Abstract
Methane-oxidizing bacteria (MOB) gain energy from the oxidation of methane and may play important roles in freshwater ecosystems. In this study, the community structure of planktonic MOB was investigated in a subtropical reservoir. Bacterial community structure was investigated through the analysis of the 16S rRNA gene. Three groups of phylogenetically distinct MOB were detected in the clone libraries of polymerase chain reaction products obtained with universal primers. The groups belonged to the class Gammaproteobacteria, the class Alphaproteobacteria, and the candidate phylum NC10. The last group, which consists of close relatives of the nitrite reducer ‘Candidatus Methylomirabilis oxyfera', was frequently detected in the clone libraries of deep-water environments. The presence of 3 groups of MOB in deep water was also shown by a cloning analysis of the pmoA gene encoding particulate methane monooxygenase. The dominance of ‘M. oxyfera'-like organisms in deep water was confirmed by catalyzed reporter deposition–fluorescence in situ hybridization, in which cells stained with a specific probe accounted for 16% of total microbial cells. This is the first study to demonstrate that close relatives of the nitrite reducer can be major component of planktonic MOB community which may affect carbon flow in aquatic ecosystems.
Methane-oxidizing bacteria (MOB) are capable of gaining energy from the oxidation of the simplest hydrocarbon, methane. This ability is observed only in restricted lineages of prokaryotes, and 4 major phylogenetic groups of MOB are currently known. Two of these MOB groups, belonging to the classes Gammaproteobacteria and Alphaproteobacteria in the phylum Proteobacteria, have been well characterized. These 2 groups are also referred to as type I and type II, respectively. Most type I MOB are members of the family Methylococcaceae, but the family Methylothermaceae, was recently proposed1. Type II MOB species belong to the families Methylocystaceae and Beijerinckiaceae. The third group, MOB in the phylum Verrucomicrobia, is extremely acidophilic, and the family Methylacidiphilaceae was proposed to encompass this group2. The fourth group has no representative isolated in pure culture. Members of this group are recognized as ‘Candidatus Methylomirabilis oxyfera' and its close relatives, belonging to the candidate phylum NC103,4. ‘M. oxyfera' has the notable ability to produce oxygen from nitrite, which enables nitrite-dependent methane oxidation without an external supply of oxygen4. Accordingly, the dominance of ‘M. oxyfera' and its relatives has been observed consistently in methane-oxidizing enrichment cultures established under anoxic nitrite-reducing conditions5,6,7,8,9. Based on these studies, ‘M. oxyfera'-like bacteria detected in natural environments have been regarded as the MOB responsible for nitrite-dependent methane oxidation9,10,11,12.
Methane oxidation by all known MOB is mediated by monooxygenases, in contrast to the reverse methanogenesis pathway used by methane-oxidizing archaea. Especially, the particulate methane monooxygenase is found in most of the known MOB. The pmoA gene encoding the α-subunit of this enzyme has been widely used as a marker to detect and identify MOB, along with the 16S rRNA gene. By using these and other markers such as phospholipid fatty acids, the community structures of MOB have been intensively investigated in various types of ecosystems, mainly targeting type I and type II MOB.
Considering their unique function, MOB may play important roles in the water columns of freshwater lake ecosystems. In the sediments of freshwater lakes, methanogenesis is a major terminal step in organic matter degradation. Methane release from sediments is consumed and assimilated by planktonic MOB, and therefore, they reduce methane emission from lakes and retrieve carbon released from the degradation of organic matter. The importance of MOB in the diets of other planktonic organisms has been discussed and evaluated in previous studies13,14,15,16,17. The community structures of planktonic MOB have been investigated mainly in temperate18,19,20,21 and subarctic17,22,23,24 areas. These studies consistently indicated the dominance of type I MOB over type II MOB. Some studies detected type I MOB as the major components of entire bacterial communities in temperate water columns21,25,26. On the other hand, dominance of type II MOB in water column was reported in a tropical dam reservoir27,28. The primary factor to determine community structure has not been identified, although the involvement of temperature is likely. One possible approach to resolving this issue is to analyze the community structure of planktonic MOB in other climate zones where dominant MOB have not been identified. Especially, studies in subtropical zone may be valuable to fill a gap between temperate and tropical zones.
In this study, the community structure of MOB inhabiting the water column of a subtropical reservoir was investigated to complement the current knowledge of planktonic MOB in freshwater environments. The study site, the Feitsui Reservoir (FTR), is located in northern Taiwan29,30. The reservoir is protected from human activities to maintain water quality.
