Abstract
BACKGROUND
Malignant transformation is often accompanied by morphological and functional alterations in subcellular organelles. The Golgi apparatus is a subcellular structure primarily involved in modification and sorting of macromolecules for secretion and transport to other cellular destinations. Molecular alterations associated with the Golgi apparatus may take place during prostate carcinogenesis but such alterations have not been documented.
METHODS
To demonstrate that the Golgi apparatus undergoes alterations during prostate carcinogenesis, we examined the expression and localization of two candidate molecules, Golgi phosphoprotein 2 (GOLPH2) and myosin VI (MYO6), both overexpressed in prostate cancer as initially identified by expression microarray analysis.
RESULTS
Elevated GOLPH2 expression in prostate cancers was validated through real-time RT-PCR, Western blot, and tissue microarray analysis, and its Golgi localization in surgical prostate cancer tissues confirmed using two-color immunofluorescence. In addition, distinctive juxtanuclear MYO6 staining pattern consistent with Golgi localization was observed in surgical prostate cancer tissues. Two-color immunofluorescence revealed intensive Golgi-specific staining for both GOLPH2 and myosin VI in prostate cancer cells but not in the adjacent normal prostate epithelium.
CONCLUSIONS
We show that the Golgi apparatus in prostate cancer cells differs from the normal Golgi by elevated levels of two molecules, GOLPH2 and MYO6. These results for the first time demonstrated consistent cancer cell-specific alterations in the molecular composition of the Golgi apparatus. Such alterations can be explored for discovery of novel prostate cancer biomarkers through targeted organellar approaches.
Keywords: prostate cancer, GOLPH2, myosin VI, Golgi
INTRODUCTION
The Golgi apparatus, first described by Camillo Golgi as the “internal reticular apparatus” in 1898, is composed of stacks of cisternae where proteins and lipids synthesized in the endoplasmic reticulum are further processed, modified, and sorted for secretion and other cellular destinations such as the cell membrane and endosomes [1]. In addition to its primary function as a sorting and packaging station [1], the Golgi apparatus may also participate in vital functions that are related to human cancer. For example, fragmentation and dispersion of the Golgi membrane, itself regulated by mitotic signals, is required for entry into mitosis [2]. The Golgi apparatus also hosts a number of proteins critical for cell signaling and apoptosis [3]. Moreover, the trans side of the Golgi is a microtubule organizing center (MTOC) where noncentrosomal, parallel microtubule arrays are originated in polarized epithelial cells, implicating a role for the Golgi in cell migration and mitotic spindle formation [4,5]. In light of these Golgi-associated cellular functions, molecular alterations in the Golgi apparatus occurring during human carcinogenesis would be expected. Yet aside from commonly observed phenotypic changes in protein glycosylation [6], little is known about the alterations in the molecular composition of Golgi in human cancer.
In the normal prostate luminal epithelium, the secretory elements, including the Golgi, are organized along the polarization axis at the apical pole. Evidence from electron micrographic analysis of prostate carcinoma suggested morphological changes of the Golgi apparatus that included loss of polarization and dispersion of hypertrophic Golgi elements [7]. Corresponding molecular alterations in the Golgi apparatus during prostate carcinogenesis might be expected but have not been definitively documented. To demonstrate that such alterations indeed take place during prostate carcinogenesis, we focused on examining the subcellular localization of two candidate Golgi-associated proteins, Golgi phosphoprotein 2 (GOLPH2) and myosin VI (MYO6), both overexpressed in human prostate cancer as initially identified by expression microarray analysis [8].
The GOLPH2 (also named GOLM1 or GP73) gene was first cloned following differential screening of a cDNA library of liver tissues from a patient with giant-cell hepatitis [9]. GOLPH2 is a type II Golgi membrane protein with a short N-terminal sequence in the cytoplasm and its expression was induced by viral infection [9]. Although GOLPH2 has been characterized as a serum marker for a number of advanced liver diseases [10,11] including hepatocellular carcinoma, and urinary detection of GOLPH2 mRNA was recently explored for the diagnosis of human prostate cancer [12], GOLPH2 protein expression and localization have not been validated in clinical cancer specimens. MYO6 is consistently overexpressed in human prostate cancer and previously implicated in cancer invasion [8,13]. Apparently a multi-functional protein involved in a number of biological processes [14], MYO6 plays a role in the maintenance of Golgi morphology and in exocytosis as characterized using mouse fibroblasts [15]. However, no definitive Golgi staining pattern of myosin VI was previously reported in clinical human cancer tissues [8,13].
