Abstract
Background:
Human exposure to antibiotic resistant bacteria (ARB) is a public health concern which could occur in a number of ways. Wastewaters seem to play an important role in the dissemination of bacteria and antibiotic resistant genes (ARGs) in our environment. The aim of this study was to evaluate the occurrence of three groups of ARB and their resistance genes in hospital and municipal wastewaters (MWs) as possible sources.
Methods:
A total of 66 samples were collected from raw MWs and hospital wastewaters (HWs) and final effluents of related wastewater treatment plants (WWTPs). Samples were analyzed for the detection of three groups of ARB including gentamicin (GM), chloramphenicol (CHL) and ceftazidime resistant bacteria and their ARGs (aac (3)-1, cmlA1 and ctx-m-32, respectively).
Results:
The mean concentration of GM, CHL and ceftazidime resistant bacteria in raw wastewater samples was 1.24 × 107, 3.29 × 107 and 5.54 × 107 colony forming unit/100 ml, respectively. There is a variation in prevalence of different groups of ARB in MWs and HWs. All WWTPs decreased the concentration of ARB. However, high concentration of ARB was found in the final effluent of WWTPs. Similar to ARB, different groups of ARGs were found frequently in both MWs and HWs. All genes also detected with a relative high frequency in effluent samples of MWs WWTPs.
Conclusions:
Discharge of final effluent from conventional WWTPs is a potential route for dissemination of ARB and ARGs into the natural environment and poses a hazard to environmental and public health.
Keywords: Antibiotic resistance genes, antibiotic resistant bacteria, ceftazidim, chloramphenicol, gentamicin
INTRODUCTION
The widespread use of antibiotics in human and veterinary medicine is often associated with increased bacterial resistance[1,2,3,4,5,6,7] to these chemical substances. The antibiotic resistant bacteria (ARB) have created serious problems in the treatment of infectious diseases. These bacteria and their antibiotic resistance genes (ARGs) enter the environment through various sources.[8] Hospital wastewaters (HWs) and municipal wastewaters (MWs) are a potential source for entry of ARB and ARGs into the natural environment. Dissemination of these bacteria and genes in the environment is a growing public health concern. Antibiotic resistance in the environment can be transferred from pathogenic bacteria to nonpathogenic bacteria, which impair water ecology through change in population dynamics and physiology.[9] A study has shown that a large number of bacteria and resistance genes particularly in hospital strains have been found in the area of hospital wastewater discharge. ARGs can be transferred among the bacteria in the hospital and environment through plasmid, integrons, and transpositions.[10]
A number of studies have reported that hospital ARB and ARGs have been identified in MW, activated sludge and water resources.[11,12] It is important to note that some of these contaminated water resources may be used as public water supplies. Some studies indicate that discharge of HWs is associated with a high rate dissemination of resistant bacteria into the natural environment.[13,14] In contrast, some studies have reported that the MW is an important route for the release of ARB and ARGs. However, many studies have shown that the removal of ARB and ARGs in MW and HW treatment plants (WWTPs) does not occur or is very low. Even some researchers have shown that resistant bacteria have increased in WWTPs.[8,15,16]
Therefore, this study was designed to investigate (1) The occurrence of three groups of ARB in hospital and MWs; (2) the effect of WWTPs on the removal of antibiotic resistance bacteria; (3) the presence/absence of specific ARGs in isolated ARB (4) the frequency of detection of specific ARGs in raw and final effluent samples of MWs and HWs. We used three more consumed antibiotics including gentamicin (GM), chloramphenicol (CHL), and ceftazidime (CAZ); and for each antibiotic, a resistance encoding gene was selected based on the frequency and importance in clinical and environmental samples.[17,18,19]
METHODS
Samples were collected from six different sites including: MW1 (a municipal WWTP with two stage activated sludge treatment system), MW2 (a municipal WWTP with activated sludge treatment system), MW3 (a municipal WWTP with stabilization pond treatment system), HW1 and HW2 (HW), HW3 (a Hospital WWTP with activated sludge treatment system) [Table 1]. A total of 66 samples from raw wastewater and final effluents were collected in 1 L sterile glasses between September 2012 and April 2013. Samples were transferred to the laboratory in an insulated box with cooling packs and were analyzed immediately after arrival at the laboratory.
