Abstract
Background
Lung cancer, including lung adenocarcinoma (adenoCa), is a heterogeneous disease, which evolves from molecular alterations in the airway epithelium. The study explores whether a subtype of lung adenoCa expresses the unique molecular features of human airway basal cell (BC), and how expression of the airway BC features correlates with the molecular, pathologic and clinical phenotype of lung adenoCa.
Methods
Three independent lung adenoCa data sets were analyzed for expression of genes that constitute the airway BC signature. Expression of the BC signature in lung adenoCa was then correlated to clinical and biologic parameters.
Results
Remarkable enrichment of airway BC signature genes was found in lung adenoCa. A subset of lung adenoCa (“BC-high adenoCa”) exhibited high expression of BC signature genes in association with poorer tumor grade, higher frequency of vascular invasion, and shorter survival than adenoCa with lower expression of these genes. At the molecular level, “BC-high adenoCa” displayed higher frequency of KRAS mutations, activation of transcriptional networks and pathways related to cell cycle, extracellular matrix organization, and a distinct differentiation pattern with suppression of ciliated-and Clara cell-related genes.
Conclusions
Activation of the airway BC program is a molecular feature of a distinct, aggressive subtype of lung adenoCa.
Keywords: Airway basal cell, lung adenocarcinoma, gene expression, cell-of-origin
Introduction
Lung cancer, the leading cause of cancer mortality worldwide, is a heterogeneous disease that evolves from molecular alterations in the airway epithelium mediated by environmental oncogenic stress, primarily cigarette smoking [1–3]. The human airway epithelium is a pseudostratified layer dominated by ciliated cells together with secretory, intermediate, basal cells (BC), and rare neuroendocrine cells [4].
The specific contribution of these individual cell types of the airway epithelium to lung cancer heterogeneity is not well understood. Airway BC, the stem/progenitor cell population of the airway epithelium [5], are considered the candidate cell-of-origin of lung squamous cell carcinoma (SqCa), in part, because airway BC are likely the source of squamous cell metaplasia, the SqCa-related potential preneoplastic lesion [1]. In contrast, the cellular origin of lung adenocarcinoma (adenoCa)is not clear [1]. Centrally located adenoCa are thought to arise from the surface or glandular epithelium of bronchi [6]. By contrast, a Clara cell-and type II pneumocyte-associated differentiation pattern, also known as “terminal respiratory unit”, has been observed in peripheral adenoCa [7, 8]. Subpopulations of Clara cells, a secretory cell type present throughout the airway epithelium in mice [9], but limited to small airways in humans [10], have been related to lung adenoCa development in murine models [11]. However, the contribution of other cell types, such as BC-like progenitors, to human lung adenoCa has also been proposed [1, 12].
In the present study, using our recent description of the human airway BC transcriptome [13], we analyzed the contribution of the unique molecular features of airway BC to the molecular and clinical phenotype of lung adenoCa. The data provides evidence for a subtype of lung adenoCa that expresses high levels of airway BC genes in association with aggressive clinical phenotype.
Methods
Additional details of methods are in the online supplement.
Lung AdenoCa Data Sets
Three independent lung adenoCa cohorts were analyzed: primary cohort (n=182 of 199 originally described by Chitale et al [14], which was re-evaluated histologically and updated with recent clinical information)and two validation cohorts, one described by Bild et al. (n=58 [15]) and one by Shedden et al. (n=327 of 442, i.e. excluding 104 subjects analyzed in Memorial Sloan Kettering Cancer Center, the majority of which are present in the primary cohort [16], and 11 large cell neuroendocrine carcinoma samples identified based on pathologic re-evaluation [17]). Patient characteristics are summarized in Supplemental Table I.
Analysis of Airway BC Signature Expression in Human Lung AdenoCa
The airway BC signature (862 genes) was previously characterized in our laboratory [13]. To analyze gene enrichment, microarray data was normalized by chip and then median expression levels for all genes across all samples was determined. Median levels for each gene were compared to the median level for all 862 airway BC signature genes, the non-BC signature which had significantly higher expression in the complete large airway epithelium vs purified BC based on the genome-wide microarray comparison (criteria for high expression: fold-change ≥5, p<0.01 with Benjamini-Hochberg correction), and 50 random 862-gene sets (selected from the Affymetrix HG-U133A genome using Excel “RAND” function).
To compare the expression of the airway BC signature among various carcinoma subtypes [18–24] with airway BC samples, the data sets were analyzed by principal component analysis (PCA) using GeneSpring version 7.3.1 (Agilent Technologies, Santa Clara, CA).
ABC index (IBC) was calculated for each individual subject as a cumulative measure of the airway BC signature expression as previously described for the complete airway epithelium [25]. Categorization of subjects was performed based on the IBC using quartile method: individuals within the bottom quartile were categorized as “BC-low” and individuals within the top quartile were categorized as “BC-high”.
To determine transcriptome differences between BC-high vs BC-low adenoCa, we performed genome-wide comparison (criteria for differentially expressed genes: fold-change >2, p<0.01 with Benjamini-Hochberg correction). Enrichment of pathways within differentially expressed genes was analyzed using the DAVID Bioinformatics Resources 6.7 analytic tool (http://david.abcc.ncifcrf.gov/). To analyze networks for the BC-high adenoCa up-regulated genes, co-expressed genes were identified in the up-regulated genes using Weighted Correlation Network Analysis (WGCNA) and identified network genes (criteria – Spearman correlation Rho>0.6, p<0.05) were then linked to the airway BC signature genes up-regulated in BC-high lung adenoCa based on the known physical protein-protein interactions and transcriptional regulation using GNC Pro analytic tool (http://gncpro.sabiosciences.com/gncpro/gncpro.php).
Expression of genes associated with the major cell types of the human airway epithelium (ciliated, mucus-secreting, Clara, and neuroendocrine cells) and epithelial-mesenchymal transition (EMT) were compared in the lung adenoCa subtypes of the primary cohort. To compare the expression of the airway BC signature in lung adenoCa to SqCa, the dataset containing 58 adenoCa and 53 SqCa described by Bild et al [15] was analyzed.
