Abstract
Prostate-specific antigen (PSA) is a serine protease secreted by both normal prostate glandular cells and prostate cancer cells. The major proteolytic substrates for PSA are the gel-forming proteins in semen, semenogelin (Sg) I and II. On the basis of the PSA cleavage map for Sg I and II, a series of small peptides (i.e., ≤ 7 amino acids) was synthesized and coupled at the COOH terminus to 7-amino-4-methyl coumarin. Using these fluorescently tagged substrates, Kms and kcats were determined for PSA hydrolysis, and the substrates were also tested for activity against a panel of purified proteases. Previously, a variety of chymotrypsin substrates have been used to assay the enzymatic activity of PSA. The present studies have identified a peptide sequence with a high degree of specificity for PSA (i.e., no detectable hydrolysis by chymotrypsin) and improved Kms and kcats over previously used substrates. On the basis of these parameters, the best peptide substrate for PSA has the amino acid sequence HSSKLQ. Using PC-82 human prostate cancer xenografts and human prostate tissues, this PSA substrate was used to document that prostate cancer cells secrete enzymatically active PSA into the extracellular fluid but that once in the blood, PSA is not enzymatically active. On the basis of this information, it should be possible to use the HSSKLQ peptide as a carrier to target peptide-coupled prodrugs for selective activation within sites of PSA-secreting, metastatic prostate cancer cells and not within the blood or other nonprostatic normal tissues.
INTRODUCTION
Currently, there is no treatment that significantly prolongs survival in men with metastatic prostate cancer (1). Androgen ablation therapy, although of substantial palliative benefit, has had little impact on overall survival, and the mortality rate from prostate cancer has increased steadily since the introduction of this therapy in the 1940s (2). Androgen ablation therapy eventually fails, because the metastatic prostate cancer within an individual patient is composed heterogeneously of clones of both androgen-dependent and -independent cancer cells (3). Due to these androgen-independent prostatic cancer cells, the patient is no longer curable using the numerous chemotherapeutic agents that have been tested over the past 30 years (1). Thus, there will be an estimated 41,000 deaths this year in the United States due to androgen-independent metastatic prostate cancer (2).
Standard antiproliferative chemotherapeutic agents may be ineffective against androgen-independent prostatic cancers because these cancers have a low proliferative rate. Berges et al. (4) demonstrated that the median daily proliferative rate of prostate cancer cells within lymph node or bone metastases was <3.0% per day. Newer agents are needed that target the >95% of prostate cancer cells within a given metastatic site that are not immediately proliferating. An example of such an agent is TG,3 a potent and selective inhibitor of the sarcoplasmic/endoplasmic reticulum Ca2+ ATPase pump (5, 6). Inhibition of the sarcoplasmic/endoplasmic reticulum Ca2+ ATPase pump by TG results in a sustained elevation of intracellular Ca2+, which induces programmed cell death/apoptosis of metastatic, androgen-independent human and rodent prostatic cancer cells in a proliferation independent manner (5–7). TG is also able to induce programmed cell death in proliferatively quiescent (i.e., Go) human prostate cancer cells in primary culture without recruitment into the proliferative cell cycle (8). Because of its unique cell proliferation-independent mechanism of cytotoxicity, TG could be a novel agent for the treatment of metastatic prostate cancer. However, TG itself is difficult to administer systemically without significant toxicity, because it is highly lipophilic, and its cytotoxicity is not prostate cancer cell specific.
As an approach to target cytotoxicity of TG specifically to metastatic prostate cancer, a prodrug could be developed consisting of a primary amine containing a TG derivative coupled via its amino group to form a peptide bond to a water-soluble peptide carrier (9). The peptide carrier may be designed so that the peptide linkage between the amino-TG derivative and the peptide is efficiently and specifically cleavable only by the proteolytic activity of PSA. PSA is a Mr 33,000 single-chain glycoprotein first characterized from human prostate tissue by Wang et al. (10). PSA is synthesized and secreted as a unique differentiation product of the prostatic glandular cells (10–12). No other normal cell types in the human secrete PSA in any significant level (10–12). Besides normal prostatic glandular cells, the only other cell type in males that secretes substantial amounts of PSA are prostate cancer cells (10–12). Low levels of PSA have also been detected in homogenates from normal breast tissue and breast cancer (13); however, serum levels of PSA are not detectable, even in women with breast cancers staining positive for PSA (14).