Results
Physicochemical factors
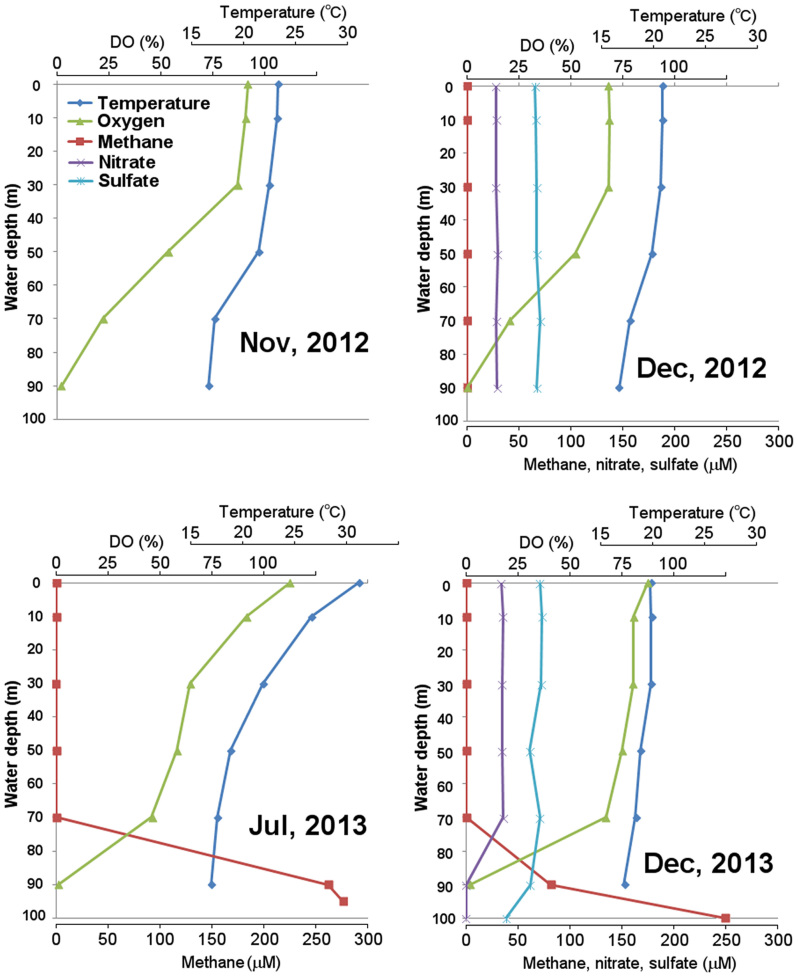
Vertical profiles of physicochemical factors measured in 4 sampling days are shown in Fig. 1. Water temperature ranged from 16.6 to 31.1°C. Depth-related changes in the availability of methane and oxygen were consistently observed. Oxygen was almost depleted at 90 m (2% or less) in all sampling days. A high concentration of methane was observed under anoxic conditions, and only a trace amount of methane was detected in the oxic water column. In Nov 2012, methane concentration was not determined on the day when samples for DNA-based analysis were obtained (Nov 13th), but measured on Oct 23rd and Nov 27th. On those days, methane concentration at 90 m was 2.9 and 7.1 μM, respectively, and did not exceed 0.1 μM at the other depths.
Figure 1. Vertical profiles of temperature, dissolved oxygen, methane, nitrate, and sulfate in FTR.
Detection of MOB in bacterial 16S rRNA gene clone libraries
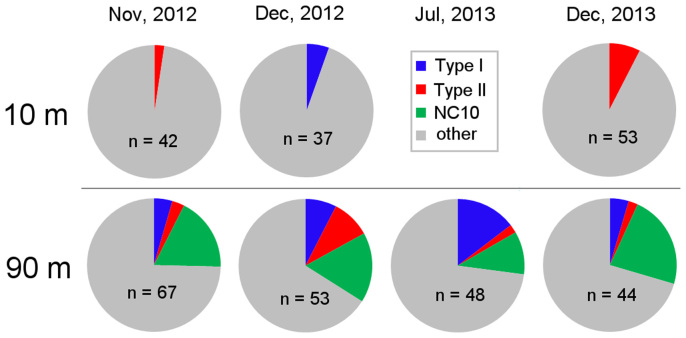
The phylogenetic affiliations of 344 clones from 7 libraries of surface- and deep-water samples were analyzed (phylum-level composition of the libraries is shown in Supplementary Table S1), and 68 clones were identified to be MOB as described below. As type I and type II MOB, 18 clones of the family Methylococcaceae and 14 clones of the family Methylocystaceae were obtained, respectively (Fig. 2). In addition, 36 clones from the deep-water samples were also identified as MOB, since they were closely related to ‘M. oxyfera', with sequence identities of 96% (show as “NC10” in Fig. 2).
