10-57 | 9×10-22 |
| rs9892651 | 17 | 61,734,255 | PRKCA | rs11658550 | 0.97 | ↑ | 2×10-41 | 0.22 | YES | 3×10-14 | NA |
Lastly, because genetic variation that influences gene expression may act through modulation of enhancers, we examined data available from the NIH Roadmap Epigenomics Program22. We specifically focused on transcriptional enhancer elements marked by combinations of histone modifications (specifically presence of H3K4me1 and absence of H3K4me3), as emerging evidence indicates that variants associated with complex traits preferentially reside in these non-coding regulatory regions and can affect gene expression23,24. We tested whether lead QT interval-associated SNPs or highly correlated variants (r2>0.8) overlapped enhancers in adult left ventricular tissue. Of 68 lead (or correlated) SNPs, 34 overlapped a left ventricular enhancer, a substantially greater proportion than randomly sampled sets of matched SNPs (z-score = 9.45, P ≪ 10-200 Supplementary Note, Supplementary Figure 3). These findings highlight specific SNPs at QT-associated loci that can be prioritized for experimental follow-up (Supplementary Table 13).
We also examined the association results for over-representation of specific pathways in QT loci compared to the genome as a whole using GO, KEGG, Panther, and Ingenuity gene set annotations (Supplementary Note). Three of the top 10 pathways in this analysis are specifically involved in calcium processes including: “regulation of the force of heart contraction” (P = 1×10-4), “cation transport” (5x10-4), “cellular calcium ion homeostasis” (1×10-3, Supplementary Note, Supplementary Table 14)25. These signals were confirmed by a secondary analysis, in which we matched the 68 QT-associated SNPs to randomly selected genome-wide SNPs to calculate statistical significance (Supplementary Note). Three GO terms involving “ion transport” were significantly enriched for QT associations (P < 0.00044) as well as a gene set based on having a cardiac phenotype in knockout mice (P < 0.00025, Supplementary Note).
Population variation in QT interval explained
The common variants at the 12 previously published common variant loci from the QTGEN and QTSCD consortia explained 3-6% of variation in QT interval, after accounting for effects of age, sex and heart rate3,4. The top SNPs at the additional novel loci increase the variance explained to 5.5-7.0%, while all 68 independent SNPs at the 35 loci explain 7.6-9.9% of variance (Supplementary Tables 15 and 16). A recent heritability analysis found that 21% of overall variance (>50% of heritable variation) in QT interval is explained by common autosomal SNPs captured on contemporary genome-wide genotyping arrays10. Because the current study was focused on identifying bona fide associations of specific loci, rather than explaining overall variance, we set a stringent p-value threshold for identifying individual SNPs. Larger studies are likely to continue to identify additional novel QT loci, as well as additional independent signals of association at the 35 loci described here.
LQTS proband mutation screening
Common variant loci found in the current and prior studies include 5 genes previously established to cause monogenic LQTS (KCNQ1, KCNH2, SCN5A, KCNE1, and KCNJ2). Given the co-existence of common QT variants at loci with established rare coding mutations in LQTS disease genes, we hypothesized that some of the novel QT loci may likewise contain previously unrecognized Mendelian LQTS genes. We selected on the basis of statistical significance, proximity to the signal of association, absence of multiple nearby genes in the associated interval and known cardiac expression or involvement in ion flux, 6 genes (ATP2A2, CAV1, CAV2, SLC8A1, SRL, and TRPM7) from 5 novel loci for coding mutation screening. We studied 298 unrelated individuals with clinically diagnosed LQTS on the basis of the Schwartz score, but genotype negative for the canonical LQTS1-3 causative genes, for rare exonic or splice site sequence variants in these 6 genes (Supplementary Note, Supplementary Table 17). We identified 13 amino acid-altering variants present in cases but not in ≥ 300 controls of the same continental ancestry (Table 3). Of these, 11 were not observed in ∼6,800 individuals whole-exome sequenced by the Exome Sequencing Project (ESP), or included on the Exome Chip array; several are predicted to be disruptive to protein function (Supplementary Note, Table 3).
Table 3. Candidate gene mutational screening.
6 genes (ATP2A2, CAV1, CAV2, SLC8A1, SRL, TRPM7) at 5 loci were screened for amino-acid altering variants in 298 LQTS cases and compared to >300 same-ancestry controls, presence on an exome chip array designed from exome sequencing of >12,000 multi-racial samples (number of alternate alleles shown) and in the Exome Sequencing Project (alternate allele counts per total number of individuals shown). Predicted function by Polyphen2 (BENign, POSSibly damaging or PROBably damaging) or SIFT (TOLerated, DAMaging) is also indicated. See Supplementary Material for details.