Table 1.
Characteristics of hospital and municipal wastewaters

The conventional heterotrophic plate count (HPC) method was used to evaluate the concentration of three groups of ARB in wastewater samples. In the study, three different antibiotics including GM, CHL and CAZ were selected. To determine the concentration of HPC bacteria, R2A agar medium along with antifungal additive nystatin was used. R2A plating media were also used for ARB enumeration and each antibiotic was individually amended into the media as follow: (1) GM 10 μg/ml, (2) CHL 16 μg/ml and (3) ceftazidime 30 μg/ml.[20,21,22] Wastewater samples were thoroughly stirred and serially diluted up to 10-6, and 0.1 ml of each serial dilution was plated on the medium. Plates were incubated at 37°C for at least 48 h.[23] All the experiments were carried out in duplicates and the mean values as colony forming unit (CFU)/100 ml were considered.
For detection of related ARG in each group of ARB, predominant ARB with regard to the apparent characteristics were isolated and subcultured onto R2A agar plates with specific amended antibiotics. Isolated colonies were then suspended in 100 μl of deionized water, and genomic DNA was extracted by boiling for 15 min and centrifugation at 13,000 rpm for 5 min. 0.1 (v/v) of 3M sodium acetate and 2.5 (v/v) of 95% ethanol were added to the supernatant, and then the suspensions were mixed and centrifuged at 13,000 rpm for 10 min. The supernatant was discarded, and 500 μl of 80% ethanol was added to the pellets. Suspensions were centrifuged at 13,000 rpm for 5 min. DNA was suspended in 50 μl of distilled water and extracted DNA was stored in a freezer at -20°C until use.
DNA was also extracted from the original wastewater samples to determine the presence/absence of selected genes in raw wastewater and effluent samples. For this purpose, 50 ml of all samples were centrifuged at 6000 rpm for 15 min. The supernatant was discarded, and the pellet was resuspended in 300 μl of distilled water. The resuspended pellets were frozen in liquid nitrogen and heated in boiling water 3 times. The DNA was extracted and purified using Promega DNA Extraction kit (Promega Wizard Genomic DNA purification kit, Madison, WI) according to the manufacturer's manual.
For each group of antibiotics, a resistance encoding gene was selected as follow: Aac (3)-1 for GM resistant bacteria, cmlA1for CHL resistant bacteria and ctx-m-32 for ceftazidime resistant bacteria Three pair of primers used for amplification of these genes. The characteristics of primers are given in Table 2.
Table 2.
Characteristics of primers used in the study

Polymerase chain reaction (PCR) amplification was done in a total volume of 25 μl containing 2.5 μl of ×10 PCR buffer, 0.2 μM of each primer, 0.2 mM of each dNTPs, 2 units of Taq DNA polymerase, and 1 μl of DNA. All assays contained a positive and a negative control. PCR process is performed using an initial denaturation at 94°C for 10 min, denaturation at 94°C for 45 s, annealing at varied for 30 s, and extension at 72°C for 45 s for 30 cycles, followed by a final extension at 72 for 10 min. PCR products were analyzed by agarose gel electrophoresis using 1.5% gels containing ethidium bromide together with a DNA molecular weight marker. Gels were viewed on an ultra violet (UV) transilluminator (UV Tech, France).
T-test was applied to compare HPC and ARB quantitative mean in raw wastewater and final effluent samples. To compare the presence and absence of genes in different sites and before and after treatment Chi-square and McNemar test were used, respectively. ANOVA statistical analysis was used to compare variables in the different sites. P value at level of 0.05 was considered as significant.