Figure 2. MOB in clone libraries of bacterial 16S rRNA gene.
Total number of clones analyzed in each library is shown by “n”.
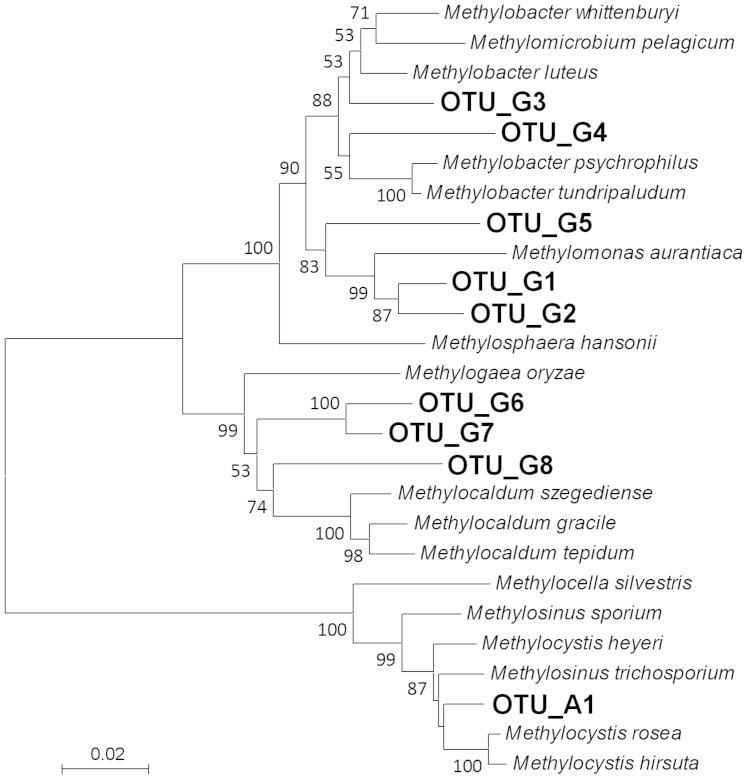
These MOB clones were further grouped into species-level OTUs (Supplementary Table S2). All clones of type II belonged to a single OTU (OTU_A1), but clones of type I were grouped into 8 OTUs (Fig. 3). The most frequently detected type I OTU was OTU_G1, which included 10 clones (Supplementary Table S2). The ‘Methylomirabilis'-like clones were very closely related to each other, and grouped into a single OTU, OTU_N1. In fact, these clones could not be differentiated even at a cutoff value of 0.01. The OTU_N1 was the most frequently detected species-level phylotype among MOB in all deep-water libraries (Supplementary Table S2). An environmental clone from the sediment of a freshwater lake, clone B84 from Lake Biwa31, had very high sequence identities with OTU_N1 clones (Fig. 4). The clone B84 was classified in the same OTU with these clones, in case that it was included in the grouping with the same methods.
Figure 3. Phylogenetic relationships of the OTUs of 16S rRNA gene clones belonging to the families Methylocystaceae and Methylococcaceae.
Numbers on nodes are percentage values of 1000 bootstrap resampling (values larger than 50 are shown).
Figure 4. Phylogenetic position of a clone belonging to the OTU_N1.
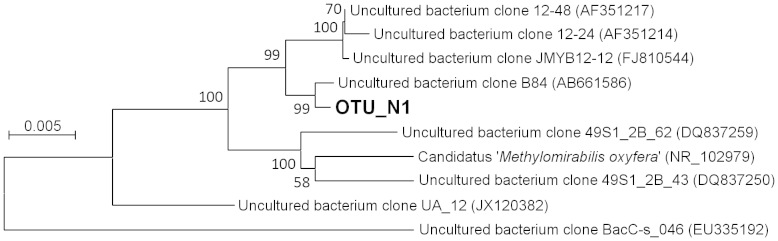
Numbers on nodes are percentage values of 1000 bootstrap resampling (values larger than 50 are shown).
Cloning analysis of pmoA genes
From the 2 libraries constructed with the primer pair A189/mb661r, 101 clones were sequenced. They were grouped into 23 OTUs, 6 of which were detected in both libraries (Supplementary Fig. S1). Phylogenetic analysis revealed that only OTU_PP22 belonged to the type II lineage and that the other OTUs were relatives of type I MOB (Supplementary Fig. S2).