PSA is a serine protease with extensive sequence identity to the glandular kallikreins (11, 12, 15). However, unlike the glandular kallikreins that have a common trypsin-like activity, PSA has chymotrypsin-like substrate specificity (15–17). The major proteolytic substrates for PSA are the gel-forming proteins in freshly ejaculated semen, Sg I and II, which are synthesized and secreted by the seminal vesicles (11, 16, 18). The ejaculatory mixing of the secretions from the seminal vesicles and the prostate results in immediate formation of a loosely connected gel structure that entraps spermatozoa. Sg I and II are the major structural proteins in the gel, and enzymatically active PSA in the seminal fluid proteolytically generates multiple soluble fragments of Sg I and II (11). Lilja et al. previously described several PSA-specific cleavage sites within Sg I(16), and recently, additional cleavage sites were defined within both Sg I and II (19).
Previously, a variety of chymotrypsin substrates have been used to assay the enzymatic activity of PSA (12, 16, 20). These substrates have relatively high Kms (i.e., 1–2 mm), low kcats for PSA, and poor specificity (i.e., 10,000-fold more rapidly cleaved by chymotrypsin). The aim of the present study was to develop a peptide substrate for PSA with improved Kms and kcats, a high degree of specificity for PSA, and stability in serum so that it could be used as a carrier to specifically target TG prodrug activation by PSA-secreting prostatic tissue. On the basis of the PSA cleavage map for Sg I and II (19), small peptides were synthesized containing the amino acid sequence proximal to the PSA proteolytic sites and having 7-AMC coupled via a peptide bond to the carboxylic residue of the terminal amino acid. This allowed hydrolysis at the COOH-terminal residue to be followed kinetically by the increase in fluorescence due to release of the AMC. Kinetics parameters (i.e., Kms and kcats) were determined for the proteolysis of these substrates by PSA, and activity against a panel of purified proteases was tested to identify the best substrate based on its efficacy and specificity for PSA.
MATERIALS AND METHODS
Materials
PSA was purified from human seminal plasma as described previously (15). Recombinant hK2 was generated as described previously (21). Plasmin, urokinase, tissue plasminogen activator, human plasma, and porcine pancreatic kallikrein and ACT were obtained from Calbiochem (La Jolla, CA); all other proteases were obtained from Sigma Chemical Co. (St. Louis, MO). All of these proteases were tested separately with the appropriate standard substrate to document that the proteins were in the active form. Mouse monoclonal IgG anti-PSA antibodies H117 and 5A10 were described previously (21–23). Mouse serum was obtained from East Acres Biologicals (South Bridge, MA). Goat and calf sera were obtained from Life Technologies, Inc. (Grand Island, NY). Rat serum and mouse plasma were obtained in-house from sacrificed animals. Human serum was obtained from patients with prostate cancer who had PSA values between 165 and 689 ng/ml by Tandem-R assay. Human control serum was obtained from male volunteers without prostate cancer with serum PSA levels <0.5 ng/ml by Tandem-R assay.
Cell Line and Collection of PSA
PC-82 androgen-responsive human prostate cancer xenografts were maintained by serial passage in nude mice (Charles River Breeding Laboratories). The origins and characteristics of this xenograft have been described previously (24–26). Blood from tumor-bearing animals was obtained by retro-orbital sinus puncture under metofane anesthesia (Mallinkrodt, Mundelein, IL). To determine the proportion of PSA in the extracellular fluid of PC-82 tumors that are enzymatically active, freshly removed PC-82 tumors (200–300 mg) were wetted with ~100 µl RPMI, and then after 10 min, fluid was wicked off the surface of the tumor and collected for additional analysis after determining the PSA concentration by Tandem-R assay (Hybritech, San Diego, CA). The PSA in the extracellular fluid was then concentrated by immunoprecipitation and assayed for its enzymatic activity as described below. PC-82 homogenates were made using a hand-held homogenizer to liquefy tumors (300–600 mg) in 1 ml of PSA assay buffer [50 mm Tris and 0.1 m NaCl (pH 7.8)].
To determine the activity of PSA in the extracellular fluid of normal human prostate tissue and prostate cancers, radical prostatectomy specimens were obtained, and normal peripheral prostate tissues and cancers were dissected separately by a pathologist. These tissues were then wetted with RPMI at room temperature as described above, and extracellular fluid was collected. PSA concentrations of this tissue extracellular fluid were determined using the Tandem-R assay, and then samples were diluted with PSA assay buffer to a final PSA concentration of 4 µg/ml prior to direct enzymatic assay.