RESULTS
Concentrations of HPC and ARB in different MWs and HWs are given in Table 3. Mean concentration of HPC in MW and HW inflow and outflow were 1.8 × 108, 9.3 × 107, 9.09 × 107 and 1.37 × 106 CFU/100 ml, respectively. Figure 1 shows the prevalence of different groups of ARB in MWs and HWs as a percentage of ARB with respect to total HPC. As shown in Figure 1, prevalence of CHL resistant bacteria was the highest in MWs. However, GM resistant bacteria had the lowest percentage in both MWs and HWs. No significant correlation was observed between the three groups of resistant bacteria.
Table 3.
Concentration of HPC and ARB as CFU/100 ml in raw wastewater from different sites
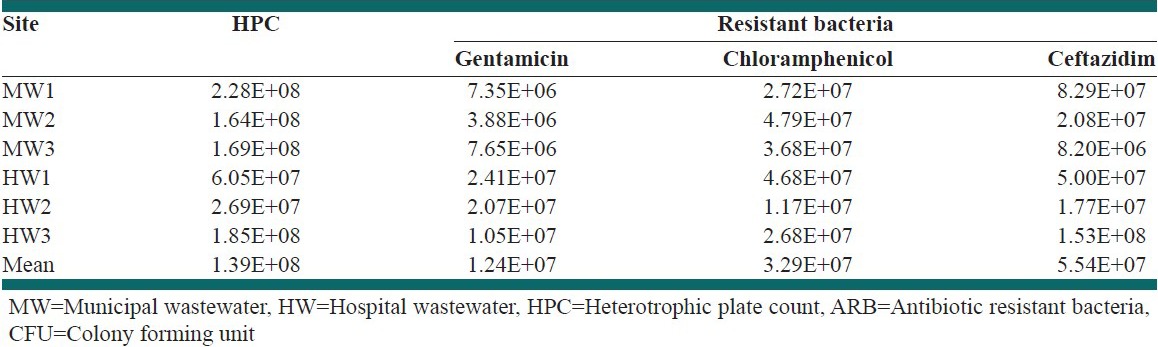
Figure 1.
Prevalence of different groups of antibiotic resistant bacteria (ARB) (as percentage of each group of ARB with respect to heterotrophic plate count) in (a) municipal wastewaters, (b) hospital wastewaters
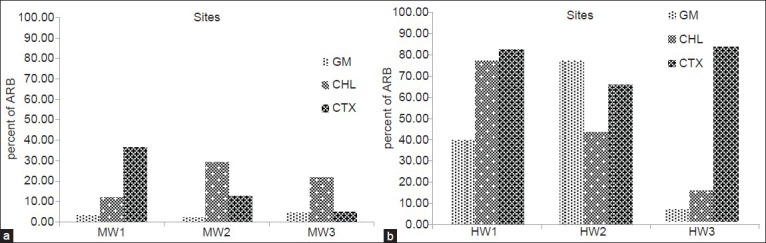
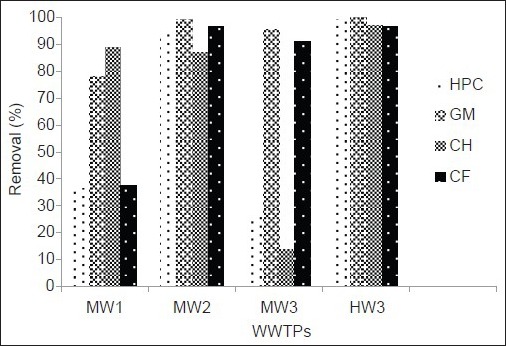
The removal efficiency of HPC and different groups of ARB by WWTPs is shown in Figure 2. As shown in this figure HW3 was the most efficient treatment plant in removing of bacteria. Concentration of GM resistant bacteria significantly reduced in this WWTP. There was also a significant difference between the inflow and outflow concentration of these bacteria in MW2 treatment plant. Figure 3 shows the concentrations of HPC and ARB in the final effluent of WWTPs.
Figure 2.
Removals (percentage) of heterotrophic plate count and different groups of antibiotic resistant bacteria in wastewater treatment plants
Figure 3.