From the libraries constructed with the primer pair A189/682_NC10, 11 clones were sequenced (4 from Nov 2012 at 90 m, 7 from Dec 2012 at 90 m). All clones sequenced were closely related to each other and to ‘M. oxyfera'. When the protein sequences of these clones were compared with those of ‘M. oxyfera', only 5–7 amino acid differences were observed in 166 amino acid positions. Further, these clones were very closely related to 21 clones from the sediment of Lake Biwa, obtained with the same pair31. In pairwise comparisons of these 32 clones (21 from FTR and 12 from L. Biwa), only 4 or fewer amino acid sequence differences were found. Phylogenetic position of a representative clone is shown in Fig. 5.
Figure 5. Phylogenetic position of PmoA sequence deduced from a pmoA gene clone obtained in this study (12Nov_NP27), with the primer pair A189/682_NC10.
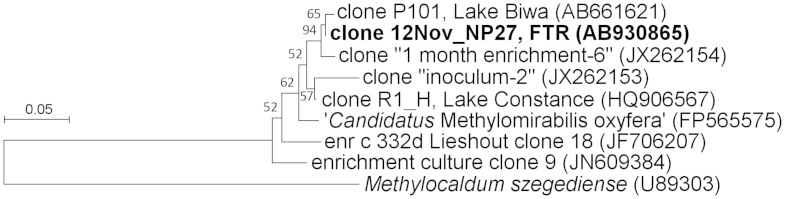
Numbers on nodes are percentage values of 1000 bootstrap resampling (values larger than 50 are shown).
Catalyzed reporter deposition–fluorescence in situ hybridization (CARD–FISH)
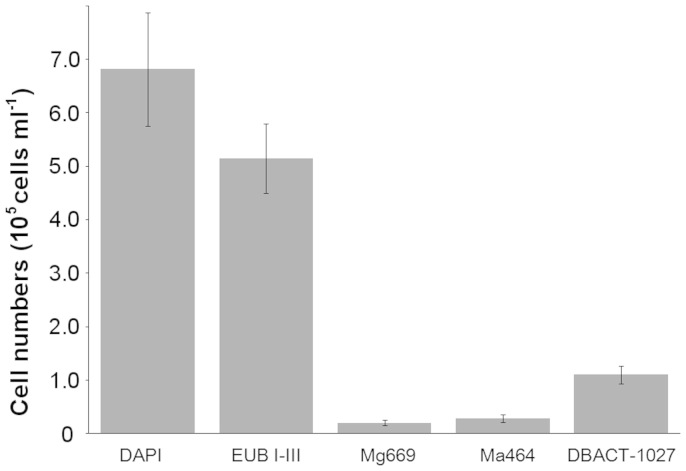
The dominance of ‘M. oxyfera'-like organisms in deep water suggested by DNA-based analyses was confirmed by CARD–FISH analysis. The water sample obtained at 90 m in Dec 2013 contained cells stained with the specific probe DBACT-1027 at a concentration of 1.1 × 105 cells ml−1 (Supplementary Fig. S3). The cells positive for DBACT-1027 comprised 16% of cells stained with DAPI and 21% of cells stained with EUB I–III, and were significantly frequently observed than cells hybridized with the probes Mg669 and Ma464 (p < 0.01, for both), targeting type I and type II MOB respectively (Fig. 6).
Figure 6. Abundance of cells hybridized with probes targeting bacteria, MOB of typeI, typeII, and NC10.
Error bars represent ± SD.
Discussion
In this study, the most predominant planktonic methane oxidizer was ‘M. oxyfera'-like bacterium belonging to NC10 in deep-water samples (Fig. 2, Fig. 6). Considering that ‘M. oxyfera'-like pmoA gene sequences were also detected from the same samples, it is reasonable to consider OTU_N1 as a methane oxidizer. At present, a mechanism for the dominance of OTU_N1 in the FTR is almost completely unknown. To our knowledge, such a high relative abundance of the ‘M. oxyfera'-like bacterium has never been reported from other aquatic environments. Further research is needed to determine whether the dominance of this lineage is specific to the study site or if it is also observed in other sites in similar environments. Water columns have not previously been considered a major habitat of ‘M. oxyfera'-like bacteria, but subtropical freshwater lakes may become important for studying the diversity and distribution of these organisms as planktonic MOB.