| gene | position hg19 | exon | nucleotide change | amino acid change | # cases | in controls (yes/no) | alt alleles in Exome Chip | in ESP | PolyPhen/SIFT |
|---|---|---|---|---|---|---|---|---|---|
| ATP2A2 | chr12:110,734,419 | 5 | c.340_A>G | p.Asn114Asp | 1 | no | no | no | BEN/TOL |
| ATP2A2 | chr12:110,765,553 - 110,765,554 | 8 | c.826_827insA | p.Ile276fsX281 | 1 | no | no | no | STOP |
| SLC8A1 | chr2:40,656,318 | 1 | c.1104_C>T | p.Ala368Val | 1 | no | no | 24/5379 | PROB/TOL |
| SLC8A1 | chr2:40,397,450 | 6 | c.2009_C>T | p.Pro670Leu | 1 | no | no | no | BEN/DAM |
| SLC8A1 | chr2:40,342,664 | 10 | c.2651_T>G | p.Val884Gly | 1 | no | no | no | PROB/DAM |
| SRL | chr16:4,256,990 | 2 | c.1177_G>T | p.Gly393Cys | 1 | no | no | no | PROB/DAM |
| SRL | chr16:4,256,754 | 2 | c.1409_G>A | p.Arg470Lys | 1 | no | no | no | BEN/TOL |
| SRL | chr16:4,256,384 | 7 | c.2566_C>T | p.Arg856Cys | 1 | no | no | 1/4915 | PROB/TOL |
| TRPM7 | chr15:50,955,189 | 2 | c.58_INS_A | p.Ile19fsX59 | 1 | no | no | no | STOP |
| TRPM7 | chr15:50,935,731 | 5 | c.341_A>T | p.Asp114Val | 1 | no | no | no | PROB/DAM |
| TRPM7 | chr15:50,884,537 | 26 | c.3895_A>C | p.Ser1299Arg | 1 | no | no | no | BEN/TOL |
| TRPM7 | chr15:50,884,406 | 26 | c.4026_A>T | p.Glu1342Asp | 2 | no | no | no | BEN/TOL |
| TRPM7 | chr15:50,884,280 | 26 | c.4152_A>T | p.Leu1384Phe | 1 | no | no | no | POSS/TOL |
Of the 13 amino-acid altering variants, two mutations in ATP2A2 (p.Ile276fsX281) and TRPM7 (p.Ile19fsX59) result in frame shifts and premature truncation of the corresponding protein product. The ATP2A2 mutation was detected in a 6-year old girl with LQTS on the basis of a QTc of 492 msec without symptoms. The proband's mother carried the mutant allele and had borderline QTc prolongation and T wave abnormalities; the proband's father lacked the mutation and had a normal QTc. The TRPM7 mutation was detected in a 14-year old girl with LQTS on the basis of a QTc of 500 msec without symptoms. The mutation was found in the proband's mother and brother, both of whom had a normal QTc, and was absent in the proband's father who had a normal QTc. Whether or not these two loss-of-function alleles contribute to LQTS pathogenesis in these individuals cannot be determined from these observations alone.
Protein-protein interaction networks
We next sought evidence that proteins encoded by genes at common variant loci interact physically with known myocardial repolarization proteins. We have constructed a protein-protein interaction network from the InWeb database (Supplementary Note)26. Using the DAPPLE algorithm27 we seeded the network with the first 12 known Mendelian LQTS genes and seven loci harboring previously-identified common QT variants (but not known Mendelian genes)3,4. Consistent with the known relationships among several of the Mendelian genes, significant interconnectivity was observed (P = 0.0006 for direct connections, P = 0.008 for indirect connections, Supplementary Note). We thus identified 606 proteins interacting directly with the seed proteins and investigated whether these protein-protein interactions could help identify candidate genes within any of the 22 novel loci identified in the current study. We found 8 interactors from 7 novel loci (ATP2A2, CAV1, CAV2, PRKCA, SLC8A1, ATXN1, ETF1, SGOL2), representing significant enrichment compared to the null expectation (hypergeometric P = 0.03, Supplementary Note, Supplementary Table 18). We hypothesized that the other proteins interacting directly with the seed network may nonetheless be enriched for association, even if not genome-wide significant. We assigned association scores to all interacting proteins (except those in the 35 loci already identified) and tested for enrichment of association in those genes compared to all genes in the genome from non-associated regions. We found that interacting proteins were more associated than chance expectation (rank-sum P = 0.00012), suggesting that they include true associations yet to be discovered (Supplementary Figure 4, Supplementary Note). The protein interaction network analysis suggests that interactors of Mendelian LQTS genes are functionally involved in QT interval duration.
This conclusion is further supported by in vivo data presented in an accompanying paper by Lundby et al. (in press, Nature Methods). We immunoprecipitated proteins encoded by the Mendelian LQTS genes, KCNQ1, KCNH2, CACNA1C, CAV3 and SNTA1 from murine cardiac tissue and identified proteins they interact with by high performance orbitrap tandem mass spectrometry. We found proteins encoded by 12 genes from 10 loci identified by our GWAS that physically interact with proteins encoded by the 5 Mendelian LQTS genes, a significant enrichment compared to random expectation (P = 1×10-6 using permutation), including ATP2A2 (SERCA2a), SRL (sarcalumenin), which regulates SERCA2a in cardiomyocytes28-30, CAV1 (caveolin 1), PLN (phospholamban), which also regulates SERCA2a, and ATP1B1 (Table 1). Molecular interactions of proteins encoded by genes at QT-associated loci with known mediators of the currents underlying myocardial repolarization strongly implicates these genes, and not others in the relevant associated intervals, as the causal genes underlying the QT interval association.
Discussion
Altogether, our integrated analysis of genomic, transcriptomic and proteomic data highlight calcium signaling as playing an important role in myocardial repolarization, the cellular process that underlies the QT interval, derangement of which is arrhythmogenic (see detailed description of genes at several loci in Supplementary Note).