Log concentration (number of colony forming unit/100 ml) of heterotrophic plate count and different groups of antibiotic resistant bacteria in final effluent of municipal and hospital wastewaters. Error bars is indicated standard errors
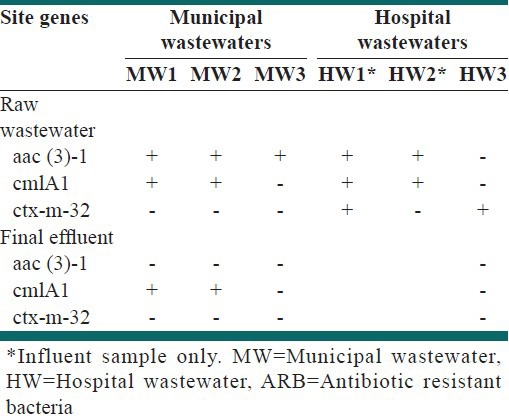
The presence/absence of specific ARGs in isolated ARB from raw wastewater and final effluent samples of different sites is presented in Table 3. Resistant genes were found mostly in isolated resistant bacteria from raw wastewater samples, while ARGs did not detected in many of effluent samples.
The percent of raw wastewater and final effluent samples which were positive for genes resistance to antibiotic groups are given in Figure 4. The results show that the studied genes were found more frequently in inflow than outflow samples. However, the frequency of resistance gene (ctx-m-32) in MW1 and MW2 treatment plants unchanged. This gene also increased in MW3 treatment plant.
Figure 4.
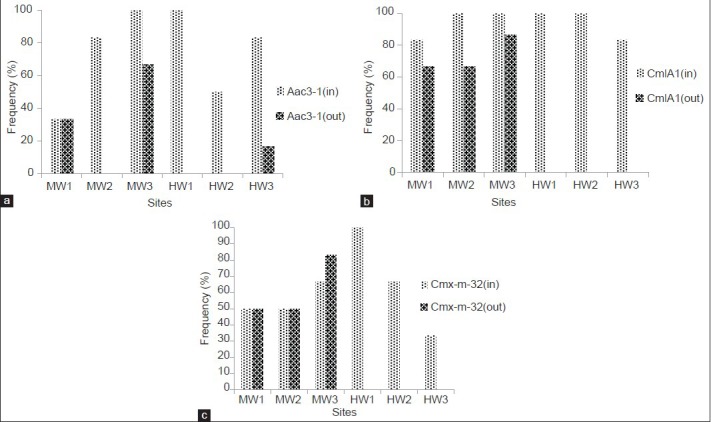
Frequency of detection of different groups of ARGs in raw wastewater (in) and final effluent (out) samples. (a) Gentamicin resistant gene, aac(3)-1; (b) chloramphenicol resistant gene, cmlA1 and (c) ceftazidime resistant gene, ctx-m-32
DISCUSSION
Human exposure to ARB is a public health concern as it has direct links with disease management. This exposure could occur in a number of ways. Hospital and MWs seem to play an important role in the dissemination of bacteria and antibiotic resistant genes in our environment. The present study showed a high concentration of ARB in both MWs and HWs. The mean concentration of HPC and GM, CHL and CAZ resistant bacteria in raw wastewater samples was 1.39 × 108, 1.24 × 107, 3.29 × 107 and 5.54 × 107 CFU/100 ml, respectively. The concentration of different groups of ARB had no significant difference for MWs and hospital raw wastewaters. Among the ARB, however, CAZ resistant bacteria had the highest concentration in HWs. Higher concentration of HPC was observed in municipal raw wastewaters compared with HWs, but this difference was not significant. The mean concentrations of ARB and HPC in municipal and hospital raw wastewaters in this study were higher than the average obtained in other studies.[16,18] Total mean of HPC and ARB in influent MWs and HWs were 1.39 × 108, 4.6 × 107, 3.35 × 107, and 4.19 × 107 CFU/100 ml, respectively, while Reinthaler et al. (2003) determined that the concentration of resistant bacteria in wastewater to be 103.9-105.45 CFU/100 ml.[25]
As shown in Figure 1, there is a variation in prevalence of different groups of ARB in MWs and HWs. This variation in concentration of different groups of ARB was also observed in other studies.[15,26] Munir et al. found higher concentration of sulfonamide resistant bacteria when compared to tetracycline resistant bacteria. It is important to note that the total percentage of ARB in any type of wastewater is higher than 100% [Figure 1], which indicates the multi resistance of heterotrophic bacteria in wastewaters. Among different groups of ARB, the concentration of CHL resistant bacteria in MWs was the highest and GM resistant bacteria were the lowest. This result is consistent with that of Huang et al. regarding higher frequency of bacteria resistance to CHL than other antibiotics.[16] The higher concentration of CHL resistant bacteria in MWs could be due to the use of this antibiotic in animal husbandry.[27] In HWs, the maximum values were for CAZ and the minimum goes for GM resistant bacteria. Beta-lactamases (e.g. ceftazidim) group of antibiotics is more used in hospitals.[28] However, GM resistant bacteria in both MWs and HWs were minimal. This could be the result of prohibited taking of this antibiotic in recent years. However, this study proved the presence of different groups of ARB in MWs and HWs in relatively high concentrations.