The clone library analyses of 2 genes suggested that type I diversity in the FTR was greater than type II diversity. In these analyses, only 1 OTU of type II was detected in the libraries of the 2 genes. Considering their phylogenetic positions, OTU_PP22 may correspond to OTU_A1 belonging to the genus Methylocystis. For type I, the correspondence between the OTUs of the 16S rRNA and pmoA genes is not easy to identify. The most frequently detected type I OTU of the 16S rRNA gene was OTU_G1, suggested to be a Methylomonas species (Fig. 3). The most frequently detected OTU of the pmoA gene, OTU_PP19, was also a close relative of a Methylomonas species (Supplementary Fig. S1, Fig. S2). These results suggest that Methylomonas species are major type I MOB in the FTR. In previous studies using methods similar to those used in this study, the dominance of Methylobacter species among planktonic MOB was shown in the water columns of freshwater environments19,20,21,24.
In the CARD–FISH analysis of the water sample obtained at 90 m in Dec 2013, there was no significant difference between numbers of cells hybridized with Ma464 and Mg669 (Fig. 6). The probe Ma464, used to estimate type II MOB abundance, perfectly matches all type II clones obtained in this study. In contrast, the probe Mg669 had 1–2 mismatches against 6 type I clones belonging to 5 OTUs. In the water sample analyzed with CARD–FISH, OTU_G1 and OTU_G2 were detected as a type I MOB in the clone library (Table S2). Although these OTUs matches the probe perfectly, the number of detected clones is obviously too small to exclude the presence of other OTUs with mismatches. Generally, the relative abundance of type I and type II was largely different among the libraries (Fig. 2), but their clone numbers in the respective libraries are too small to discuss quantitatively. Further experiments including quantification by CARD–FISH with other probes will complement the results of the present study.
In freshwater lake sediments, methane is produced in the main terminal process of organic matter degradation. Therefore, planktonic MOB have been considered a key link between sediment and pelagic carbon flow in freshwater lakes. Their contributions to carbon dynamics have been assessed by isotopic signature, assuming that MOB cell material is composed of carbon originated from methane13,14,15,16,17. Considering some recent findings, however, this basal assumption may not always be applicable. Verrucomicrobial MOB are known to oxidize methane but do not assimilate it as a carbon source32,33. They fix inorganic carbon via the Calvin–Benson–Bassham cycle at the expense of energy derived from methane oxidation. In addition, recent research suggests that methane-dependent autotrophy is also a physiological feature of ‘M. oxyfera'34. In the ecosystems dominated by these recently recognized MOB, the re-entry of methane to the organic carbon flow may be difficult to estimate. On the other hand, another carbon flow path via type I MOB was also suggested recently35. In that report, a methane-oxidizing bacterium was shown to excrete organic acids and hydrogen under oxygen-limited conditions, and genes responsible for this process are shared by various type I MOB35. In the present study, type I MOB were detected in samples characterized with low oxygen concentrations (Fig. 1, Fig. 2) and they were also detected in oxygen-depleted zones of other freshwater water columns21,24,26. These MOB might play a particular role in carbon flow without the assimilation of methane by converting it to carbon dioxide and organic acids available for other functional microbe groups.
Methods
Sample collection and chemical analyses
Samples were obtained from the Feitsui Reservoir (FTR) located in northern Taiwan (121° 34′ E, 24° 54′ N)29. Water sampling was performed at the same site as that of a previous study30 using a 5-L Go-Flo bottle. In the present study, samples collected on the following 4 sampling days were analyzed: 13 Nov 2012, 11 Dec 2012, 30 Jul 2013, and 24 Dec 2013. Water samples were collected from 6 depths (0, 10, 30, 50, 70, and 90 m from the water surface) and were subjected to chemical analyses. Additional samples were obtained from a depth of 100 m in Dec 2013 and from a depth of 95 m in Jul 2013.
Dissolved methane concentration was determined with headspace equilibration methods36,37. Briefly, portions of the water samples were poured into glass vials. The vials were immediately sealed with butyl rubber stoppers leaving no headspace. The vials were stored in a cooling box (around 4°C) until addition of HgCl2 solution conducted within 3–4 h after the sampling. He gas (>99.999% purity) was injected into the bottles containing the fixed water to create headspace, and the vials were shaken vigorously to equilibrate. Methane in the headspace was quantified using a gas chromatograph (GC-2014; Shimadzu) equipped with a flame ionization detector to calculate dissolved methane concentration in the water. The concentrations of nitrate and sulfate were measured with an ion chromatograph (DX-120; Dionex), and dissolved oxygen concentration was determined by Winkler titration. Exceptionally, in the sampling day of Dec 2012, dissolved oxygen was measured with an oxygen sensor calibrated by the titration.