Survival Analysis
To assess the relationship of expression of the airway BC signature on survival of patients with lung adenoCa, we first identified poor survival -associated genes by genome-wide comparison between adenoCa patients with less than 2-year overall survival (“poor survivors”) vs those with more than 5 -year overall survival in primary cohort (criteria for differentially expressed genes: p<0.05 with Benjamini-Hochberg correction). All survival analyses were performed using the Kaplan-Meier method. Survival between the adenoCa subtypes was compared using log-rank test. Multivariate analysis was performed using Cox proportional hazard model.
Immunohistochemical Analysis
Biopsy samples were independently collected from adenoCa patients undergoing lung resection according to the protocol and informed consent approved by the MSKCC Institutional Review Board. Categorization of the lung adenoCa samples used in immunohistochemistry into “BC-high” and “BC-low” was made using the index method, as described above, based on the TaqMan PCR analysis (Applied Biosystems, Foster City, CA) of the expression of top 10 genes with >85% sensitivity for BC-high adenoCa (in all 3 independent lung adenoCa data sets; gene list is presented in Supplemental Table II). Immunohistochemical analysis was performed to validate differential expression of selected proteins between BC-low adenoCa and BC-high adenoCa [13]. The only modification was that the samples were incubated with primary antibodies against tumor protein 63 (TP63, 2 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA), and antithyroid transcription factor-1 (TTF-1, 3 μg/ml; DAKO, Carpinteria, CA) for 2 hr, 37°C. Commercially available normal lung and lung SqCa tissue samples (US Biomax Inc., MD) were used for comparative analysis.
Statistical Analysis
All analyses, except for the microarray data, were performed using the SPSS statistical package (SPSS Inc, Chicago, IL). Relationship between the IBC and the NKX2-1 gene expression was analyzed in the primary adenoCa cohort using Pearson correlation analysis. The relationship between the groups was assessed using Chi-square test or Mann-Whitney test. Analysis of the microarray data was performed as specified above using GeneSpring version 7.3.1 (Agilent Technologies).
Results
Airway BC Signature is Enriched in Lung AdenoCa
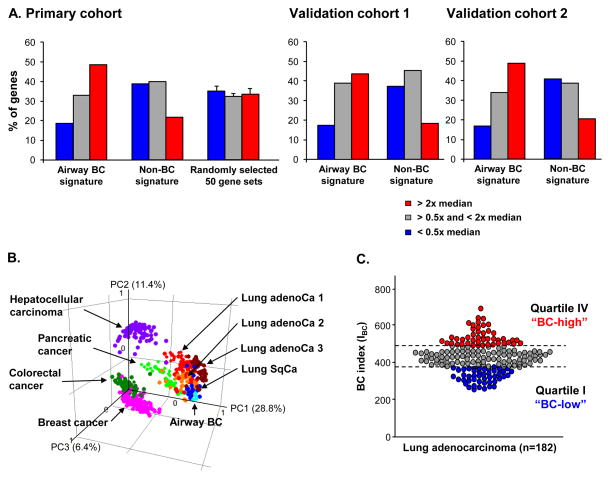
To provide comprehensive view on the expression of airway BC molecular features in lung adenoCa, expression of the 862-gene airway BC signature (Supplemental Gene List I) was analyzed. Of the 862 airway BC signature genes, 420 (48.7%) were among the highly expressed lung adenoCa genes (expression level > two-fold median for all genes), whereas only 118 of 544 (22%) non-BC signature genes and <35% of genes in randomly selected gene sets were among highly expressed lung adenoCa genes (Figure 1A, left panel). By contrast, <20% of the BC-signature genes contributed to the genes with low expression in lung adenoCa (expression level <0.5x median for all genes) as compared to >35% non-BC genes and the genes from the randomly selected data sets. The enrichment of the BC signature genes in lung adenoCa was validated using 2 independent cohorts (Figure 1A, middle and right panels). Combined analysis of all 3 cohorts revealed statistically significant enrichment of the airway BC signature genes among the highly expressed lung adenoCa genes vs non-BC genes (p<0.0006) and vs randomly selected gene sets (p<0.02) (Supplemental Table III).
Figure 1.
Expression of the airway basal cell (BC) signature genes in human lung adenocarcinoma (adenoCa). A. Frequency of the airway BC signature genes, non-BC signature genes and the genes of the 50 random 862-gene sets contributing to the genes with high expression in lung adenoCa (expression level > 2x median for all expressed genes; red bars); genes with low expression in lung adenoCa (expression level < 0.5x median for all expressed genes; blue bars) and genes with intermediate expression in lung adenoCa (remaining lung adenoCa-expressed genes; grey bars) in the primary cohort [14], validation cohorts 1 [15]and 2 [16]; see Supplemental Table III for details. B. Principal component analysis (PCA) comparing various types of human carcinomas and airway BC based on expression of the airway BC signature. Analyzed data sets include lung adenoCa1 from Ding et al. [22] (red circles; n=68); lung adenoCa2 from Hou et al. [21] (dark red circles; n=40); adenoCa 3 from Kuner et al. [20] (orange circles; n=40); lung squamous cell carcinoma (SqCa) from Kuner et al. [20] (dark blue circles; n=18); colorectal cancer from Smith et al. [18] (dark green circles; n=55); breast cancer from Lu et al. [19] (pink circles; n=129); hepatocellular carcinoma from Chiang et al. [23] (purple circles; n=91); pancreatic cancer from Badea et al. [24] (light green circles; n=39); and airway BC samples from healthy nonsmokers (light blue circles; n=4). Each circle represents an individual sample. The % contributions of the first 3 principal components (PC) to the observed variability are indicated. C. Categorization of lung adenoCa into high and low airway BC gene expressors. Y-axis – BC index (IBC) based on a number of airway BC signature genes expressed above the median level in lung adenoCa subjects(n=182) [14]. AdenoCa subjects were divided into BC-high (quartile IV; red circles), BC-intermediate (quartile II–III; grey circles)and BC -low (quartile I; blue circles) subtypes.