Electrical activation and relaxation of the ventricular myocyte on average once per second requires the interplay of multiple coordinated ion channel fluxes. Cellular depolarization begins with Na+ influx and is sustained by Ca2+ influx, which triggers Ca2+ release from the sarcoplasmic reticulum leading to myocardial contraction (Supplementary Figure 5). Prolonged inward (depolarizing) Ca2+ current during the plateau phase of the cardiac action potential leads to delays in ventricular myocyte repolarization, a subsequent prolonged QT interval on electrocardiogram, and a highly arrhythmogenic and potentially lethal substrate. In fact, gain-of-function mutations in the L-type Ca2+ channel lead to the highly arrhythmogenic Timothy syndrome (LQT8) that is associated with extremely prolonged QT intervals31.
Normal myocyte repolarization results from efflux of potassium and less so Ca2+; Ca2+ is actively taken up by the sarcoplasmic reticulum to halt myocardial contraction. The Na+ that enters the myocyte is counterbalanced by an active Na+/K+ ATPase (a beta subunit of which is encoded by ATP1B1, at a common variant QT locus). The Ca2+ that enters the myocyte is counterbalanced by a Na+/Ca2+ exchanger (NCX1, encoded by SLC8A1) to ensure net even cation balance, at the expense of a net depolarizing effect (potentially prolonging repolarization). Disruption of this delicate cation balance, and in particular Ca2+ homeostasis, can have a profound impact on action potential duration, formation of early afterdepolarizations (EADs), and triggered activity, leading to potentially lethal arrhythmias including torsade de pointes and ventricular fibrillation. In fact, administration of an inhibitor of the Na+/Ca2+ exchanger is associated with reduced arrhythmia and shortened action potential duration in models of LQTS32 and heart failure33 and its over-expression delays myocardial repolarization and leads to ventricular arrhythmias34.
ATP2A2 encodes the SERCA2a cardiac sarcoplasmic reticulum calcium pump and by alternative splicing a ubiquitously expressed SERCA2b calcium pump (Supplementary Figure 5). The protein is negatively regulated by phospholamban (PLN), also a QT-interval associated locus3,4,6. In turn, PLN is negatively regulated by PRKCA, a gene in a newly discovered QT-interval associated locus35. SERCA2a is responsible for Ca2+ sequestration by the cardiac sarcoplasmic reticulum and its dysregulation is implicated in heart failure due to the centrality of calcium cycling to excitation-contraction coupling. Dominant SERCA2 mutations are a cause of keratosis follicularis Darier-White disease (OMIM #124200)36. No study that we are aware of has described electrocardiographic or other cardiac changes in affected humans but detailed investigation of heterozygous Serca2 (+/-) mice show a reduction in Serca2 protein by about a third with deficits in myocardial relaxation and contractility, and a reduced Ca2+ transient by haploinsufficiency37 as well as upregulation of transient receptor potential canonical 1 (TRPC1) channel38. Moreover, overexpression of SERCA2a in a rat model of heart failure demonstrated a substantial reduction in arrhythmias39.
TRPM7 encodes the widely expressed transient receptor channel melastatin 7 protein, a 6 transmembrane molecule which is Mg2+ and Ca2+ permeable and has protein kinase function40,41. The touchtone/nutria zebrafish TRPM7 mutant demonstrates defective skeletogenesis, kidney stones42 and abnormal melanophores43. Trpm7 (-/-) deletion in mice is embryonic lethal; targeted deletion disrupts normal thymogenesis40. Targeted cardiac deletion in cultured embryonic ventricular myocytes leads to down-regulation of several genes involved in calcium cycling, including SERCA2a44. In migrating human embryonic lung fibroblasts, TRPM7 mediates transduction of mechanical stretch into calcium influx underlying calcium flickers (focally high intracellular calcium microdomains), involved in steering cell migration45. In human atrial fibroblasts, atrial fibrillation is associated with increased TRPM7-mediated Ca2+ influx while TRPM7 knockdown results in loss of spontaneous Ca2+ influx46. More recently, targeted Trpm7 deletion in mice has been shown to result in lethal cardiomyopathy in early cardiogenesis; cardiomyopathy, delayed repolarization and heart block in mid cardiogenesis; and no recognizable phenotype in late cardiogenesis47. In total, this prior work raises the possibility that TRPM7 in humans leads to altered myocardial repolarization through developmental differences or through ongoing functional effects in adulthood, potentially involving calcium signaling.
Potassium flux has long been recognized through rare mutations underlying LQTS as a critical effector of myocardial repolarization. Ca2+ has been recognized as a central mediator in excitation-contraction coupling. However, our studies of common and rare genetic variation now place Ca2+ as a central modulator of repolarization given the role of the proteins encoded by the Mendelian Timothy syndrome gene (LQT8) CACNA1C, as well as the following genes at common variant QT interval loci: ATP2A2, PLN, PRKCA and SRL and SLC8A1. How the Mg2+/Ca2+ channel TRPM7 might contribute to repolarization is unclear but its involvement in Ca2+ flickers45 suggests a potential role in localized Ca2+ fluxes or indirect effects on Ca2+-sensitive potassium channels or the Na+/Ca2+ exchanger.