Polymerase chain reaction (PCR) of 16S rRNA genes
Planktonic microorganisms in the water samples were collected on 0.22-μm filters (Sterivex filter cartridges; Millipore). The filters were immersed in RNA-later (Ambion) and kept at 4°C until DNA extraction. DNA samples were prepared using the bead-beating method. Prior to DNA extraction, the filters were washed with Tris–EDTA (TE) buffer to remove RNA-later. The washed filters were cut into small pieces and then placed into screw-cap tubes containing 600 μl of TE buffer, 30 μl of 20% sodium dodecyl sulfate, and 0.5 g of glass beads (equal-weight mixture of 0.1- and 0.05-mm-diameter beads). After adding 600 μl of phenol–chloroform–isoamyl alcohol (25:24:1, v/v/v), the samples were shaken twice with a bead beater (FastPrep-24; MP Biomedicals) at 4.0 m s−1 for 30 s, followed by centrifugal separation. DNA in the aqueous supernatant was precipitated by adding NaCl and isopropanol, and excess salt was removed by rinsing the pellet with 70% ethanol.
From 7 DNA samples at 2 water depths (10 and 90 m on 4 sampling days, except 10 m in Jul 2013), fragments of bacterial 16S rRNA genes were amplified using the universal primer pair 27F and 1492R. PCR amplification was initiated with 2 min of denaturation at 94°C, followed by 28 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C. Final extension was carried out for 10 min at 72°C. Clone libraries were constructed from the resulting PCR products from the 7 samples using the pCR2.1-TOPO vector and competent TOP10 cells (Invitrogen). For clones randomly selected from the libraries, inserts of the 16S rRNA gene were fully sequenced. After exclusion of chimeras and other anomalous sequences using Mallard software38, all sequences were classified using the Ribosomal Database Project classifier to sort out MOB clones. MOB clones were grouped into operational taxonomic units (OTUs) using mothur software39 at a cutoff value of 0.03. The clone sequences representing OTUs were aligned with related sequences using the program ClustalX40. Phylogenetic trees were constructed using the neighbor-joining method in the program MEGA541.
Cloning analysis of pmoA genes
Fragments of the pmoA genes were amplified with 2 primer pairs from 2 DNA samples of water obtained from a depth of 90 m in 2012. One of the primer pairs, A18942 and mb661r43, was used to detect proteobacterial MOB. PCR was initiated with 5 min of denaturation at 94°C, followed by 34 cycles of 30 s at 94°C, 30 s at 56°C, and 45 s at 72°C. Final extension at 72°C was carried out for 10 min. The other primer pair, A189 and 682_NC1031, was used to detect pmoA genes of ‘Methylomirabilis'-like organisms. PCR conditions for this primer pair were almost the same as those for A189/mb661r, but initial denaturation was shortened to 2 min, and annealing temperature was 55°C. Four clone libraries (2 samples, 2 primer pairs) were constructed as described above. The resulting sequences were translated into amino acid sequences, and distance matrices were calculated with Poisson model using MEGA5. The distance matrices generated were imported into mothur to group the clones into OTUs at a cutoff value of 0.03. Phylogenetic trees were constructed with amino acid sequences of the clones representing respective OTU, using the neighbor-joining method.
CARD–FISH
CARD–FISH analysis was performed on the water sample obtained from a depth of 90 m in Dec 2013. The lake water sample of lake water was mixed with 4% paraformaldehyde solution in phosphate-buffered saline to obtain a final concentration of 0.9%. After fixation at 4°C for ca. 4 hour, fixed cells were trapped onto track-etched polycarbonate membrane filters (Cyclopore, 0.22-μm pore size, 25-mm diameter; Whatman) and stored at −20°C until further processing. CARD–FISH analysis methods were similar to those described previously44,45. Prior to successive treatments in liquids, filters were treated with 0.1% (w/v) low-melting-point agarose (SeaKem, FMC BioProducts) to hold the cells. The filters were cut into small pieces, and one of them was used for direct count of total cell number by staining with 4′,6-diamidino-2-phenylindole (DAPI). Other pieces for CARD–FISH analysis were incubated in lysozyme solution (10 μg/mL lysozym in 0.1 M Tris–HCl, 0.05 M EDTA [pH 8.0]) for 25 min at 37°C to permeabilize the cells. To eliminate endogenous peroxidase activity, the filter pieces were treated with 0.01 M HCl with 0.3% H2O2 for 10 min at room temperature. The samples were hybridized using probes labeled with horseradish peroxidase. The specific probe DBACT-10273 was used to detect ‘Methylomirabilis'-like organisms. It perfectly matches the sequences of NC10 from the FTR and does not match the other sequences obtained in this study. Probe mix EUB I–III46,47 was used to estimate bacterial abundance. Two probes, Mg669 and Ma46448, were also used to detect type I and II MOB, respectively. Hybridization was performed for 2 h at 46°C. Formamide concentrations in the hybridization buffers were 20% (for Mg669 and Ma464), 35% (for EUB I–III), or 40% (for DBACT-1027). After washing at 48°C, signal amplification was performed with Alexa Fluor 488 tyramide at 46°C for 15 min. CARD-FISH counts were calculated as means from 15–31 randomly chosen microscopic fields (on average, approximately 1200 stained cells were counted for each probe). Results from the group-specific probes were compared by Tukey-Kramer test.