Airway BC Signature is Up-regulated in a Subset of Lung AdenoCa
Next, we asked whether the pattern of airway BC signature expression in lung adenoCa is shared by other carcinomas or relatively unique to this type of lung cancer. The PCA revealed that lung SqCa [20] and all 3 lung adenoCa data sets [20–22] exhibited similar patterns, with clustering closer to airway BC samples than colorectal [18], breast [19], hepatocellular [23] and pancreas [24] cancers (Figure 1B). The majority of the lung SqCa samples displayed similarity to the airway BC gene expression pattern, whereas the lung adenoCa was more heterogeneous.
To further explore the heterogeneity of lung adenoCa based on the airway BC signature expression, a BC index (IBC) was developed as a cumulative gene expression parameter. Consistent with the PCA data above, the analysis revealed remarkable heterogeneity of lung adenoCa patients based on the airway BC signature expression (Figure 1C). Based on the IBC, “BC-high” (top quartile) and “BC-low” (bottom quartile) adenoCa subtypes were identified (Figure 1C).
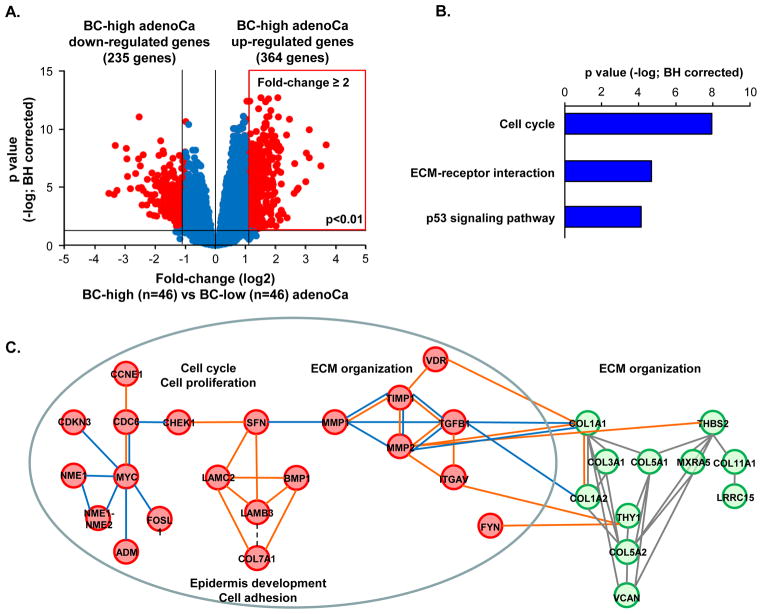
BC-high Lung AdenoCa Exhibits Distinct Biologic Phenotype
To determine biological pathways and patterns enriched BC-high adenoCa, we first performed genome-wide comparison of the BC-high vs. BC-low adenoCa (Figure 2A, Supplemental Gene List II). Among the 364 genes up-regulated in BC-high adenoCa, there was significant enrichment of the biologic pathways related to cell cycle, extracellular matrix (ECM)-receptor interaction and p53 signaling pathway (Figure 2B). Consistent with the pathway analysis, the network analysis of the BC-high adenoCa up-regulated genes revealed enrichment of the transcriptional network elements related to the ECM organization (Figure 2C). The BC-high adenoCa-enriched co-expressed ECM network components were interaction partners of the BC signature genes regulating epithelial-mesenchymal interactions and lung tissue homeostasis, including transforming growth factor beta (TGFB1), metalloproteases (MMP) 1 and -2, tissue inhibitor of metalloproteases (TIMP) 1, integrin alpha V (ITGAV), vitamin D receptor (VDR) (Figure 2C).
Figure 2.
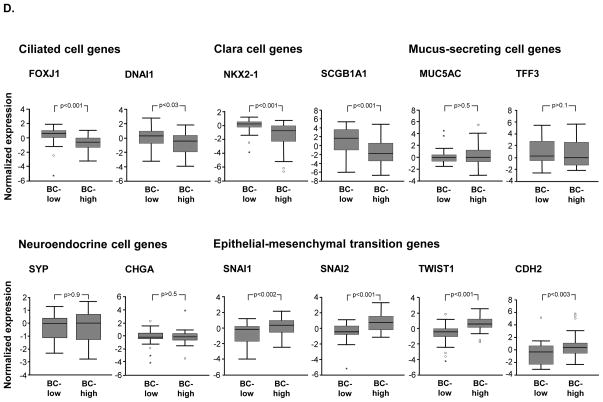
Differentially expressed genes between BC-high lung adenocarcinoma (adenoCa) and BC-low adenoCa. A. Volcano plot of genome-wide comparison between BC-high adenoCa (n=46) vs BC-low adenoCa (n=46) in primary lung adenoCa cohort (n=182). Y-axis -negative log of p value; x-axis -log2 -transformed fold-change. Red dots -significant genes (fold change ≥2; p<0.01 with Benjamini-Hochberg (BH) correction); blue dots -genes with no significant difference between the groups. B. KEGG pathway analysis of significantly (p<0.05 with Benjamini-Hochberg (BH) correction)enriched in BC -high adenoCa up-regulated genes. C. Molecular networks enriched within BC -high adenoCa up-regulated genes identified using the Weighted Correlation Network Analysis (WGCNA) (green circles represent co-expressed BC-high lung adenoCa genes within the primary data set) and connected to the BC signature genes up-regulated in BC-high adenoCa (red circles) using GCNPro analytic tool (orange lines – transcriptional regulation; blue lines – physical protein-protein interactions; see Methods for details). D. Examples of expression of differentiation-associated molecular patterns in BC-high adenoCa compared to BC-low adenoCa. Ciliated cell genes: forkhead box J1 (FOXJ1) and dynein axonemal intermediate chain 1 (DNAI1); Clara cell genes: NK2 homeobox 1 (NKX2-1) and secretoglobin 1A1 (SCGB1A); Mucin producing secretory cell-related genes: mucin 5AC (MUC5AC) and trefoil factor 3 (TFF3); Neuroendocrine cell genes: synaptophysin (SYP) and chromogranin A (CHGA); Genes associated with epithelial -mesenchymal transition: snail homolog 1(SNAI1), snail homolog 2 (SNAI2), twist homolog 1 (TWIST1), and N-cadherin (CDH2). In all panels, log2-transformed normalized gene expression levels based on the microarray analysis are shown; n=46 in each group. Outliers are indicated on the basis of interquartile range (IQ R); ° −1.5 x IQR to 3 x IQR, * -more or less than 3 x IQR.