PSA Substrates
PSA substrates were custom synthesized by Enzyme Systems Products (Dublin, CA), and, after high-performance liquid chromatography separation and purification to >95%, the molecular weights were confirmed by mass spectroscopy. All substrates had AMC attached via an amide bond to the carboxyl group of the COOH-terminal amino acid. The NH2 terminus of the substrates was protected by coupling to a 4-morpholinecarbonyl blocking group (formula weight 115). 4-Morpholinecarbonyl was chosen as the NH2-terminal blocking group because of its stability and to enhance substrate solubility. Substrates were first dissolved in distilled H20 (5 mm) and then diluted into PSA assay buffer. The glutamine-AMC (Q-AMC) substrate had the NH2 terminus and side chain amine unprotected and was obtained from Bachem (Torrance, CA)
Kinetic Analysis of Substrate Hydrolysis by PSA
Substrate hydrolysis was studied by measuring fluorescence change secondary to AMC release using a Fluoroskan II 96-well fluorometric plate reader (ICN Biomedicals, Costa Mesa, CA; excitation, 355 nm; emission, 460 nm) connected to a Macintosh computer. Data were collected and analyzed using Deltasort III software (Biometallics, Princeton, NJ). All reactions were performed at room temperature by the addition of substrate and proteases into PSA assay buffer, described previously, in a final volume of 200 µl. The results were determined from the initial linear increase of fluorescence and were expressed as the pmol of AMC released per min based on comparison to a standard curve of the fluorescence of known amounts of 7-AMC (i.e., fluorescence was linear from 20 to 3000 pmol of AMC). The data were analyzed by Lineweaver-Burke reciprocal plots to determine the Michaelis-Menten constant (i.e., Km), expressed as the amount of substrate needed to saturate half of the enzyme, and the catalytic rate constant (i.e., kcat), expressed as the amount of substrate converted to product per time per amount of enzyme.
PSA Immunoprecipitation
PSA levels in the serum and extracellular fluid from PC-82 tumor-bearing animals were measured by Tandem-R assay, and samples were then diluted to 600–1000 ng PSA/ml of PSA buffer. After incubation with 100 µl of 50% protein A-Sepharose (Pharmacia Biotech, Uppsala, Sweden) beads for 30 min at 4°C, the mixture was centrifuged at 10,000 rpm for 5 min to preclear the supernatant. To the resultant supernatant was added mouse mAb H117, which recognizes both PSA and glandular kallikrein (hK2; Ref. 21), to a final concentration of 5 µg/ml. Samples were rotated at 4°C overnight, and then 100 µl of protein A-Sepharose were added with rotation for 3 h at 4°C. The mixture was centrifuged to pellet the captured PSA, and then the pellets were washed three times with PSA buffer at room temperature. Aliquots of the washed pellets were then analyzed for enzymatic activity with PSA substrates in 96-well plates by measuring fluorescent change over time. Analysis of supernatants by Tandem-R assay after immunoprecipitation demonstrated >95% capture of PSA (final concentration, <5 ng/ml). Purified PSA from human seminal plasma was used as a positive control and immunoprecipitated as above. The specificity of captured PSA was determined by Western blotting and by inhibition of enzymatic activity of immunoprecipitated PSA by a second mouse mAb, mAb 5A10 (10 µg/ml), that is specific for free PSA (i.e., uncomplexed to serum protease inhibitors), does not cross-react with hK2, and inhibits the enzymatic activity of PSA (21–23).
Western Blotting
The protein A-Sepharose-immunoprecipitated pellets were suspended in loading buffer [50 mm Tris (pH 6.8), 100 mm DTT, 2% SDS, 0.1% bromphenol blue, and 10% glycerol], and the samples were heated to 100°C for 5 min. The Sepharose beads were spun down, and aliquots of the supernatant containing 25 ng of PSA (i.e., based on prepelleted determination of PSA concentration by Tandem-R assay) were separated on a 12% SDS-PAGE gel. The proteins were then electroblotted to Immobilon membranes (Millipore, Bedford, MA). The membranes were processed by standard Western blot procedures and incubated with a polyclonal rabbit antihuman PSA antibody (DAKO Corp., Carpinteria. CA) diluted 1:1000. The immune complexes were visualized with the enhanced chemiluminescence detection system (Amersham Corp., Arlington Heights, IL) according to the manufacturer’s specifications.