Nucleotide sequence accession numbers
Nucleotide sequences obtained in this study have been assigned the DDBJ/EMBL/GenBank accession numbers AB930495–AB930950.
Author Contributions
H.K., F.K.S., N.O. and M.F. conceived the research. H.K., R.T., K.K., Y.K. and M.I. performed the experiments. H.K. wrote the manuscript. F.K.S. and M.F. edited the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Kyoko Kubo for her help in CARD-FISH. This study was supported by JSPS KAKENHI Grant Numbers 24405007 and 24770010. This study was carried out under the Joint Research Program of the Institute of Low Temperature Science, Hokkaido University.
References
- Hirayama H. et al. Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov. Int J Syst Evol Microbiol 64, 989–999 (2014). [DOI] [PubMed] [Google Scholar]
- Op den Camp H. J. et al. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1, 293–306 (2009). [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., van Alen T., van de Pas-Schoonen K. T., Jetten M. S. & Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75, 3656–3662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig K. F. et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464, 543–548 (2010). [DOI] [PubMed] [Google Scholar]
- Ettwig et al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol 10, 3164–3173 (2008). [DOI] [PubMed] [Google Scholar]
- Hu S. H. et al. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Env Microbiol Rep 1, 377–384 (2009). [DOI] [PubMed] [Google Scholar]
- Luesken F. A. et al. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera': kinetic and transcriptional analysis. Environ Microbiol 14, 1024–1034 (2012). [DOI] [PubMed] [Google Scholar]
- Kampman C. et al. Enrichment of denitrifying methanotrophic bacteria for application after direct low-temperature anaerobic sewage treatment. J Hazard Mater 227–228, 164–171 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu B. et al. Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite-dependent methane-oxidizing bacteria. Appl Environ Microbiol 78, 8657–8665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutzmann J. S. & Schink B. Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater lake. Appl Environ Microbiol 77, 4429–4436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. L. et al. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc Natl Acad Sci USA 111, 4495–4500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. D. et al. Distribution and diversity of nitrite-dependent anaerobic methane-oxidising bacteria in the sediments of the Qiantang River. Microb Ecol 67, 341–349 (2014). [DOI] [PubMed] [Google Scholar]
- Bastviken D., Ejlertsson J., Sundh I. & Tranvik L. Methane as a source of carbon and energy for lake pelagic food webs. Ecology 84, 969–981 (2003). [Google Scholar]
- Kankaala P., Huotari J., Peltomaa E., Salorant a. T. & Ojala A. Methanotrophic activity in relation to methane efflux and total heterotrophic bacterial production in a stratified, humic, boreal lake. Limnol Oceanogr 51, 1195–1204 (2006). [Google Scholar]
- Kankaala P. et al. Experimental δ13C evidence for a contribution of methane to pelagic food webs in lakes. Limnol Oceanogr 51, 2821–2827 (2006). [Google Scholar]
- Taipale S., Kankaala P. & Jones R. I. Contributions of different organic carbon sources to Daphnia in the pelagic foodweb of a small polyhumic lake, results from mesocosm DI13C-additions. Ecosystems 10, 757–772 (2007). [Google Scholar]
- Jones S. E. & Lennon J. T. Evidence for limited microbial transfer of methane in a planktonic food web. Aquat Microb Ecol 58, 45–53 (2009). [Google Scholar]
- Ross J. L., Boon P. I., Ford P. & Hart B. T. Detection and quantification with 16S rRNA probes of planktonic methylotrophic bacteria in a floodplain lake. Microb Ecol 34, 97–108 (1997). [DOI] [PubMed] [Google Scholar]
- Kojima H., Iwata T. & Fukui M. DNA-based analysis of planktonic methanotrophs in a stratified lake. Freshw Biol 54, 1501–1509 (2009). [Google Scholar]
- Tsutsumi M., Iwata T., Kojima H. & Fukui M. Spatiotemporal variations in an assemblage of closely related planktonic aerobic methanotrophs. Freshw Biol 56, 342–351 (2011). [Google Scholar]
- Biderre-Petit C. et al. Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol Ecol 77, 533–545 (2011). [DOI] [PubMed] [Google Scholar]