BC-high adenoCa displayed significant down-regulation of genes associated with differentiation of the major cell types of the small airway epithelium, including ciliated cells (forkhead box J1 [FOXJ1] and dynein axonemal intermediate chain 1 [DNAI1]) and Clara cells (NK2 homeobox 1 [NKX2-1] and secretoglobin 1A1 [SCGB1A]). Expression of genes typical for mucus-secreting cells and neuroendocrine cells was not different between these 2 subtypes. There was a negative correlation between the IBC and the NKX2-1 gene expression (Supplemental Figure 2). Consistent with this observation, there was a trend of a lower expression of the NKX2-1-encoded thyroid transcription factor-1 (TTF-1)in BC -high adenoCa, although it was detectable in both adenoCa subtypes, by contrast to the TTF-1-negative SqCa (Supplemental Figure 3A). By contrast to differentiation genes, the expression of genes related to EMT, such as SNAI1, SNAI2, TWIST1 and CDH2 [26], was up-regulated in BC-high adenoCa compared to BC-low adenoCa (Figure 2D). Genes related to other lung cancer subtypes, including small cell lung carcinoma, such as those encoding tumor protein TP53, retinoblastoma (RB) 1 and L-MYC, were not differentially expressed between the BC-high and BC-low adenoCa subtypes (Supplemental Figure 4).
BC-high AdenoCa Exhibits Aggressive Clinical Phenotype
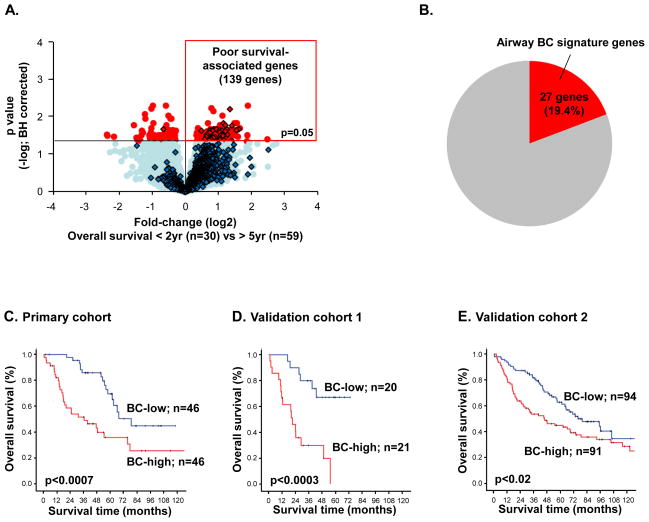
Up-regulation of cell cycle-related and EMT genes and suppression of the differentiation-related genes expression program in BC-high adenoCa suggest that high expression of the BC signature in lung adenoCa may be associated with more aggressive tumor phenotype. The analysis revealed that among 139 genes associated with poor survival in lung adenoCa (Figure 3A), ~20% genes are the BC signature genes(Figure 3B; Supplemental Gene List III).
Figure 3.
Relationship between airway BC signature expression and lung adenocarcinoma (adenoCa) patient survival. A. Genome-wide comparison between adenoCa patients with <2 -yr overall survival (“poor survivors”; n=30) vs those with >5 -year overall survival (n=59) in primary lung adenoCa cohort (n=182). Y-axis-negative log of p value; x -axis -log2 -transformed fold-change. Red dots -significant genes (p<0.05 with Benjamini-Hochberg [BH] correction); blue dots -genes with no significant difference between the group s; diamonds – 862 airway BC signature genes. B. Overlap between the 139 poor survival-associated genes(identified as described above) and airway BC signature genes. C–E. Kaplan -Meier analysis-based estimates of overall survival of BC-high adenoCa patients (red) vs BC-low adenoCa patients (blue) from: C. primary cohort of Chitale et al [14], and the validation cohorts of D. Bild et al [15] and E. Shedden et al [16]. p values were determined by the log-rank test; the number of individuals in each group are indicated.
Compared to BC-low adenoCa, BC-high adenoCa was characterized by poorer differentiation (p<0.001), lower frequency of prognostically favorable adenoCa with lepidic pattern (formerly BAC; p<0.001), and higher frequency of vascular invasion (p<0.004). At the molecular level, BC-high adenoCa exhibited significantly higher frequency of KRAS mutations and lower frequency of EGFR mutations. Consistent with KRAS mutation status, known to be more characteristic for smoking-associated adenoCa [1, 3], there were significantly more smokers among BC-high-compared to BC-low adenoCa patients (Table I).
Table I.
Clinicopathological Characterization of the Basal Cell (BC)-high and BC-low Adenocarcinoma
| BC-low (n=46) | BC-high (n=46) | p value7 | |
|---|---|---|---|
| Age, mean ± S.D. | 65.9±12.2 | 68.5±8.9 | >0.2 |
| Gender | >0.8 | ||
| Male | 24 | 24 | |
| Female | 22 | 22 | |
| Smoking history | <0.04 | ||
| Never 1 | 14 | 6 | |
| Ever | 31 | 40 | |
| COPD co-morbidity | >0.1 | ||
| No | 40 | 34 | |
| Yes | 6 | 12 | |
| Pathological stage 2 | <0.05 | ||
| I | 36 | 27 | |
| II/III | 5/5 | 5/14 | |
| Tumor size, mean ± S.D. | 3.0±1.4 | 4.1±2.9 | <0.05 |
| Node metastasis | <0.04 | ||
| No (N0) | 37 | 28 | |
| Yes (N1/N2) | 6/3 | 8/10 | |
| Pathological grade | <0.001 | ||
| Well/Moderate | 19/19 | 1/21 | |
| Poor | 5 | 21 | |
| Lepidic pattern 3 | <0.001 | ||
| No | 17 | 39 | |
| Yes | 2/27 | 2/5 | |
| Vascular invasion | <0.004 | ||
| No | 35 | 18 | |
| Yes | 8 | 18 | |
| NKX2-1 (TTF-1) expression, mean ± S.D.4 | 1.20±0.55 | 0.63±0.48 | <0.001 |
| EGFR mutations 5 | <0.03 | ||
| Wild-type | 32 | 41 | |
| Mutant | 14 | 5 | |
| TP53 mutations | >0.2 | ||
| Wild | 38 | 33 | |
| Mutant | 8 | 13 | |
| KRAS mutations 6 | <0.04 | ||
| Wild-type | 39 | 30 | |
| Mutant | 7 | 16 |
Never smokers were defined as subjects who have never had a smoking habit.