As shown in Figure 2, all the WWTPs decreased the concentration of HPC and ARB. However, among different WWTPs, the highest removal of HPC and ARB was observed in HW3 which is an activated sludge process with a UV disinfection system. Likewise, this treatment plant also benefits from high speed sand filtration after disinfecting. Although, activated sludge utilities are also used as the biological wastewater treatment processes in MW1 and MW2, but the significant removal efficiency of ARB in HW3 could be related to the UV disinfection as well as high speed sand filtration system. After HW3, the MW2 plant has the highest efficiency in removing HPC and ARB. This treatment plant become equipped with an integral operation system and in comparison with other MW plants in the study is in a better condition from the view point of biochemical oxygen demand and suspended solids removal.
Furthermore, removal efficiencies by each WWTPs and among different plants varied for different groups of ARB [Figure 2]. The lowest removal rate of CHL resistant bacteria was observed in MW3 which is a stabilization pond treatment process. Similar finding was also reported by Munir et al. which observed higher removal of tetracycline resistant bacteria in comparison to sulfonamide resistant bacteria by activated sludge process. They proposed that multiple selective pressures in the environment might be attributed to these variations.[23] Given the selection pressure, bacteria with appropriate mechanism of resistance may have a better chance of survival.
Overall, we observed that even though the concentrations of ARB in raw wastewater are significantly reduced by WWTPs, high concentration of ARB could be released in the environment through discharge of WWTP effluents [Figure 3].
Resistant genes were found mostly in isolated resistant bacteria from raw wastewater samples [Table 4]. Except for CHL (cmlA1) resistant gene in MW effluents, other genes were not found in either the final effluent isolated colonies at municipal WWTPs or in HW. Since, antibiotic resistance to each group of antibiotics could be induced by several genes; it is possible that other genes which not detected in the present study are responsible for antibiotic resistance.
Table 4.
Presence/absence of selected antibiotic resistance genes in isolated ARB from raw wastewater and final effluent samples
Similar to ARB, different groups of antibiotic resistant genes were found frequently in both MWs and HWs [Figure 4]. Studies conducted by Chee-Stanford et al. (2001),[29] Mackie et al. (2001)[30] and Heuer et al. (2004)[31] showed that these genes are found in abundance in various environments. Except the HW3, all genes detected with a relative high frequency in effluent samples of municipal WWTPs. This result shows that the role of conventional wastewater treatment processes for the removal of ARGs is very little [Figure 4]. These findings are consistent with the results of other studies.[26,32,33] However, CHL (CmlA1) and CAZ (Ctx-m-32) resistance genes were not found in the outflow of HW3, and GM resistant gene (Aac(3)-1) were found in very lower frequency than inflow. The observed decrease could be due to the UV disinfection process or high speed sand filtration after disinfecting. As mentioned above, significant decreases were also observed in the concentration of ARB especially GM resistant bacteria (100%) at this treatment plant. However, we found GM resistance gene in one effluent sample which could be related to the viable, but non-culturable GM resistant bacteria as a result of the disinfection process.