Much work will be needed to understand the normal physiologic contribution to repolarization of these Ca2+-regulating proteins, as well as the pathophysiologic consequences arising from their derangements. While anti-arrhythmic agents targeting the IKr (LQT2/KCNH2) channel have a relatively limited contribution to clinical management of some arrhythmias due to their propensity to cause other arrhythmias, targeting the newly identified proteins that contribute to myocardial repolarization could potentially treat some arrhythmias without pro-arrhythmia. Conversely, existing therapies that inadvertently target some of the newly discovered proteins could in fact contribute to arrhythmogenesis.
We have identified 22 novel QT interval loci, bringing to a total 35 common variant loci. We have used diverse approaches to highlight specific genes at these loci likely to mediate the repolarization effects. While we cannot say with certainty which gene underlies the QT trait at every locus, these complementary experiments represent a quantum leap in our understanding of this critical electrophysiologic process. The elucidation of fundamental mechanisms of arrhythmogenesis promises to expose new approaches to predict and prevent death from lethal ventricular arrhythmias in the general population.
Online Methods
Study cohorts
Cohorts for QT interval association analyses included individuals largely with population- or community-based ascertainment and a few with case-control sampling for traits not strongly associated with QT interval. Mandatory exclusions included presence of atrial fibrillation or a trial flutter, and presence of QRS duration > 120 msec or presence of right or left bundle branch block. Optional exclusions included use of QT-altering medications, presence of a pacemaker or implantable cardioverter defibrillator, or pregnancy. All studies were reviewed by local ethics committees and all participants provided informed consent.
Genotyping, imputation, quality control
GWAS studies used a variety of genome-wide genotyping arrays. All studies used hidden Markov model approaches to impute genotypes at unmeasured HapMap SNPs so that a common set of 2.5M SNPs were available across all discovery samples. Monomorphic SNPs and SNPs with beta estimates larger than 100,000 were removed from all results. Cohort-specific SNP filters on minimum minor allele frequency, imputation quality metric, call rate and Hardy-Weinberg equilibrium p-value were selected to minimize any test statistic distortion of the quantile-quantile plot or genomic inflation factor (λ). Replication genotyping was performed using a variety of arrays.
Association analyses, meta-analysis
Genomic control was applied to genome-wide results from each cohort prior to meta-analysis. Meta-analyses were performed in parallel at two analytic sites using MANTEL3 or METAL48 using inverse variance weighted, fixed effects meta-analysis. Genome-wide significance was set at P < 5×10-8, a threshold accounting for the effective number of independent common variant tests in the genome of European-derived populations.49
Expression in cardiac samples
Samples of cardiac tissue were acquired from patients in the Myocardial Applied Genomics Network. Left ventricular free-wall tissue was harvested at the time of cardiac surgery from subjects with heart failure undergoing transplantation or from unused donor hearts. DNA samples were genotyped using the Affymetrix 6.0 genome-wide array and RNA expression measured using the Affymetrix Genechip ST1.2 array. Imputation to SNP genotypes in 1000 Genomes was performed. Analyses were restricted to samples with genetically inferred European ancestry. SNP genotype was tested for association with log2 transformed expression level, after adjustment for age, sex, study site, disease status and batch. Association of each QT-associated SNP with all transcripts within 1Mb of the SNP was examined for 63 QT-associated SNPs (5 SNPs were not available due to poor imputation). SNP-transcript associations meeting experiment-wide significance (P < 4.4×10-5 = 0.05/1,146 tests) were examined after additional adjustment for the best cis eSNP for the transcript in question. We inferred that the SNP-transcript association could explain the SNP-QT association when the SNP-QT association was substantially attenuated after additional adjustment for the best cis eSNP.
Cardiac enhancer analyses
Enhancer annotations were generated by integrating combinations of histone modifications obtained from the Roadmap Epigenomics project using ChromHMM23,50. We identified SNPs in LD (r2> 0.8) with each of the 68 QT interval-associated loci using genotype data from the 1000 Genomes Project (CEU population) and computed overlap with ChromHMM-annotated enhancer elements in the left ventricle tissue sample (BC Left Ventricle N41) in the NIH Roadmap Epigenomics Program22 using the intersect BED command in BED Tools (v2.12.0). To assess significance of the overlap, we compared the set of SNPs at 68 QT interval-associated loci against 100,000 sets of randomly sampled control SNPs. Control SNPs were chosen from the Affymetrix 660W genotyping array and were matched for size of the LD block (+/- 5 SNPs), MAF of the lead SNP (+/- 0.1) and distance to the nearest gene (+/- 25 kb if outside a gene).