RESULTS
Properties of PSA Substrates
In freshly ejaculated semen, the proteolytic activity of PSA is directed mainly against the major gel-forming proteins Sg I and II. A map of PSA cleavage sites within the Sg proteins has been generated (16, 19), and peptides of seven amino acids representing the sequence proximal to the PSA cleavage site (P7 − P1) were synthesized with AMC attached to the COOH terminus of the P1 amino acid in the P−1, position. A series of 12 sequences obtained from the Sg cleavage map were synthesized and tested initially as substrates for purified PSA and a panel of other purified extracellular proteases (Table 1). To allow valid comparisons, each assay contained 200 pmol of the particular protease and 0.2 mm concentration of the particular substrate (i.e., 40,000 pmol/200 −l of assay volume). Those substrates containing tyrosine in the P1 position were hydrolyzed efficiently by PSA; however, proteolysis rates were 10–20-fold higher with chymotrypsin and elastase. Substrates with glutamine in the P1 position were less efficiently hydrolyzed but demonstrated better specificity for PSA (Table 1).
Table 1.
PSA substrate hydrolysis by purified extracellular proteasesa
| Substrate group |
Peptide sequenceb |
Pmol of substrate hydrolyzed/min/100 pmol of indicated protease |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PSA | Chymotrypsin | Elastase | Trypsin | Urokinase | Plasmin | TPA | Thrombin | hK1 | hK2 | ||
| 1 | KGISSQY | 510 | 4290 | 3068 | 658 | UD | ND | ND | UD | ND | UDc |
| SRKSQQY | 234 | 3630 | 2788 | 260 | 0.4 | 0.4 | UD | UD | 2.0 | UDc | |
| GQKGQHY | 49.6 | 4274 | 922 | 122 | ND | UD | 0.6 | UD | ND | UDc | |
| 2 | EHSSKLQ | 31.6 | UD | UD | UD | UD | UD | UD | UD | UD | UDc |
| QNKISYQ | 16.8 | 9.0 | 9.0 | UD | ND | UD | 0.4 | 0.4 | ND | UDc | |
| ENKISYQ | 14.4 | 3.6 | 2.6 | 1.8 | UD | 0.6 | 0.4 | 0.3 | UD | ND | |
| ATKSKQH | 16.0 | 38.8 | UD | UD | UD | UD | UD | 0.2 | UD | ND | |
| 3 | KGLSSQC | 13.6 | 12.2 | 950 | UD | UD | UD | UD | UD | UD | ND |
| LGGSQQL | 6.0 | 314 | 94.0 | UD | UD | UD | UD | UD | ND | ND | |
| 4 | QNKGHYQ | 1.8 | 0.2 | 0.6 | UD | UD | UD | UD | UD | UD | ND |
| TEERQLH | 1.2 | 48.8 | 1.0 | UD | 0.4 | 3.8 | UD | 1.0 | UD | UDc | |
| GSFSIQH | UD | 3.6 | 0.4 | 0.4 | ND | 0.2 | UD | 0.4 | ND | ND | |
| 2 | HSSKLQ | 62.7 | UD | UD | UD | UD | UD | UD | UD | UD | UD |
| SKLQ | 29.6 | UD | UD | UD | UD | UD | ND | ND | UD | ND | |
| KLQ | UD | 10.6 | 0.4 | UD | ND | ND | ND | ND | UD | ND | |
| LQ | UD | UD | UD | UD | ND | UD | UD | UD | ND | ND | |
| Q | UD | UD | 0.7 | UD | UD | UD | ND | ND | ND | ND | |
TPA, tissue plasminogen activator; UD, undelectable (<0.1 pmol of substrate hydrolysis/min/100 pmol of protease); ND, not done; hK1, human plasma kallikrein; hK2, human glandular kallikrein.
Substrates at 0.2 mm concentration in PSA assay buffer [50 mm Tris-0.1 m Nacl (pH 7.8)], except LGGSQQL, GSFSIQH, SKLQ, KLQ, and LQ, which were in 1.4% acetonitrile-PSA buffer and Q-AMC, which was in 0.2% formic acid-PSA buffer (pH 7.8).
hK2 tested at 17 pmol/assay except HSSKLQ-AMC at 100 pmol.