Pathological stage was based on 6th edition TNM staging.
Lepidic pattern was formerly described as bronchioloalveolar carcinoma (BAC).
Normalized expression based on the microarray gene expression analysis as described in Methods; S.D., standard deviation
The patients with lung adenocarcinoma had epidermal growth factor receptor (EGFR) mutations such as deletion in exon 19 (n=24) and a point mutation (L858R) in exon 21 (n=14).
KRAS mutants included G12C (n=17), G12V (n=16), G12D (n=6), G12A (n=4), and G13D (n=1).
p values were calculated by Pearson’s chi -square test (for categorical variables) and Mann-Whitney test (for continuous values).
Consistent with the remarkable contribution of the BC signature to the poor survival-associated gene set (Figure 3B), individuals with BC-high adenoCa had shorter overall survival compared to those with BC-low adenoCa (median survival 36 months vs 79 months; log rank p<0.0007; Figure 3C). Multivariate survival analysis demonstrated that high expression of the airway BC signature was an independent prognostic factor associated with shorter survival (hazard ratio; 1.59, 95% confidence interval; 1.14–2.22, p<0.008; Table II).
Table II.
Multivariate Cox Regression Analyses Including the Category Associated with the Airway Basal Cell (BC) Signature
| HR | 95% C.I. | p | |
|---|---|---|---|
| Age | 1.07 | 1.03–1.11 | <0.002 |
| Gender | 1.14 | 0.58–2.22 | >0.7 |
| Smoking status | 1.27 | 0.46–3.51 | >0.6 |
| Pathological stage | 3.81 | 2.01–7.22 | <0.001 |
| Lepidic pattern | 1.23 | 0.25–6.09 | >0.8 |
| Adjuvant therapy | 0.87 | 0.36–2.15 | >0.7 |
| Airway BC signature | 1.59 | 1.14–2.22 | <0.008 |
Note: In the multivariate analyses, age (continuous variable), gender (male vs female), smoking status (never vs ever smoker), pathological stage (I vs II to III) based on 6th edition TNM staging [36], pathological feature (adenocarcinoma with lepidic pattern (formarly bronchioloalveolar carcinoma) vs other adenocarcinoma), adjuvant therapy (no vs yes) and airway BC signature (BC-low vs BC-high adenocarcinoma) were included as factors. Adjuvant chemotherapy referred to systemic chemotherapy performed in pre- and/or post surgery, including adjuvant chemotherapy (n=20), adjuvant chemoradiotherapy (n=3), induction chemotherapy (n=4), and induction chemotherapy and adjuvant chemoradiotherapy (n=1).
Abbreviations: HR: hazard ratio for overall survival, CI; confidence interval, N.A: not available
The prognostic relevance of the airway BC signature expression was validated using 2 independent lung adenoCa cohorts [15, 16]. The proportion of BC-high adenoCa cases ranged between 36% and 28%, compared to 25% in the primary cohort. Similar to the primary cohort, the BC-high adenoCa cases had significantly shorter overall survival compared to BC-low adenoCa (Figures 3D, E).
BC-high Lung AdenoCa is Distinct from Lung SqCa
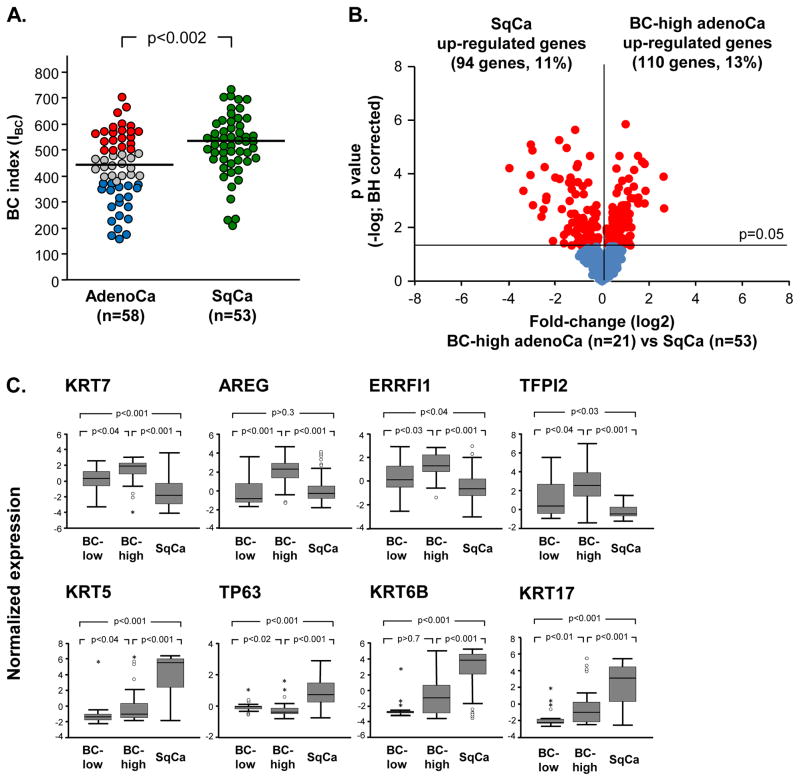
Abnormal activation of some airway BC genes has been previously linked to lung SqCa [12]. Consistent with these prior observations, the overall expression of the airway BC signature was significantly higher in SqCa compared to the adenoCa cohort (Figure 4A). However, there was no significant difference in the overall survival between BC-high and BC-low SqCa individuals (Supplemental Figures 6A and B). Notably, despite that the overall expression of the BC genes was higher in SqCa compared to adenoCa, the overall survival of the SqCa patients was longer compared to the BC-high adenoCa in the analyzed cohort (Supplemental Figure 6C).