LQTS mutation analysis
A cohort of 298 unrelated, LQT1-3 mutation negative patients with LQTS [191 females (64%), average age= 27 ± 20 years, average QTc = 529 ± 58 ms], who satisfied the case inclusion criteria of QTc ≥ 480 msec (n= 261, 86%) or Schwartz score51 ≥3.0 (n=298, 100%), was derived from 7 international congenital LQTS recruitment centers [l'Institut du Thorax, Nantes, France (n=91), Mayo Clinic, Rochester, Minnesota, United States (n=72), University of Pavia, Pavia, Italy (n=38), Academic Medical Centre, Amsterdam, Netherlands (n=30), The Hospital for Sick Children, Toronto, Ontario, Canada (n=24), Munich Medical International GmbH, München, Germany (n=23), and St. George's Hospital, London, England (n=20)]. Of the 265 patients with a documented clinical history, 175 (66%) were symptomatic with ≥ 1 LQTS-related cardiac event (i.e. syncope or cardiac arrest). Six genes (ATP2A2, CAV1, CAV2, SLC8A1, SRL, TRPM7), derived from 5 genome-wide significant novel loci, were selected for comprehensive open-reading frame/splice-site mutation analysis. These 6 candidate genes were chosen based on nominal statistical significance, proximity to the signal of association, absence of multiple nearby genes in the associated interval, and known cardiac expression or involvement in ion channel macromolecular complexes. For each gene, mutational analysis was performed using either direct Sanger-based DNA sequencing of all patient samples or using an intermediate mutation detection platform (i.e. denaturing high performance liquid chromatography [DHPLC]) followed by direct DNA sequencing of only samples showing an aberrant DHPLC elution profile.
Protein-protein interaction in silico analyses
We used a public database of protein-protein interactions26. This database contains 428,430 interactions, 169,810 of which are high-confidence interactions across 12,793 proteins. All human interactions were pooled and interactions in orthologous protein pairs passing a strict threshold for orthology were included. Each interaction was assigned a probabilistic score based on the neighborhood of the interaction, the scale of the experiment in which the interaction was reported and the number of different publications in which the interaction had been sited. We used a published algorithm called DAPPLE (Disease Association Protein-Protein Link Evaluator) to build and analyze a network of seed genes27. We seeded the network with 12 known Mendelian LQTS proteins (KCNQ1, KCNH2, SCN5A, KCNE1, KCNE2, CAV3, SNTA1, KCNJ2, CACNA1C, ANK2, AKAP9, SCN4B) as well as genes from 7 previously associated common variant QT loci3,4. We considered direct connections among the seed proteins as well as indirect connections through other proteins, filtering on connections between proteins from different loci. DAPPLE evaluates the significance of the network and individual proteins within it by comparing it to 10,000 random, matched networks that are generated using a within-degree node-label permutation27. We considered the ability of protein-protein interactions to identify proteins newly associated in the QT-IGC meta-analysis. We translated the novel loci into genes, identifying 124 genes in total, 85 of which were in the In Web database.
Supplementary Material
Acknowledgments
A full listing of acknowledgments is provided in the Supplementary Note.
Author contributions
Author contributions are indicated by cohort/group. All co-authors revised and approved the manuscript.
Writing Group. C.N.-C. takes overall responsibility for the QT-IGC study. The study design was developed by M.J.A., D.E.A., A.C., L.C., P.I.W.d.B., T.T.K., P.B.M., C.N.-C., A.P., S.L.P., P.J.S., N.S. in consultation with the respective study groups. The manuscript was written by C.N.-C. The manuscript was critically revised in detail by members of the writing team before circulation to all co-authors.
GWAS cohorts. AGES: Phenotyping: V.G. Data analysis: A.V.S. Oversight: T.B.H., L.J.L., V.G. Amish studies: Clinical data collection, genotyping and oversight: B.D.M., A.R.S. EKG data collection: W.P. Analysis: A.Parsa, J.R.O. Interpretation: A.Parsa, W.S.P. ARIC: Study design: A.A., D.E.A, A.C., W.H.L.K. Analyses: D.E.A., J.S.B., A.C., G.E., H.H. Steering: D.E.A, A.C. Writing: D.E.A., A.C. BLSA: Analysis: T.T. Phenotype collection: J.B.S. Overall project supervision: L.F. BRIGHT: Phenotyping: M.B., M.J.C., P.W.M., P.B.M., N.J.S. Genotyping: P.B.M., S.J.N. Analysis: S.J.N. Overall study supervision: M.B., M.J.C., P.B.M., N.J.S. Carlantino: Sample/data collection: M.C., L.Z. Overall study supervision: P.G.. Data collection/statistical analysis: S.U. CHS: Study design: J.B., S.R.H., B.P., N.S. Data collection: S.R.H., B.P., D.S.S. Genotyping: J.I.R. Analysis, interpretation: J.B., N.S. Supervision of analyses: B.P. Funding for GWAS: B.P. Croatia-Korcula& Croatia-Split: GWAS analysis: C.H. Data collection, phenotype measurement, data entry, and field work supervision: I.K., O.P. Study design, funding: I.R., A.F.W. DCCT/EDIC: Analyses: D.W. Supervision of analyses: A.D.P. deCODE: Data collection: D.O.A., H.H. Study design: K.S., U.T., H.H., D.F.G., D.O.A. Data alignment, imputation and statistical analysis: D.F.G. Additional analysis, interpretation of results: K.S., U.T., H.H., D.F.G., D.O.A. eMERGE: Data curation, GWAS analysis: R.L.Z., Y.B. Supervision of QC/analysis of dataset: M.D.R. Study conception, analysis framework: D.R. Algorithm for case ascertainment: J.C.D. ERF: Analysis: A.I. Data acquisition: C.v.D., A.I., B.A.O., J.A.K., A.G.U. Overall study PIs: C.v.D., B.A.O. FHS: Analysis plan development: C.N.