In this study, we show that in clinical prostate cancer tissues, overexpressed GOLPH2 and MYO6 are predominantly detected in the Golgi apparatus following the use of suitable antibodies, thus providing two examples of previously unappreciated molecular alterations of the Golgi apparatus in human prostate cancer. Given the importance of the Golgi apparatus in the secretory export pathway, such alterations can be further explored for the development of novel prostate cancer markers through targeted organellar approaches, and may help to decipher the mechanisms that relate Golgi structure and function in both normal and cancer cells.
MATERIALS ANDMETHODS
Human Prostate Tissue Samples
The formalin-fixed paraffin-embedded prostate samples for immunohistochemical analysis were obtained from radical prostatectomy specimens from the Department of Pathology at Johns Hopkins University. Fresh frozen prostate tissue samples for real-time RT-PCR analysis (30 normal/tumor pairs) and Western blot analysis (5 normal/tumor pairs) were collected at the time of prostate surgery from 1993 to 2000 at the Johns Hopkins Hospital. Fresh human prostate specimens were trimmed and sectioned to enrich for the target epithelium (normal or cancer) as previously described [8]. The use of surgical specimens for molecular analysis was approved by the Institutional Review Board at Johns Hopkins University.
Real-Time RT-PCR Analysis
Total RNA was isolated as described previously [8]. The quality and concentration of the isolated RNA was determined using the Agilent 2100 Bioanalyzer Total RNA Nano Series II assay (Agilent, Santa Clara, CA). First Strand cDNA Synthesis was performed using 500 ng total RNA, 0.5 μg oligo (dT), and 200 units of SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) in a volume of 20μl. Real-time PCR amplifications were performed, in triplicate for each reaction, using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) with 0.125% of the total cDNA product and gene specific primers. Primers pairs with validated amplification specificity for GOLPH2 are: 5′-ATCCGAGTG-CTGCAAGACCAGTTA-3′ (forward) and 5′-TCTGA-TTGATGCACTGGCTCAGGT-3′ (reverse). Primers pairs with validated amplification specificity for GAPDH are: 5′-TCGACAGTCAGCCGCATCTTCTTT-3′ (forward) and 5′-ACCAAATCCGTTGACTCCGACC-TT-3′ (reverse). Following validation of equal efficiencies for both target (GOLPH2) and control (GAPDH) amplifications, the average threshold cycle (Ct) numbers from the triplicates for each sample were used for comparative threshold analysis [16], in which relative GOLPH2 transcript abundance in each sample was derived by normalizing against GAPDH in the same sample.
Western Blot
Frozen human prostate tissues and cell line were subjected to standard Western blot analysis as described [17]. For GOLPH2, 1:100 dilution of goat polyclonal anti-GP73 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used for Western blot. β-actin was detected using a monoclonal antibody (AC-15) (Sigma, St. Louis, MO) at 1:1,000 dilution for loading control. Horseradish peroxidase-conjugated secondary antibody to goat IgG (Sigma) and rabbit IgG (Bio-Rad) at 1:1,000 dilution were used, followed by signal detection with SuperSignal West Pico Chemiluminescent reagents (Pierce, Rockford, IL).
Immunohistochemical and Immunofluorescent Staining
Standard formalin-fixed paraffin-embedded (FFPE) prostate tissue blocks were processed for immunohistochemical and 2-color immunofluorescent staining of GOLPH2, MYO6, and TGN46. The hydrated FFPE sections were placed into Tris–EDTA Buffer (10 mM Tris Base, 1 mM EDTA Solution, pH 9.0) and steamed for 40 min for antigen retrieval. Endogenous peroxidase activity was quenched by incubating the slides for ten minutes with 3% H2O2. Incubations with primary antibodies were performed at the following dilutions: rabbit polyclonal anti-myosin VI antibody (a gift from Denise Montell, Johns Hopkins University, Baltimore, MD) at 1:500, mouse monoclonal anti-GOLPH2 (Abnova Corp., Taipei, Taiwan) at 1:50, rabbit polyclonal anti-TGN46 (Sigma) at 1:500. The slides were incubated with peroxidase-based EnVision™+ (DAKO Corp., Carpinteria, CA) reagents for 30 min, and staining was developed with 3,3′-diaminobenzidine (DAB) and counterstained with Mayer’s Hematoxylin (DAKO). For two-color immunofluorescent staining, FFPE sections were processed similarly but incubated with two primary antibodies from different host species simultaneously, followed by incubation with Fluorescein (FITC)-conjugated AffiniPure Goat Anti-Rabbit IgG and Rhodamine Red-X-AffiniPure Goat Anti-Mouse IgG (Jackson immunoresearch, West Grove, PA) secondary antibodies. Prolong Gold anti-fade reagent with DAPI (Invitrogen) was used for DAPI nuclear staining and mounting.