Figure 4.
Comparative analysis of the airway basal cell (BC) signature expression in lung adenocarcinoma (adenoCa) and lung squamous cell carcinoma (SqCa). A. Lung adenoCa (n=58)and SqCa (n=53) cases from the Bild et al [15] data set were analyzed. The BC index (IBC) was calculated based on median levels of adenoCa subjects for each gene. Red circles – BC-high adenoCa, grey circles – BC-intermediate adenoCa, blue circles – BC-low adenoCa, green circles – SqCa. The median IBC for both types of cancer (436 - adenoCa, 529 - SqCa) are highlighted with a horizontal lines; p value indicated was determined by Mann-Whitney test. B. Volcano plot comparing expression of the airway BC signature genes [13] in BC -high lung adenoCa (n=21) to SqCa(n=53) from data set of Bild et al. [15]. Y-axis corresponds to the negative log of p value and the x-axis corresponds to the log2-transformed fold-change. Red dots -significant genes (p<0.05 with Benjamini-Hochberg correction); blue dots -genes with no significant difference between the groups. C. Airway BC signature gene examples differentially expressed in BC-high adenoCa (n=21) vs SqCa (n=53) from dataset of Bild et al [15]. In all panels, log2-transformed normalized gene expression levels are based on the microarray analysis. Outliers were indicated on the basis of interquartile range (IQR); ° −1.5 x IQR to 3 x IQR, * -more or less than 3 x IQR.
Next we asked whether BC-high adenoCa shares airway BC-related molecular features of SqCa. Comparison of expression of airway BC signature genes in BC-high adenoCa vs SqCa identified 13% of the airway BC genes with higher expression in BC-high adenoCa and 11% in SqCa (Figure 4B, Supplemental Gene List IV). This indicates that BC-high adenoCa is characterized by a distinct pattern of airway BC genes that distinguishes this subtype of lung cancer from SqCa. Among the airway BC genes predominantly up-regulated in BC-high adenoCa were keratin (KRT) 7, the EGFR ligand amphiregulin (AREG), ERBB receptor feedback inhibitor 1 (ERRFI1), and tissue factor pathway inhibitor 2 (TFFPI2; Figure 4C, upper panel). By contrast, the classical BC markers KRT5, TP63, KRT6B and KRT17 had significantly higher expression in SqCa compared to BC-high adenoCa (Figure 4C, lower panel). Consistent with this observation, immunohistochemical analysis revealed that TP63 protein, normally expressed in airway BC population, was overexpressed in SqCa, but not in both adenoCa subtypes (Supplemental Figure 3B).
Discussion
There is a growing body of evidence that biological heterogeneity of human malignancies is determined by specific populations of tissue resident “cells-of-origin” that, under the influence of a distinct set of oncogenic alterations, contribute to particular clinically relevant phenotypes within each individual histologic n type of cancer [27]. Based on this concept, we have assessed the molecular and clinical heterogeneity of human lung adenoCa using our recent characterization of the transcriptome of human airway BC, the stem/progenitor cell population of the airway epithelium [5, 13]. This analysis led us to the identification of a novel biologic subtype of lung adenoCa, designated as “BC-high adenoCa”, characterized by up-regulation of a distinct set of airway BC signature genes in association with clinical and pathological features of tumor aggressiveness.
Lung cancer originates from molecular alterations in the airway epithelium [1, 3, 8], a cell population comprised of ciliated, intermediate, secretory (goblet cells in the large airways and Clara cells in the small airways), BC, and rare neuroendocrine cells [4]. Depending on the unique morphologic features of individual subtypes of lung cancer, candidate cell types as the origins of each histologic subtype have been proposed. Small cell lung carcinoma and large cell neuroendocrine carcinoma are thought to originate from the neuroendocrine cells [1]. Airway BC have been considered putative cells-of-origin of lung SqCa based on the knowledge that airway BC serve as a source of squamous metaplasia, a histologic lesion associated with the early steps of the development of lung SqCa [1], as well as overexpression of selected BC markers such as KRT5 and TP63 in lung SqCa [28]. For lung adenoCa, the cell-of-origin is not known, although Clara cells and type II pneumocytes have been proposed as cellular origins of peripheral adenoCa, and cells of the surface and glandular bronchial epithelium as the source of more proximal adenoCa [6]. In the mouse, “bronchioalveolar stem cells”, a unique stem cell population at the bronchioalveolar duct junction sharing features of Clara and alveolar type II cells, have been implicated in initiation and propagation of lung adenoCa [11]. However, cellular composition of the human airway epithelium is different from that in mice. Inhuman s, BC are present throughout the airways, whereas they are virtually absent in the small airways of mice [29], impeding investigation of the role of airway BC in lung adenoCa development using mouse models.
In the present study, we assessed the biologic heterogeneity of lung adenoCa at the transcriptional level by hypothesizing that a subtype of lung adenoCa may be derived from airway BC. Based on the expression of the airway BC signature genes, the data demonstrates that lung adenoCa can be categorized into BC-high and BC-low subtypes, which exhibit remarkably different biological, pathological and clinical characteristics. The data provides insights into the biology of lung adenoCa by demonstrating that phenotypic diversity of human lung adenoCa can be explained, at least in part, by persistent activation to a greater or lesser degree of the gene expression program associated with airway BC.