-C., C.O.D., P.A.N., M.G.L. GWAS analysis: X.Y.Y., M.G.L. Wrote manuscript: C.N.-C. Secured funding: C.N.-C., C.J.O. FVG: Data collection: M.B. Primary analysis: A.I. Statistical analysis: A.P.d'A. Overall study supervision: G.S. Health2000: Data analysis, replication genotyping, QC: A.M.L. Primary data analysis: A.Marjamaa. Phenotyping, including ECGs: A.J. Electrocardiographic measurements: K.P. GWAS and replication genotyping: M.P. Design of ECG study, analysis, interpretation: L.O. Genetic data collection, analysis: K.K.K. PI, supervision: V.S. HealthABC: Data collection, supervision: S.R.C., Y.L. Data analysis: D.S.E., M.A.N. HNR: Data collection: H.K. Data generation: H.K., T.W.M. Genetic data generation: M.N.N., P.H., T.W.M. Data analysis: L.E., P.H., T.W.M., M.N.N. Overall study design, PIs: R.E., K.-H.J. KORA-F3/S4: Overall QT project supervision: A.Pfeufer. Genotyping oversight: T.M. ECG collection, measurement and interpretation: M.F.S., S.P., B-M.B, E.M. Primary genetic analysis: M.M.-N. Interpretation of results: A.Pfeufer, S.Kaab, T.M., M.W. Overall study PI: A.Peters. LifeLines: Phenotyping: R.A.d.B., P.A.v.d.V. Genotyping: L.F. Analyses: I.M.N. and L.F. MICROS: Sample recruitment, overall study PI: P.P. Study supervision, genotyping, data coordination: A.A.H. Data analysis: F.D.G., C.F. ORCADES: Phenotype collection: S.H.W. Genotype generation: H.C., J.F.W. GWAS analysis: P.N. Raised funding: J.F.W. Overall study supervision: J.F.W. PopGen: Recruitment, phenotyping: N.E.E.M., N.F. Genotyping, data preparation: A.F. Data preparation, analysis: D.E. PREVEND: Phenotyping: M.P.v.d.B., D.J.v.V, G.N. Genotyping, data-analysis: F.W.A., I.M.L., P.v.d.H. Obtained funding: G.N., D.J.v.V., F.W.A., P.v.d.H. Rotterdam Study-I and II: Study concept, design: M.E., B.H.C.S. Data acquisition: M.E., B.P.K., J.A.K., A.H., J.C.M.W., B.H.C.S., A.G.U. Statistical analysis: M.E. Interpretation: M.E., B.H.C.S. Obtained funding: A.H., J.C.M.W., A.G.U., B.H.C.S. Study supervision: B.H.C.S. SardiNIA: Phenotyping: M.O. Genotyping, data analysis: G.A., E.G.L., A.Mulas, M.O., S.S., D.S., K.V.T., and M.U. Overall study supervision, PIs: D.S., M.U. SHIP: Data acquisition: M.D., M.R.P.M., U.V., S.B.F. Statistical analysis: U.V., M.D., M.R.P.M. Interpretation: U.V., M.D., S.B.F. Obtained funding: S.B.F., U.V. TwinsUK: Study concept, design: H.S., Y.J. Data acquisition: T.D.S. Statistical analysis, interpretation: I.M.N., H.S., Y.J. Obtained funding: Y.J., T.D.S. Young Finns Study: Data collection: T.L., O.R., M.Kähönen, J.V. Genotyping: T.L., N.M. Genotyping: T.L. Phenotype preparation: O.R., M.Kähönen, J.V. Analysis: T.L., O.R., M.Kähönen, J.V., L.-P.-L. Obtained funding: T.L., O.R., M.Kähönen, J.V.
Directly genotyped SNP replication cohorts. (Author contributions for cohorts that contributed to both GWAS and replication genotyping are shown under the GWAS entry above) BRHS: Analysis: R.W.M. Custodian of genetic resource: R.W.M., P.H.W. Data collection for genetic resource: P.H.W. Development of genetic resource: A.D.H. Overall study supervision, PIs: P.H.W., R.W.M. ECG analyses: P.W.M. Bruneck: Data analysis, interpretation, writing: S.K. DNA preparation: F.K., C.L. ECG measurement, database: M.Knoflauch. Supervision, funding, administration, PI: J.W. Carla: Study concept, design: K.H.G., K.W. Genotyping: H.M.z.S. Supervision: K.W. Study design, analysis: A.K. Study concept, supervision, PI: J.H. Cyprus: Study concept, funding, supervision, analysis: A.N. Data acquisition, analysis, interpretation: M.G. Genetic, biochemical data acquisition, statistical analysis: A.G.P. Czech Post-MONICA: Data collection, submission: J.A.H., V.A. Galicia: Cohort collection: M.B. Study design: M.B. Genotyping platform management: A.C. Genotyping: M.T. Analysis: M.T. Interpretation: M.B., A.C. Financial support: A.C. Intergene: Genotyping, data analysis, epidemiology expertise: F.N. Genotyping, genetic expertise: A.T.N. Study design, data collection, disease area knowledge: D.S.T. MIDSPAN Family Study: Data acquisition, statistical analysis, interpretation: S.P. Genotyping: W.K.L. Overall study supervision, data collection, funding: A.F.D., G.C.M.W. PIVUS: Genotyping: A.-C.S. Phenotyping: L.L., J.A., J.S. Data analysis: S.G. Overall supervision, PI: E.I. SAPHIR: DNA preparation: F.K., C.L. Data collection: L.K., B.P., B.S. Data analysis: L.K., F.K., C.L., B.P., B.S. Study design, PI: B.P. ULSAM: Genotyping: A.-C.S. Phenotyping: L.L., J.A., J.S. Data analysis: S.G. Overall supervision, PI: E.I. Whitehall II: Data collection, submission: M.Kumari. Overall supervision: M.Kivimaki. Funding: A.H.