Tissue Microarray
Tissue microarrays (TMAs) were constructed as previously described [17]. To assess the protein expression differences of GOLPH2 between normal and cancer tissues, a TMA containing four pairs of matched normal and tumor cores (0.6 mm cores) from each of the 40 radical prostatectomy cases was processed and stained using the same optimized antibody dilutions (1:50) as described above for IHC staining in standard tissue sections. Scanning, processing of stained sections and analysis of scanned images were modified based on previously described methods [8]. Each TMA spot was reassigned a diagnosis and staining data analyzed using the Software FRIDA (FRamework for Image Dataset Analysis, available at http://sourceforge.net/projects/fridajhu/). The FRIDA software allows the use of lasso masks to select one tissue type (either normal or cancerous epithelium) from each TMA spot for subsequent automated image analysis. The analysis yielded two values for each TMA spot, pixel ratio and intensity score. Pixel ratio was the number of brown pixels (DAB staining) divided by the total pixels of the normal or cancer epithelium as selected by the lasso masks in each TMA spot. Intensity score was the total intensity of brown color divided by the total pixels of the normal or cancer epithelium as selected by the lasso masks in each TMA spot. Comparison between normal and cancer tissues for both scored values were analyzed statistically using Wilcoxon–Mann–Whitney test in SPSS (SPSS Inc., Chicago, IL).
RESULTS
GOLPH2 Overexpression in Human Prostate Cancer
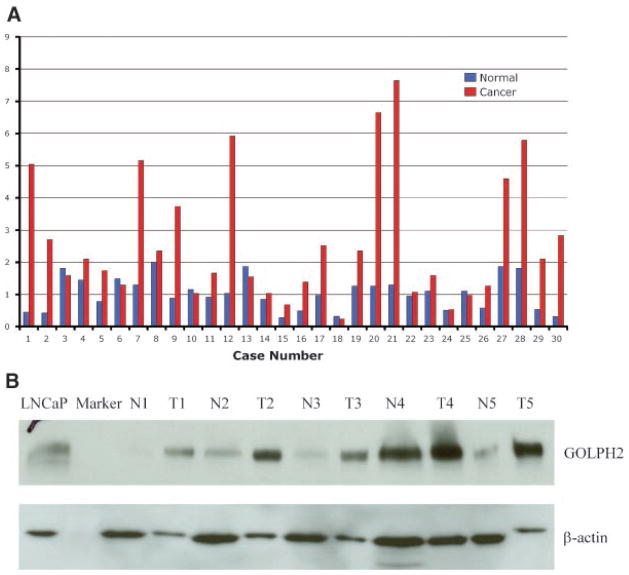
In a previous study that focused on the novel role of myosin VI (MYO6) in human prostate cancer, we also revealed elevated mRNA expression of GOLPH2 in human prostate cancers following unsupervised clustering analysis of gene expression data derived from surgical prostate cancer specimens [8]. GOLPH2 was shown in a cluster of 21 overexpressed genes that included well-characterized prostate cancer markers such as PCA3, AMACR, SIM2, HPN, TARP, and MYO6 [8]. Although a recent study included GOLPH2 in a multiplexed mRNA assay for the detection of prostate cancer cells in urine sediments, GOLPH2 as a putative prostate cancer marker has not been characterized beyond mRNA levels. We first performed real-time RT-PCR in 30 radical prostatectomy cases, each represented by paired prostate tumor and adjacent normal prostate epithelial samples, to validate the extent of GOLPH2 mRNA overexpression in human prostate cancer. As shown in Figure 1A, GOLPH2 expression was higher by at least 1.8-fold in the majority of cancer samples when compared with their normal counterparts (63.3%, 19 out of 30), and relatively unchanged (less than 1.8-fold change) in the remaining normal/tumor pairs (36.7%, 11 out of 30). In average, GOLPH2 demonstrated 3.04-fold higher expression in cancer samples than in the paired normal samples when all pairs were included (Z = −4.174; P <0.001, Wilcoxon Signed Ranks test). The real-time RT-PCR results likely represented an underestimate of the extent of GOLPH2 expression in prostate cancer because for the lack of a perfect control gene, GAPDH, which may be higher in cancer tissues [18], was used to normalize the data. No statistically significant differences were observed between the extent of cancer-specific overexpression with any of the clinical and pathological variables in this sample set (e.g., Gleason score, pathological stage, data not shown). GOLPH2 protein expression was subsequently examined by Western blot analysis in five randomly selected normal/tumor pairs of human prostate specimens, using a goat polyclonal anti-GOLPH2 antibody (see Materials and Methods Section). As shown in Figure 1B, though we selectively overloaded protein lysates from the normal tissues as indicated by the elevated -actin levels in the normal samples, GOLPH2 protein levels were still higher than those in the paired normal prostate tissues (Fig. 1B), providing definitive evidence for increased GOLPH2 protein levels in human prostate cancer.