The molecular patterns associated with BC-high vs BC-low adenoCa also provide insights into the mechanisms that could lead to activation of airway BC program in a subset of lung adenoCa. First, there is a higher frequency of KRAS mutations in BC-high adenoCa. The association of KRAS mutations with stem/progenitor cells has been reported with regard to lung cancer development. Endoderm progenitors with KRAS mutations exhibit increased proliferation, and fail to differentiate and maintain stem cell characteristics in vitro [30]. These data support the concept that the BC-high adenoCa is potentially derived from airway BC carrying KRAS mutations in association with smoking [1, 3]. By contrast, BC-low adenoCa was characterized by higher frequency of EGFR mutations. This is consistent with previous observations that EGFR mutations are more frequent in nonsmoking adenoCa patients [3] and are associated with the differentiation pattern known as “terminal respiratory unit” (TRU), with high expression of genes typical for Clara cells and alveolar type II cells, such as those encoding TTF1 and surfactant proteins [7].
Second, BC-high lung adenoCa was enriched in transcriptional pathways and networks related to the ECM organization interacting with various BC signature genes encoding important regulators of homeostatic processes in the lung tissue, including TGFB1, MMP1, MMP2, TIMP2, ITGAV, and VDR, as well as the networks associated with epidermis development, cell adhesion, cell cycle and proliferation. Given the anatomic location of BC immediately above the basement membrane, it is possible that BC might contribute to the pathogenesis of lung adenoCa by regulation of the ECM homeostasis and epithelial-mesenchymal interactions. Activation of distinct growth factor signaling mechanisms including those related to TGFB1 and/or mediated via activation of MMP and integrin signaling at the epithelial-mesenchymal interface may be responsible for enrichment of the cell adhesion-and cell cycle-related networks and contribute to more aggressive tumor characteristics observed in BC-high lung adenoCa.
Third, comparative analysis of the differentiation pattern of BC-high- vs BC-low lung adenoCa revealed that down-regulation of genes related to ciliated and Clara cell differentiation in BC-high adenoCa is accompanied by activation of the EMT transcriptional program, including induction of transcription factors, such as SNAI1/SNAIL, SNAI2/SLUG, and TWIST1 and CDH2, a gene encoding mesenchymal adherent junction protein N-cadherin [26]. Activation of the EMT program is believed to bean important process promoting cancer invasion and metastasis [31]. Consistent with this concept, BC-high adenoCa exhibited higher frequency of vascular invasion and lymph node metastasis. Furthermore, EMT has been reported to generate cells with tumorigenic characteristics [32]. The present study reinforces the relationship between EMT and tissue stem cells in the context of lung adenoCa development.
Finally, by comparison to the SqCa, in which BC genes are also highly expressed, we identified that BC-high lung adenoCa exhibits up-regulation of a distinct set of the BC signature genes, including the genes related to the EGFR pathway, such as AREG and ERRFI1. The EGFR pathway is enriched in airway BC [13] and smoking is known to activate EGFR signaling in the lung epithelial cells without EGFR mutation inducing cellular processes relevant to lung cancer development [33].
Although the genes contributing to BC-high adenoCa-enriched molecular pathways are not known as classic cancer-driving oncogenes, understanding their interaction is important for the development of novel therapeutic strategies aimed at regulation of tumor cell survival and growth, for example, using the synthetic lethality approach. Such strategies may be particularly beneficial for BC-high lung adenoCa, associated with high frequency of KRAS mutations, for which no effective specific targeted approaches have been developed so far [3]. Targeting interaction between some of the BC-high adenoCa-enriched genes, such as the oncogene MYC and cyclin-dependent kinases, has been recently shown to induce therapeutically-relevant synthetic lethality in aggressive basal cell-like breast cancer [34]. In addition, based on the knowledge that survival, growth and differentiation of BC is dependent on their adhesion to the extracellular matrix components (ECM) [35] and ECM-related genes are enriched in the BC-high adenoCa, it is possible that targeting ECM genes may represent an additional approach to induce synthetic lethality interactions in BC-high adenoCa.
Together, the present study identifies a novel, BC-high subtype of human lung adenoCa, associated with activation of a distinct set of airway BC signature genes and provides transcriptome-based evidence supporting the concept that this aggressive subset of human lung adenoCa is likely derived from the airway BC population.
Supplementary Material
Key messages.
What is the key question?
Human airway epithelium is composed of various cell types, including ciliated, intermediate, secretory, neuroendocrine, and basal cells (BC). The specific contribution of these individual cell types of the airway epithelium to lung adenocarcinoma heterogeneity is not well understood. The key question of the present study was whether unique gene expression features of airway BC, the stem/progenitor cells of the human airway epithelium, contribute to a distinct molecular and clinical phenotype of lung adenocarcinoma in humans.
What is the bottom line and why read on?
Activation of a unique airway BC program contributes to human lung adenocarcinoma heterogeneity and is a characteristic feature of a biologically and clinically distinct, more aggressive subtype of human lung adenocarcinoma. The data supports the concept that airway BC represent a cellular source of molecular changes associated with the development of a subset of aggressive lung adenocarcinomas in humans.
Acknowledgments
We thank M. Ladanyi for providing the Memorial Sloan-Kettering Cancer Center adenocarcinoma samples to us; J. Salit for supporting microarray analysis; and N. Mohamed for help in preparing this manuscript. These studies were supported, in part, by P50 HL084936, 1T32HL094284 and UL1-RR024996; and the Starr Foundation/Starr Cancer Consortium. RS is supported, in part, by the Parker B. Francis Foundation.
Footnotes
Conflict of interest: The authors have declared that no competing interests exist.