As shown in Figure 4, the frequency of CAZ (ctx-m-32) resistance gene in the outflow of MW3 (stabilization pond) increased. The finding of this research do not match with some results that show the direct effect of solar radiation as an important factor in reducing of resistant genes and also the high performance of stabilization ponds in reducing of these agents.[8] However, the low intensity and duration of solar radiation in the period of study (September 2012 to March 2013), might contribute to this difference.
CONCLUSIONS
The results of this study showed high concentrations of ARB in both MWs and HWs. ARGs were also frequently found in wastewater samples. We observed that the concentration of ARB decreased from inflow to outflow of all WWTPs, nevertheless conventional wastewater treatment processes couldn’t significantly reduce ARB and high concentration of these bacteria was entered into effluent. Thus, discharge of final effluent from WWTPs is a potential route for dissemination of ARB and ARGs into the natural environment and poses a hazard to public health.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S, Aga DS. Potential ecological and human health impacts of antibiotics and antibiotic-resistant bacteria from wastewater treatment plants. J Toxicol Environ Health B Crit Rev. 2007;10:559–73. doi: 10.1080/15287390600975137. [DOI] [PubMed] [Google Scholar]
- 3.Kümmerer K. The presence of pharmaceuticals in the environment due to human use – Present knowledge and future challenges. J Environ Manage. 2009;90:2354–66. doi: 10.1016/j.jenvman.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto AS, Fiiho RL, Rocha TS, Menconi A, Marietto-Gonçalves GA. Detection and transfer of antimicrobial resistance gene integron in Salmonella enteritidis derived from avian material. Braz J Poult Sci. 2009;11:191–201. [Google Scholar]
- 5.Laroche E, Pawlak B, Berthe T, Skurnik D, Petit F. Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France) FEMS Microbiol Ecol. 2009;68:118–30. doi: 10.1111/j.1574-6941.2009.00655.x. [DOI] [PubMed] [Google Scholar]
- 6.Tao R, Ying GG, Su HC, Zhou HW, Sidhu JP. Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl rivers in South China. Environ Pollut. 2010;158:2101–9. doi: 10.1016/j.envpol.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Aminov RI, Mackie RI. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol Lett. 2007;271:147–61. doi: 10.1111/j.1574-6968.2007.00757.x. [DOI] [PubMed] [Google Scholar]
- 8.Bouki C, Venieri D, Diamadopoulos E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol Environ Saf. 2013;91:1–9. doi: 10.1016/j.ecoenv.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut. 2009;157:2893–902. doi: 10.1016/j.envpol.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 10.Rahube TO, Yost CK. Antibiotic resistance plasmids in wastewater treatment plants and their possible dissemination into the environment. Afr J Biotechnol. 2010;9:9183–90. [Google Scholar]
- 11.Fuentefria DB, Ferreira AE, Corção G. Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water: Are they genetically related? J Environ Manage. 2011;92:250–5. doi: 10.1016/j.jenvman.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Tuméo E, Gbaguidi-Haore H, Patry I, Bertrand X, Thouverez M, Talon D. Are antibiotic-resistant Pseudomonas aeruginosa isolated from hospitalised patients recovered in the hospital effluents? Int J Hyg Environ Health. 2008;211:200–4. doi: 10.1016/j.ijheh.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Czekalski N, Berthold T, Caucci S, Egli A, Bürgmann H. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into lake geneva, Switzerland. Front Microbiol. 2012;3:106. doi: 10.3389/fmicb.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright GD. Antibiotic resistance in the environment: A link to the clinic? Curr Opin Microbiol. 2010;13:589–94. doi: 10.1016/j.mib.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Guardabassi L, Lo Fo Wong DM, Dalsgaard A. The effects of tertiary wastewater treatment on the prevalence of antimicrobial resistant bacteria. Water Res. 2002;36:1955–64. doi: 10.1016/s0043-1354(01)00429-8. [DOI] [PubMed] [Google Scholar]