Fig. 1.
Validation of GOLPH2 mRNA and protein overexpression in human prostate cancer. A: Real-time RT-PCR analysis of GOLPH2 mRNA in prostate cancer tissues (n =30) and paired normal prostate tissues (n =30) from surgical prostate cancer specimens (X-axis). Normalized (against GAPDH) Ct values were converted to fold expression changes (Y-axis) relative to the median value of the normal samples. B: Western blot analysis of GOLPH2 protein in five paired normal/cancer prostate tissue samples collected fresh from five surgical cases. β-Actin is a loading control. LNCaP cell lysate was used as a positive control. N1–N5: normal prostate epithelial tissues, T1–T5: prostate cancer tissues.
Immunohistochemical Analysis of GOLPH2
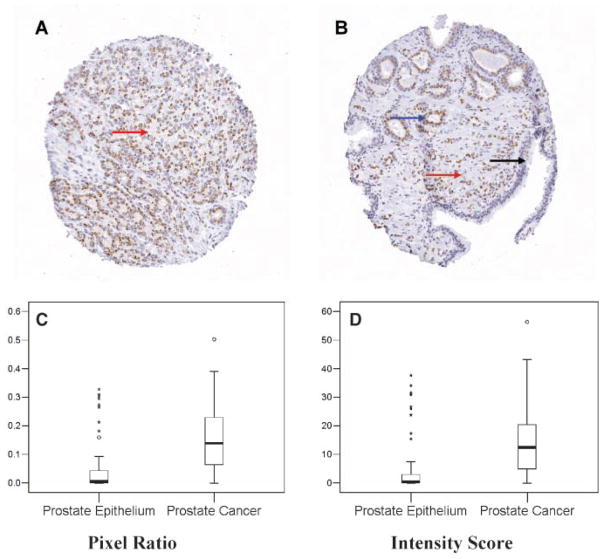
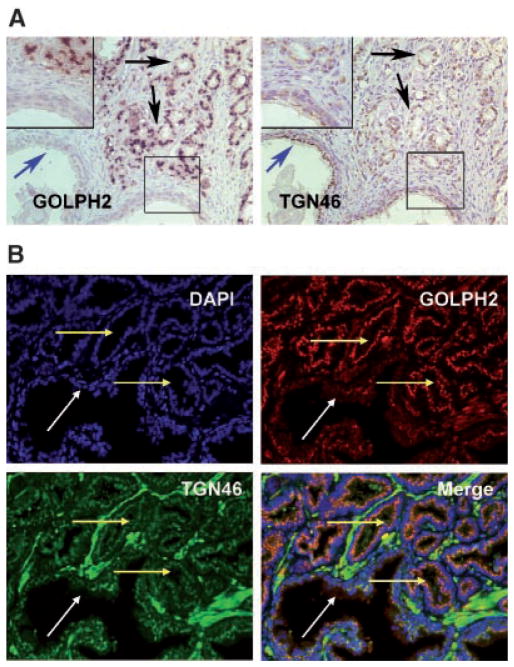
GOLPH2 has been characterized as a secreted protein and a serum marker for a number of advanced liver diseases [10,11]. Immunohistochemical analysis (IHC) of GOLPH2 in clinical cancer specimens has not been performed and its subcellular localization pattern has not been described previously in prostate cancer specimens. Following optimization of the IHC protocol using different anti-GOLPH2 antibodies, we determined that a mouse monoclonal anti-GOLPH2 antibody yielded optimal results (data not shown) under the conditions specified in Materials and Methods Section. A tissue microarray (TMA) containing 320 paired normal and prostate cancer TMA spots from 40 patients allowed us to examine GOLPH2 protein expression level in multiple samples without inter-assay variations introduced in the IHC staining process. TMA results confirmed the strongly positive GOLPH2 staining in the majority of cancerous epithelial cells but generally negative or weak staining in normal epithelium and negative staining in stromal components (Fig. 2A,B). Both the pixel ratios (Fig. 2C) and intensity scores (Fig. 2D) showed significant differences between normal prostate epithelium and prostate cancer (Z = −8.931; P <0.001 and Z = −8.765; P <0.001, respectively, Wilcoxon–Mann–Whitney test). To determine whether the IHC values can be used to discern prostate cancer lesions from the normal prostate tissues, we calculated the sensitivity and specificity for detection of cancer lesions among the 225 scored TMA spots each with a single diagnosis of either “normal” or “cancer”. At a threshold value of 0.05 for the pixel ratios (Fig. 2C), the sensitivity and specificity of detecting cancer lesions were 81.9% and 75.6%, respectively. At a threshold value of 4.0 for the intensity scores (Fig. 2D), the sensitivity and specificity of detecting cancer lesions were 79.8% and 75.6%, respectively. The predominant juxtanuclear staining pattern (Fig. 2A,B) mainly located at the luminal side (when acini lumen is present) of prostate cancer epithelium is consistent with Golgi localization. IHC analysis for GOLPH2 was also performed on standard formalin-fixed, paraffin embedded sections from 15 cases to confirm its cancer cell specific overexpression and juxtanuclear localization (data not shown). To further illustrate the elevated levels of GOLPH2 protein specifically in the cancer Golgi, the Golgi marker TGN46 (Trans-Golgi network protein, 46 kDa) [19] was stained on the adjacent serial section from the same tissue block. While identical TGN46 Golgi staining patterns was found in both