- Taipale S., Jones R. I. & Tiirola M. Vertical diversity of bacteria in an oxygen-stratified humic lake, evaluated using DNA and phospholipid analysis. Aquat Microbial Ecol 55, 1–16 (2009). [Google Scholar]
- Sundh I., Bastviken D. & Tranvik L. J. Abundance, activity, and community structure of pelagic methane-oxidizing bacteria in temperate lakes. Appl Environ Microbiol 71, 6746–6752 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blees J. et al. Micro-aerobic bacterial methane oxidation in the chemocline and anoxic water column of deep south-Alpine Lake Lugano (Switzerland). Limnol Oceanogr 59, 311–324 (2014). [Google Scholar]
- Biderre-Petit C. et al. Identification of sulfur-cycle prokaryotes in a low-sulfate lake (Lake Pavin) using aprA and 16S rRNA gene markers. Microb Ecol 61, 313–327 (2011). [DOI] [PubMed] [Google Scholar]
- Kojima H., Watanabe T., Iwata T. & Fukui M. Identification of major planktonic sulfur oxidizers in stratified freshwater lake. PLoS One 9, e93877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumestre J.-F., Vaquer A., Gosse P., Richard S. & Labroue L. Bacterial ecology of a young equatorial hydroelectric reservoir (Petit Saut, French Guiana). Hydrobiologia, 400, 75–83 (1999). [Google Scholar]
- Dumestre J.-F., Casamayor E. O., Massana R. & Pedrós-Alió C. Changes in bacterial and archaeal assemblages in an equatorial river induced by the water eutrophication of Petit Saut dam reservoir (French Guiana). Aquat Microb Ecol 26, 209–221 (2001). [Google Scholar]
- Tseng C. H. et al. Microbial and viral metagenomes of a subtropical freshwater reservoir subject to climatic disturbances. ISME J 7, 2374–2386 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y. F. et al. Typhoon effects on DOC dynamics in a phosphate-limited reservoir. Aquat Microb Ecol 67, 247–260 (2010). [Google Scholar]
- Kojima H. et al. Distribution of putative denitrifying methane oxidizing bacteria in sediment of a freshwater lake, Lake Biwa. Syst Appl Microbiol 35, 233–238 (2012). [DOI] [PubMed] [Google Scholar]
- Khadem A. F. et al. Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J Bacteriol 193, 4438–4446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp C., Stott M. & Dunfield P. Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front Microbiol 3, 1–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasigraf O. et al. Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph Methylomirabilis oxyfera. Appl Environ Microbiol 80, 2451–2460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G. et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun 4, 2785 (2013). [DOI] [PubMed] [Google Scholar]
- McAullife C. GC determination of solutes by multiple phase equilibration. Chem Tech 1, 46–51 (1971). [Google Scholar]
- Itoh M. et al. Hydrologic effects on methane dynamics in riparian wetlands in a temperate forest catchment. J Geophys Res 112, G1, G01019 (2007). [Google Scholar]
- Ashelford K. E., Chuzhanova N. A., Fry J. C., Jones A. J. & Weightman A. J. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72, 5734–5741 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 24, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 26, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A., Costello A., Lidstrom M. & Murrell J. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett 132, 203–208 (1995). [DOI] [PubMed] [Google Scholar]
- Costello A. M. & Lidstrom M. E. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol 65, 5066–5074 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A., Pernthaler J. & Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68, 3094–3101 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teira E., Reinthaler T., Pernthaler A., Pernthaler J. & Herndl G. J. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl Environ Microbiol 70, 4411–4414 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environmen Microbiol 56, 1919–1925 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H., Brühl A., Amann R., Schleifer K. H. & Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22, 434–444 (1999). [DOI] [PubMed] [Google Scholar]
- Eller G., Stubner S. & Frenzel P. Group-specific 16S rRNA targeted probes for the detection of type I and type II methanotrophs by fluorescence in situ hybridisation. FEMS Microbiol Lett 198, 91–97 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information