- 16.Huang JJ, Hu HY, Lu SQ, Li Y, Tang F, Lu Y, et al. Monitoring and evaluation of antibiotic-resistant bacteria at a municipal wastewater treatment plant in China. Environ Int. 2012;42:31–6. doi: 10.1016/j.envint.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. 2004;28:519–42. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Szczepanowski R, Linke B, Krahn I, Gartemann KH, Gützkow T, Eichler W, et al. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology. 2009;155:2306–19. doi: 10.1099/mic.0.028233-0. [DOI] [PubMed] [Google Scholar]
- 19.Fernández A, Gil E, Cartelle M, Pérez A, Beceiro A, Mallo S, et al. Interspecies spread of CTX-M-32 extended-spectrum beta-lactamase and the role of the insertion sequence IS1 in down-regulating bla CTX-M gene expression. J Antimicrob Chemother. 2007;59:841–7. doi: 10.1093/jac/dkm030. [DOI] [PubMed] [Google Scholar]
- 20.Figueira V, Vaz-Moreira I, Silva M, Manaia CM. Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 2011;45:5599–611. doi: 10.1016/j.watres.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Xi C, Zhang Y, Marrs CF, Ye W, Simon C, Foxman B, et al. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol. 2009;75:5714–8. doi: 10.1128/AEM.00382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson RW, Osborne K, Barnes G, Jolliff C, Zamani D, Roll B, et al. Multiregional evaluation of the SimPlate heterotrophic plate count method compared to the standard plate count agar pour plate method in water. Appl Environ Microbiol. 2000;66:453–4. doi: 10.1128/aem.66.1.453-454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munir M, Wong K, Xagoraraki I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011;45:681–93. doi: 10.1016/j.watres.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 24.Heuer H, Krögerrecklenfort E, Wellington EM, Egan S, van Elsas JD, van Overbeek L, et al. Gentamicin resistance genes in environmental bacteria: Prevalence and transfer. FEMS Microbiol Ecol. 2002;42:289–302. doi: 10.1111/j.1574-6941.2002.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 25.Reinthaler FF, Posch J, Feierl G, Wüst G, Haas D, Ruckenbauer G, et al. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003;37:1685–90. doi: 10.1016/S0043-1354(02)00569-9. [DOI] [PubMed] [Google Scholar]
- 26.Fraise AP. Susceptibility of antibiotic-resistant cocci to biocides. J Appl Microbiol. 2002;92(Suppl):158S–62. [PubMed] [Google Scholar]
- 27.Marni S, Malintan NT, Faridah I, Mustafa AM. Chloramphenicol in Malaysia waste water and its residues in animal husbandaries products. Health Environ J. 2010;1:41–5. [Google Scholar]
- 28.Noorbakhsh AJ, Mehrabani D, Japoni S. Multi-drug resistance bacteria in Qom Hospitals, Central Iran. Iran Red Crescent Med J. 2010;12:501–3. [Google Scholar]
- 29.Chee-Sanford JC, Aminov RI, Krapac I, Garrigues-Jeanjean N, Mackie RI. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl Environ Microbiol. 2001;67:1494–502. doi: 10.1128/AEM.67.4.1494-1502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackie RI, Koike S, Krapac I, Chee-Sanford J, Maxwell S, Aminov RI. Tetracycline residues and tetracycline resistance genes in groundwater impacted by swine production facilities. Animal Biotechnol. 2006;17:157–76. doi: 10.1080/10495390600956953. [DOI] [PubMed] [Google Scholar]
- 31.Heuer H, Szczepanowski R, Schneiker S, Pühler A, Top E, Schlüter A. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiol. 2004;150:3591–9. doi: 10.1099/mic.0.27304-0. [DOI] [PubMed] [Google Scholar]
- 32.Murray IA, Shaw WV. O-Acetyltransferases for chloramphenicol and other natural products. Antimicrob Agents Chemother. 1997;41:1–6. doi: 10.1128/aac.41.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rutala WA, Stiegel MM, Sarubbi FA, Weber DJ. Susceptibility of antibiotic-susceptible and antibiotic-resistant hospital bacteria to disinfectants. Infect Control Hosp Epidemiol. 1997;18:417–21. doi: 10.1086/647641. [DOI] [PubMed] [Google Scholar]