normal (blue arrow) and cancer cells (black arrow) (Fig. 3A, TGN46), this TGN46 staining pattern is in sharp contrast with that of GOLPH2 (Fig. 3A, GOLPH2), which showed a Golgi staining pattern that was prominent and distinctive in the cancer epithelium (black arrow) but not in the normal epithelium (blue arrow). This distinction between the staining patterns for the two Golgi proteins was further validated by double immunofluorescence staining for GOLPH2 and TGN46 in the same section. As shown in Figure 3B, while GOLPH2 (red staining) and TGN46 (green staining) were localized to the juxtanuclear areas in both prostate cancer (yellow arrow) and adjacent normal (white arrow) cells, higher GOLPH2 expression was only seen in the cancerous acini as marked by predominantly red fluorescent signals at the apical side of the nuclei (blue staining). These results confirmed the cancer-cell specific GOLPH2 protein overexpression and its localization in the Golgi apparatus. In addition, elevated levels of GOLPH2 in the Golgi was not accompanied by that of TGN46, suggesting that elevated GOLPH2 levels did not result from enrichment or expansion of the Golgi membrane.
Fig. 2.
Tissue microarray (TMA) analysis of GOLPH2 expression in human prostate cancer. A: TMA spot containing predominantly Gleason 4 (red arrow) prostate cancer tissue. B: TMA spot containing mixed Gleason 3 (blue arrow) and Gleason 5 (red arrow) prostate cancer tissues and normal prostate epithelium (black arrow). Note that only cancer cells are positive for GOLPH2 and the staining pattern is consistent with Golgi localization. C,D: Box plots showing automated TMA image analysis results for GOLPH2 staining in normal and prostate cancer tissues. Altogether 132 normal and 93 cancer lesions were evaluated excluding array spots with poor quality and ambiguous diagnoses. Box plots showed differences in positively stained areas (by pixel ratios, C) and intensity scores (D) between the normal and cancerous prostate epithelium. Note that pixel ratios (see Materials and Methods Section) cannot reach high percentages due to Golgi-confined staining for GOLPH2. Each box is lined at lower quartile, median, and upper quartile values for each group. The whiskers show the 1.5× inter-quartile range of the data. The circles and asterisks indicate outliers and extreme outliers beyond the whiskers.
Fig. 3.
Immunohistochemical analysis of GOLPH2 and TGN46 in human prostate cancer tissues. A: Two adjacent serial sections of surgical prostate cancer tissues stained with GOLPH2 and TGN46, respectively, as indicated. Black arrows indicate cancer tissues, blue arrows indicate a normal prostate gland. Insets show high power view of the boxed areas. Note strong GOLPH2 staining in cancer cells and weak or negative staining in the normal prostate epithelium, and no difference for TGN46 staining in normal and cancer cells. B: Immunofluorescence double staining for TGN46 and GOLPH2 in the same formalin fixed paraffin embedded surgical prostate cancer section. Prostate cancer tissues were stained for nuclei (blue, by DAPI staining), TGN46 (green), GOLPH2 (red), and the images merged, as indicated. Note the nearly identical, relatively weak TGN46 staining in both normal and cancer cells, and cancer cell-specific overexpression of GOLPH2. White arrow indicates a normal duct and yellow arrows indicate cancerous acini.