Meta-analysis of GWAS + replication. D.E.A. and S.L.P. independently performed quality control and meta-analysis of GWAS and replication association results. P.I.W.d.B., A.P. and C.N.-C. supervised the analyses.
Non-QT trait lookups. CARe-COGENT: Meta-analysis, lookup: J.G.S. HRGEN: Meta-analysis, lookup: M.d.H. Overall study supervision: R.J.F.L. QRS GWAS: Study supervision: N.S. Meta-analysis: D.E.A., P.I.W.d.B. Results lookup: S.L.P.
eQTL Analyses. Dataset acquisition: A.S.P., V.E. Analysis, interpretation: A.D.J. Cell-type enrichment tests: S.R.
Left ventricle eSNP analyses. Overall supervision: T.P.C. Recruitment, sample collection: K.M., C.M. Sample processing, expression analysis: J.B. Statistical analysis: M.M.
Left ventricle enhancer analyses. Analysis: X.W. Overall supervision: L.A.B., M.Kellis
Mouse knockout. Enrichment tests: K.S.
DAPPLE analysis. Concept, design, analysis: E.J.R. conceived, designed and performed the DAPPLE analyses. Supervision: K.L., M.J.D.
LQTS mutation screening. Amsterdam: SLC8A1 Sequencing: T.K.T. Clinical data collection: A.B., N.H., A.A.M.W. Study supervision: C.R.B., A.A.M.W. London: Recruitment, phenotyping, strategy: E.R.B. Screening for mutations in LQT1,2,3 and sample management: C.D. Mayo: LQTS cohort characteristic organization: D.J.T. TRPM7 mutation analysis, interpretation: D.J.T., A.M.D., J.R.G. Patient collection, study design, data review, overall supervision: M.J.A. Munich: Study oversight: S.Kääb, A.Pfeufer. Patient collection: B.M.B., E.M. Nantes: Scientific management: J-J.S Clinical, genetic information collection: S.C. ATP2A2 sequencing: S.C. Screening for mutations in LQT1,2,3, and clinical data collection: J.B. LQTS gene diagnosis management: F.K. Patient enrollment: V.P. Pavia: Patient collection, patient selection, molecular screening supervision: L.C., P.J.S. SRL mutation screening: A.G., R.I. Toronto: Identification of LQTS patients free of LQT1,2,3 mutations: R.M.H. Program co-development: S.W.S.
Immunoprecipitation experiments. Proteomic experiments, analysis:A.L. Overall study supervision: J.V.O.
Footnotes
Competing Financial Interests: The authors declare competing financial interests: details are available in the online version of the paper.
H.H. is a former and D.O.A., D.F.G., K.S., and U.T. are current full or part-time employees of deCODE Genetics/Amgen, Inc. A.S.P. was previously an employee of Merck Research Laboratory and is a current employee of Sanofi. F.N. is an employee of AstraZeneca. B.P. serves on the DSMB for a clinical trial funded by the manufacturer (Zoll Life Cor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. J.S. serves on the advisory board of Itrim.
References
- 1.Schwartz PJ, Crotti L, Insolia R. Long-QT syndrome: from genetics to management. Circ Arrhythm Electrophysiol. 2012;5:868–77. doi: 10.1161/CIRCEP.111.962019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newton-Cheh C, et al. QT interval is a heritable quantitative trait with evidence of linkage to chromosome 3 in a genome-wide linkage analysis: The Framingham Heart Study. Heart rhythm : the official journal of the Heart Rhythm Society. 2005;2:277–84. doi: 10.1016/j.hrthm.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Newton-Cheh C, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nature genetics. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeufer A, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–14. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arking DE, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nature genetics. 2006;38:644–51. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 6.Nolte IM, et al. Common genetic variation near the phospholamban gene is associated with cardiac repolarisation: meta-analysis of three genome-wide association studies. PloS one. 2009;4:e6138. doi: 10.1371/journal.pone.0006138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–22. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 8.Noseworthy PA, et al. Common genetic variants, QT interval, and sudden cardiac death in a Finnish population-based study. Circulation. Cardiovascular genetics. 2011;4:305–11. doi: 10.1161/CIRCGENETICS.110.959049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JW, et al. A common variant in SLC8A1 is associated with the duration of the electrocardiographic QT interval. Am J Hum Genet. 2012;91:180–4. doi: 10.1016/j.ajhg.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–25. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voight BF, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JG, et al. Impact of Ancestry and Common Genetic Variants on QT Interval in African Americans. Circ Cardiovasc Genet. 2012;5:647–655. doi: 10.1161/CIRCGENETICS.112.962787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.den Hoed M, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013 doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sotoodehnia N, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nature genetics. 2010;42:1068–76. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeufer A, et al. Genome-wide association study of PR interval. Nature genetics. 2010;42:153–9. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elks CE, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nature genetics. 