Author contributions: TF: participated in study design, performed experiments, analyzed data, wrote the manuscript; RS: participated in study design and data analysis, wrote the manuscript; FAP: participated in data analysis; JM: participated in data analysis; RD: contributed to sample collection, critically reviewed the manuscript; WT: critically reviewed data and the manuscript; RGC: participated in study design and data analysis, oversaw the study, wrote the manuscript
References
- 1.Wistuba II, Gazdar AF. Lung Cancer Preneoplasia. Annu Rev Pathol. 2006;1:331–348. doi: 10.1146/annurev.pathol.1.110304.100103. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, et al. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung Cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crystal RG, Randell SH, Engelhardt JF, et al. Airway Epithelial Cells: Current Concepts and Challenges. Proc Am Thorac Soc. 2008;5:772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rock JR, Onaitis MW, Rawlins EL, et al. Basal Cells As Stem Cells of the Mouse Trachea and Human Airway Epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travis WD, Brambilla E, Muller-Hermelink HK, et al. Pathology and Genetics: Tumors of the Lung, Pleura, Thymus and Heart. Lyon: IARC; 2004. [Google Scholar]
- 7.Yatabe Y. EGFR Mutations and the Terminal Respiratory Unit. Cancer Metastasis Rev. 2010;29:23–36. doi: 10.1007/s10555-010-9205-8. [DOI] [PubMed] [Google Scholar]
- 8.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pack RJ, Al-Ugaily LH, Morris G. The Cells of the Tracheobronchial Epithelium of the Mouse: a Quantitative Light and Electron Microscope Study. J Anat. 1981;132:71–84. [PMC free article] [PubMed] [Google Scholar]
- 10.Boers JE, Ambergen AW, Thunnissen FB. Number and Proliferation of Clara Cells in Normal Human Airway Epithelium. Am J Respir Crit Care Med. 1999;159:1585–1591. doi: 10.1164/ajrccm.159.5.9806044. [DOI] [PubMed] [Google Scholar]
- 11.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of Bronchioalveolar Stem Cells in Normal Lung and Lung Cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 12.Ooi AT, Mah V, Nickerson DW, et al. Presence of a Putative Tumor-Initiating Progenitor Cell Population Predicts Poor Prognosis in Smokers With Non-Small Cell Lung Cancer. Cancer Res. 2010;70:6639–6648. doi: 10.1158/0008-5472.CAN-10-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackett NR, Shaykhiev R, Walters MS, et al. The Human Airway Epithelial Basal Cell Transcriptome. PLoS One. 2011;6:e18378. doi: 10.1371/journal.pone.0018378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chitale D, Gong Y, Taylor BS, et al. An Integrated Genomic Analysis of Lung Cancer Reveals Loss of DUSP4 in EGFR-Mutant Tumors. Oncogene. 2009;28:2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bild AH, Yao G, Chang JT, et al. Oncogenic Pathway Signatures in Human Cancers As a Guide to Targeted Therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 16.Shedden K, Taylor JM, Enkemann SA, et al. Gene Expression-Based Survival Prediction in Lung Adenocarcinoma: a Multi-Site, Blinded Validation Study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryant CM, Albertus DL, Kim S, et al. Clinically Relevant Characterization of Lung Adenocarcinoma Subtypes Based on Cellular Pathways: an International Validation Study. PLoS One. 2010;5:e11712. doi: 10.1371/journal.pone.0011712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JJ, Deane NG, Wu F, et al. Experimentally Derived Metastasis Gene Expression Profile Predicts Recurrence and Death in Patients With Colon Cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X, Lu X, Wang ZC, et al. Predicting Features of Breast Cancer With Gene Expression Patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 20.Kuner R, Muley T, Meister M, et al. Global Gene Expression Analysis Reveals Specific Patterns of Cell Junctions in Non-Small Cell Lung Cancer Subtypes. Lung Cancer. 2009;63:32–38. doi: 10.1016/j.lungcan.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 21.Hou J, Aerts J, den HB, et al. Gene Expression-Based Classification of Non-Small Cell Lung Carcinomas and Survival Prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding L, Getz G, Wheeler DA, et al. Somatic Mutations Affect Key Pathways in Lung Adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang DY, Villanueva A, Hoshida Y, et al. Focal Gains of VEGFA and Molecular Classification of Hepatocellular Carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badea L, Herlea V, Dima SO, et al. Combined Gene Expression Analysis of Whole-Tissue and Microdissected Pancreatic Ductal Adenocarcinoma Identifies Genes Specifically Overexpressed in Tumor Epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 25.Tilley AE, O’Connor TP, Hackett NR, et al. Biologic Phenotyping of the Human Small Airway Epithelial Response to Cigarette Smoking. PLoS One. 2011;6:e22798. doi: 10.1371/journal.pone.0022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalluri R, Weinberg RA. The Basics of Epithelial-Mesenchymal Transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visvader JE. Cells of Origin in Cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 28.Camilo R, Capelozzi VL, Siqueira SA, et al. Expression of P63, Keratin 5/6, Keratin 7, and Surfactant-A in Non-Small Cell Lung Carcinomas. Hum Pathol. 2006;37:542–546. doi: 10.1016/j.humpath.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 29.Rock JR, Randell SH, Hogan BL. Airway Basal Stem Cells: a Perspective on Their Roles in Epithelial Homeostasis and Remodeling. Dis Model Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlan MP, Quatela SE, Philips MR, et al. Activated Kras, but Not Hras or Nras, May Initiate Tumors of Endodermal Origin Via Stem Cell Expansion. Mol Cell Biol. 2008;28:2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiery JP, Acloque H, Huang RY, et al. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Mani SA, Guo W, Liao MJ, et al. The Epithelial-Mesenchymal Transition Generates Cells With Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filosto S, Becker CR, Goldkorn T. Cigarette Smoke Induces Aberrant EGF Receptor Activation That Mediates Lung Cancer Development and Resistance to Tyrosine Kinase Inhibitors. Mol Cancer Ther. 2012;11:795–804. doi: 10.1158/1535-7163.MCT-11-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horiuchi D, Kusdra L, Huskey NE, et al. MYC Pathway Activation in Triple-Negative Breast Cancer Is Synthetic Lethal With CDK Inhibition. J Exp Med. 2012;209:679–696. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coraux C, Roux J, Jolly T, et al. Epithelial Cell-Extracellular Matrix Interactions and Stem Cells in Airway Epithelial Regeneration. Proc Am Thorac Soc. 2008;5:689–694. doi: 10.1513/pats.200801-010AW. [DOI] [PubMed] [Google Scholar]
- 36.UICC. TNM Classification of Malignant Tumors. 6. New York: Wiley-Liss, Inc; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.