GOLPH2 and MYO6 Co-Localization in Human Prostate Cancer
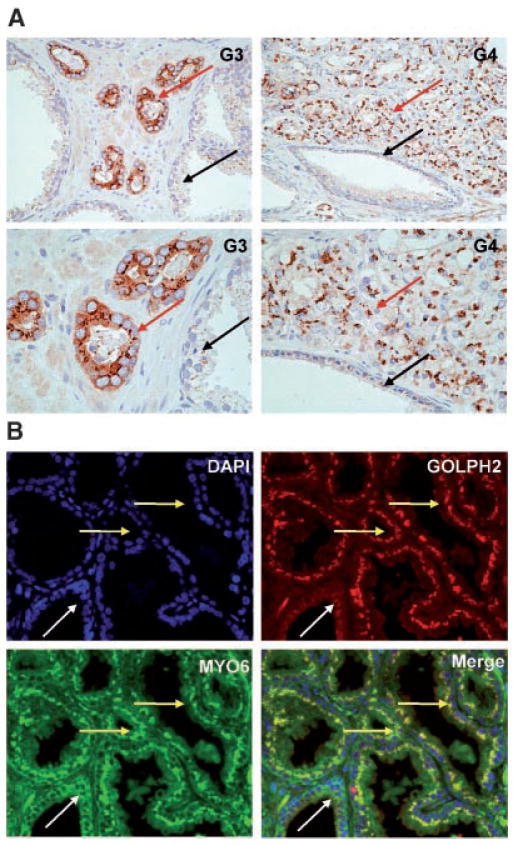
Myosin VI (MYO6) is consistently overexpressed in human prostate cancer and previously implicated in cancer invasion [8,13]. MYO6 is a multi-functional protein, with its specific cellular function linked to its subcellular localization [14]. Although MYO6 has a putative Golgi-associated function [15,20], our previous IHC study using a polyclonal rabbit antibody derived from the porcine antigen did not reveal a definitive Golgi staining pattern [8]. The rabbit polyclonal antibody myosin VI used in the present study was a different antibody produced using the C-terminal tail of human myosin VI [13]. As part of our follow-up study on MYO6 in human prostate cancer, we performed IHC analysis using this antibody in standard FFPE sections derived from 15 cases of radical prostatectomy specimens. Consistent with our previous study [8], we observed strong diffuse cytoplasmic staining of MYO6 in cancer cells, and weak or absent staining for myosin VI in the adjacent normal prostate tissues (Fig. 4A), in the majority of cases examined (14 out of 15 cases). Surprisingly, the new antibody also stained the well-defined perinuclear area consistent with Golgi localization (Fig. 4A), in nearly all cancer lesions with elevated cytoplasmic myosin VI levels, regardless of the Gleason grade of the tumor lesions (Fig. 4A). Immunofluorescence co-staining of myosin VI and GOLPH2 detected highly overlapped staining patterns at the juxtanuclear areas in the prostate cancer epithelium (Fig. 4B, yellow arrow), but not in the normal prostate epithelial cells (Fig. 4B, white arrow). The specificity and suitability of this antibody for immunohistochemical staining was determined by detecting only one prominent band of approximate 150 kDa that disappeared upon myosin VI gene knock down (data not shown). The Golgi apparatus in prostate cancer tissues therefore displayed alterations in its molecular composition as reflected by elevated levels of both GOLPH2 and MYO6 when compared to the normal Golgi.
Fig. 4.
Myosin VI and GOLPH2 both localize to the Golgi apparatus. A: Prostate cancer tissues of varying histological grade (as indicated) stained with the myosin VI polyclonal antibody. Higher power views of a restricted area in top panels were presented immediately below. Note the distinctive perinuclear staining pattern present in the cancer epithelium (red arrows) regardless of the Gleason grades, and weak or absent staining in the normal prostate epithelium (black arrows). G3: Gleason 3; G4: predominantly Gleason 4. B: Myosin VI and GOLPH2 Immunofluorescence double labeling in formalin fixed paraffin embedded surgical prostate cancer tissues. Prostate cancer tissues were stained for nuclei (blue, by DAPI staining), MYO6 (green), GOLPH2 (red), and the images merged, as indicated. Note the distinctive Golgi staining pattern for GOLPH2 and MYO6 specifically in the cancer epithelium (yellow arrow), and the negative or weak signals for both proteins in the normal appearing prostate epithelium (white arrow).