2010;42:1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zabaneh D, Balding DJ. A genome-wide association study of the metabolic syndrome in Indian Asian men. PloS one. 2010;5:e11961. doi: 10.1371/journal.pone.0011961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaitre RN, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS genetics. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambers JC, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nature genetics. 2011;43:1131–8. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am J Hum Genet. 2011;89:496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segre AV, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 27.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS genetics. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida M, et al. Impaired Ca2+ store functions in skeletal and cardiac muscle cells from sarcalumenin-deficient mice. J Biol Chem. 2005;280:3500–6. doi: 10.1074/jbc.M406618200. [DOI] [PubMed] [Google Scholar]
- 29.Shimura M, et al. Sarcalumenin alleviates stress-induced cardiac dysfunction by improving Ca2+ handling of the sarcoplasmic reticulum. Cardiovasc Res. 2008;77:362–70. doi: 10.1093/cvr/cvm019. [DOI] [PubMed] [Google Scholar]
- 30.Jiao Q, et al. Sarcalumenin is essential for maintaining cardiac function during endurance exercise training. Am J Physiol Heart Circ Physiol. 2009;297:H576–82. doi: 10.1152/ajpheart.00946.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Splawski I, et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8089–96. doi: 10.1073/pnas.0502506102. discussion 8086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milberg P, et al. Inhibition of the Na+/Ca2+ exchanger suppresses torsades de pointes in an intact heart model of long QT syndrome-2 and long QT syndrome-3. Heart rhythm : the official journal of the Heart Rhythm Society. 2008;5:1444–52. doi: 10.1016/j.hrthm.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Milberg P, et al. Acute inhibition of the Na(+)/Ca(2+) exchanger reduces proarrhythmia in an experimental model of chronic heart failure. Heart rhythm : the official journal of the Heart Rhythm Society. 2012;9:570–8. doi: 10.1016/j.hrthm.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Pott C, et al. Proarrhythmia in a non-failing murine model of cardiac-specific Na(+)/Ca (2+) exchanger overexpression: whole heart and cellular mechanisms. Basic research in cardiology. 2012;107:1–13. doi: 10.1007/s00395-012-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braz JC, et al. PKC-alpha regulates cardiac contractility and propensity toward heart failure. Nat Med. 2004;10:248–54. doi: 10.1038/nm1000. [DOI] [PubMed] [Google Scholar]
- 36.Sakuntabhai A, et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nature genetics. 1999;21:271–7. doi: 10.1038/6784. [DOI] [PubMed] [Google Scholar]
- 37.Ji Y, et al. Disruption of a single copy of the SERCA2 gene results in altered Ca2+ homeostasis and cardiomyocyte function. J Biol Chem. 2000;275:38073–80. doi: 10.1074/jbc.M004804200. [DOI] [PubMed] [Google Scholar]
- 38.Pani B, et al. Up-regulation of transient receptor potential canonical 1 (TRPC1) following sarco(endo) plasmic reticulum Ca2+ ATPase 2 gene silencing promotes cell survival: a potential role for TRPC1 in Darier's disease. Mol Biol Cell. 2006;17:4446–58. doi: 10.1091/mbc.E06-03-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyon AR, et al. SERCA2a gene transfer decreases sarcoplasmic reticulum calcium leak and reduces ventricular arrhythmias in a model of chronic heart failure. Circulation. Arrhythmia and electrophysiology. 2011;4:362–72. doi: 10.1161/CIRCEP.110.961615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin J, et al. Deletion of Trpm7 disrupts embryonic development and thymopoiesis without altering Mg2+ homeostasis. Science. 2008;322:756–60. doi: 10.1126/science.1163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Runnels LW, Yue L, Clapham DE. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science. 2001;291:1043–7. doi: 10.1126/science.1058519. [DOI] [PubMed] [Google Scholar]
- 42.Elizondo MR, et al. Defective skeletogenesis with kidney stone formation in dwarf zebrafish mutant for trpm7. Curr Biol. 2005;15:667–71. doi: 10.1016/j.cub.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 43.Arduini BL, Henion PD. Melanophore sublineage-specific requirement for zebrafish touchtone during neural crest development. Mech Dev. 2004;121:1353–64. doi: 10.1016/j.mod.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Sah R, et al. Ion channel-kinase TRPM7 is required for maintaining cardiac automaticity. Proc Natl Acad Sci U S A. 2013;110:E3037–46. doi: 10.1073/pnas.1311865110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei C, et al. Calcium flickers steer cell migration. Nature. 2009;457:901–5. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du J, et al. TRPM7-mediated Ca2+ signals confer fibrogenesis in human atrial fibrillation. Circ Res. 2010;106:992–1003. doi: 10.1161/CIRCRESAHA.109.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sah R, et al. Timing of myocardial trpm7 deletion during cardiogenesis variably disrupts adult ventricular function, conduction, and repolarization. Circulation. 2013;128:101–14. doi: 10.1161/CIRCULATIONAHA.112.000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 50.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome. An update. Circulation. 1993;88:782–4. doi: 10.1161/01.cir.88.2.782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.