DISCUSSION
While previous studies have identified cancer-specific alterations through posttranslational modifications occurring in the Golgi apparatus [21], this study for the first time provided definitive evidence for cancer-specific overexpression of two molecules that are structurally linked to the Golgi apparatus. The functional significance of these alterations remains to be fully characterized. GOLPH2 is an integral Golgi membrane protein with unknown functions, though the expression of GOLPH2 is potently induced by viral infection [9]. MYO6 is a unique member of the myosin (i.e., actin motor) superfamily in that is the only myosin that has been shown to move ‘backwards’ towards the minus end of actin filaments [14]. MYO6 is involved in multiple cellular processes such as vesicular transport, cell migration, and maintenance of Golgi morphology; each may be linked to its distinctive intracellular locations [22]. Our findings established a solid link between these two Golgi-associated molecules and human prostate cancer, and may spur future interests in the roles of GOLPH2 and MYO6 in Golgi functions during human carcinogenesis.
The involvement of organellar changes in human carcinogenesis is extensive and can be exemplified by prominent changes in the nucleus [23] and mitochondria [24]. In these two subcellular organelles, the normal biochemical composition, structure, and function are disrupted at multiple levels in human cancer. These cancer-associated organellar changes reflect the altered genome and its microenvironment, as well as the accompanying metabolic demands by the cancer cells. These alterations can be extensively investigated, through organellar proteomic approaches, for the development of novel cancer markers [25]. By the same token, the altered molecular composition of the cancer Golgi as established in this study provides a strong rationale for pursuing similar approaches, even in the absence of mechanistic insights. Given its canonical function in protein secretion, the Golgi apparatus has added value if targeted for the discovery of novel biomarkers for the detection and prognosis of human cancers. This targeted approach for biomarker discovery is particularly relevant since many Golgi resident proteins can be transported to the surface for proteolytic cleavage and secretion as a result of disruption in the pH values of the Golgi apparatus, or simply due to overexpression of the target protein [26]. Indeed, GOLPH2 is secreted through this well-characterized mechanism [27]. A recent study explored urinary detection of GOLPH2 mRNA for diagnosis of human prostate cancer [12]. The utility of protein based assays for GOLPH2 or other cancer Golgi markers in the diagnosis of human prostate cancer awaits further optimization and testing in well-annotated patient cohorts.
In summary, the present study for the first time provided definitive evidence that in human prostate carcinoma, the Golgi apparatus underwent molecular alterations as reflected in elevated levels of two Golgi associated proteins, GOLPH2 and MYO6. Both genes were initially identified through unbiased approaches (i.e., unsupervised clustering) [8] for the analysis of gene expression data, highlighting the extent of such previously unrecognized alterations in the cancer Golgi. Such alterations can be further explored for the development of novel prostate cancer markers through targeted organellar approaches, and may help to decipher the mechanisms underlying altered Golgi structure and function in human cancer.
Acknowledgments
Grant sponsor: US Department of Defense; Grant number: W81XWH-04-1-0873; Grant sponsor: NIH/NCI Specialized Program in Research Excellence in Prostate Cancer (Johns Hopkins University); Grant number: P50CA58236.
This work was supported by the US Department of Defense Grant W81XWH-04-1-0873 (to J.L.) and NIH/NCI Specialized Program in Research Excellence in Prostate Cancer P50CA58236 (Johns Hopkins University). The authors would like to thank Dr. Denise Montell for providing the anti-MY06 antibody.
References
- 1.Marsh BJ, Howell KE. The mammalian Golgi complex debates. Nat Rev Mol Cell Biol. 2002;3:789–795. doi: 10.1038/nrm933. [DOI] [PubMed] [Google Scholar]
- 2.Sütterlin C, Hsu P, Mallabiabarrena A, Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- 3.Colanzi A, Suetterlin C, Malhotra V. Cell-cycle-specific Golgi fragmentation: How and why? Curr Opin Cell Biol. 2003;15:462–467. doi: 10.1016/s0955-0674(03)00067-x. [DOI] [PubMed] [Google Scholar]
- 4.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR, III, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z, Vong QP, Zheng Y. CLASPing microtubules at the trans-Golgi network. Dev Cell. 2007;12:839–840. doi: 10.1016/j.devcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Kellokumpu S, Sormunen R, Kellokumpu I. Abnormal glycosylation and altered Golgi structure in colorectal cancer: Dependence on intra-Golgi pH. FEBS Lett. 2002;516:217–224. doi: 10.1016/s0014-5793(02)02535-8. [DOI] [PubMed] [Google Scholar]
- 7.Kirchheim D, Brandes D, Bacon RL. Fine Structure and cytochemistry of human prostatic carcinoma. In: Brandes D, editor. Male accessory sex organs: Structure and function in mammals. San Diego: Academic Press; 1974. pp. 397–423. [Google Scholar]
- 8.Dunn TA, Chen S, Faith DA, Hicks JL, Platz EA, Chen Y, Ewing CM, Sauvageot J, Isaacs WB, De Marzo AM, Luo J. A novel role of myosin VI in human prostate cancer. Am J Pathol. 2006;169:1843–1854. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kladney RD, Bulla GA, Guo L, Mason AL, Tollefson AE, Simon DJ, Koutoubi Z, Fimmel CJ. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53–65. doi: 10.1016/S0378-1119(00)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, Tennant BC, London WT, Evans AA, Blumberg BS, Dwek RA, Mattu TS, Mehta AS. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci USA. 2005;102:779–784. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, Comunale MA, D’Amelio A, Lok AS, Block TM. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43:1007–1012. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;8:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida H, Cheng W, Hung J, Montell D, Geisbrecht E, Rosen D, Liu J, Naora H. Lessons from border cell migration in the Drosophila ovary: A role for myosin VI in dissemination of human ovarian cancer. Proc Natl Acad Sci USA. 2004;101:8144–8149. doi: 10.1073/pnas.0400400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buss F, Kendrick-Jones J. How are the cellular functions of myosin VI regulated within the cell? Biochem Biophys Res Commun. 2008;369:165–175. doi: 10.1016/j.bbrc.2007.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner CL, Stewart A, Luzio JP, Steel KP, Libby RT, Kendrick-Jones J, Buss F. Loss of myosin VI reduces secretion and the size of the Golgi in fibroblasts from Snell’s waltzer mice. EMBO J. 2003;22:569–579. doi: 10.1093/emboj/cdg055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39(1):75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 17.Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM. Alpha-methylacyl-CoA racemase: A new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 18.Rondinelli RH, Epner DE, Tricoli JV. Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in late pathological stage human prostate cancer. Prostate Cancer Prostatic Dis. 1997;1:66–72. doi: 10.1038/sj.pcan.4500208. [DOI] [PubMed] [Google Scholar]
- 19.Ponnambalam S, Girotti M, Yaspo ML, Owen CE, Perry AC, Suganuma T, Nilsson T, Fried M, Banting G, Warren G. Primate homologues of rat TGN38 primary structure, expression and functional implications. J Cell Sci. 1996;109:675–685. doi: 10.1242/jcs.109.3.675. [DOI] [PubMed] [Google Scholar]
- 20.Sahlender DA, Roberts RC, Arden SD, Spudich G, Taylor MJ, Luzio JP, Kendrick-Jones J, Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivinoja A, Kokkonen N, Kellokumpu I, Kellokumpu S. Elevated Golgi pH in breast and colorectal cancer cells correlates with the expression of oncofetal carbohydrate T-antigen. J Cell Physiol. 2006;208:167–174. doi: 10.1002/jcp.20653. [DOI] [PubMed] [Google Scholar]
- 22.Spudich G, Chibalina MV, Au JS, Arden SD, Buss F, Kendrick-Jones J. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 25.Andersen JS, Mann M. Organellar proteomics: Turning inventories into insights. EMBO Rep. 2006;7:874–879. doi: 10.1038/sj.embor.7400780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puri S, Bachert C, Fimmel CJ, Linstedt AD. Cycling of early Golgi proteins via the cell surface and endosomes upon lumenal pH disruption. Traffic. 2002;3:641–653. doi: 10.1034/j.1600-0854.2002.30906.x. [DOI] [PubMed] [Google Scholar]
- 27.Bachert C, Fimmel C, Linstedt AD. Endosomal cis Golgi Protein GP73 Produces Marker for Hepatocellular Carcinoma. Traffic. 2007;8:1415–1423. doi: 10.1111/j.1600-0854.2007.00621.x. [DOI] [PubMed] [Google Scholar]