These assays identified four groups of substrates based on PSA activity and specificity. On the basis of these kinetic parameters and specificities of the substrates, four groups of PSA substrates were identified. The first group consisted of those sequences (KGISSQY, SRKSSQY, and GQKGQHY) with the highest PSA activity but that were even better substrates for other serine proteases, such as chymotrypsin and elastase. The second group (EHSSKLQ, QNKISYQ, ENKISYQ, and ATKSKQH) had moderate PSA activity but showed greater specificity for PSA. The third group (KGLSSQC and LGGSQQL) also had moderate PSA activity but were less specific for PSA than was the second group. The fourth group (QNKQHYQ, TEERQLH, and GSFSIQH) had little activity with any of the proteases tested. All substrates lacking tyrosine at the cleavage site had undetectable activity with those serum protease such as thrombin, plasmin, and urokinase that have trypsin-like specificity for cleavage primarily after arginine residues. PSA is a member of the kallikrein family of proteases (i.e., PSA is hK3) and shares substantial sequence homology with human plasma kallikrein (hK1) and human glandular kallikrein (hK2; Ref. 21). hK1 and hK2 also have trypsin-like substrate specificity (21) but demonstrated no detectable activity with the majority of substrates (Table 1).
On the basis of these preliminary studies concerning solubility and specificity, a selected subset of substrates were studied further to determine their Km and kcat kinetic parameters (Table 2). Because the kcat reflects the efficiency of the conversion of the substrate to products per enzyme whereas the Km reflects the concentration dependence for enzymatic activity, higher kcat:Km ratios denote better substrates, and these ratios were calculated to compare the various substrates.
Table 2.
Kinetic parameters of PSA peptide substrates
| Group | PSA substratea |
Km (µm) |
kcat (s−1) |
kcat:Km (s−1 m−1) |
|---|---|---|---|---|
| 1 | KGISSQY | 160 | 0.043 | 270 |
| SRKSQQY | 90 | 0.023 | 260 | |
| 2 | EHSSKLQ | 1165 | 0.012 | 10.6 |
| HSSKLQ | 470 | 0.011 | 23.6 | |
| SKLQ | 813 | 0.020 | 24.6 | |
| ATKSKQH | 1310 | 0.0091 | 6.9 | |
| 3 | KGLSSQC | 300 | 0.0017 | 5.6 |
| LGGSSQL | 900 | 0.0037 | 4.1 |
PSA-mediated hydrolysis of substrates in buffer [50 mm Tris and 0.1 m NaCI (pH 7.8)].
Stability of PSA Substrates in Sera
Because the prodrug would need to be given systemically via the blood, additional analyses were performed to determine the stability of the substrates in sera from a variety of species (Table 3). Hydrolytic activity varied widely between substrates and between sera. All of the substrates with tyrosine in the P1 position and two with glutamine (ENKISYQ and QNKISYQ) were hydrolyzed to a greater extent than the others, especially in mouse and fetal calf sera, whereas the remaining substrates showed good stability in human sera for periods up to 24 h.
Table 3.
Stability of PSA substrates in various seraa
| pmol of AMC released/min |
|||||||
|---|---|---|---|---|---|---|---|
| Group | Sequenceb | Humanc | Mouse | Rat | Fetal calf | Calf | Goat |
| 1 | SRKSQQY | UD | 3.52 | 0.52 | 0.98 | 1.05 | 0.36 |
| GQKGQHY | 0.09 | 12.86 | 2.74 | 2.90 | 0.78 | 1.78 | |
| 2 | EHSSKLQ | UD | 2.17 | 0.06 | UD | UD | UD |
| QNKISYQ | 1.54 | 1.67 | 10.7 | 11.60 | UD | 5.54 | |
| ENKISYQ | 1.27 | 3.71 | 11.2 | 9.59 | UD | 3.18 | |
| ATKSKQH | UD | 0.60 | 4.31 | 1.19 | UD | 0.35 | |
| 3 | KGLSSQC | UD | 0.66 | 0.80 | 0.03 | UD | UD |
| LGGSQQL | UD | 13.92 | 0.17 | 1.10 | UD | 0.11 | |
| 4 | QNKQHYQ | 0.24 | 9.57 | 7.03 | 4.53 | UD | 3.47 |
| TEERQLH | UD | 10.71 | 0.66 | 0.02 | UD | UD | |
| 2 | HSSKLQ | UD | 0.81 | 1.04 | 0.31 | UD | UD |
| SKLQ | UD | 1.70 | UD | 0.21 | 0.02 | ND | |
UD, undetectable (<0.01 pmol/min change); ND, not done.
Each substrate at 0.2 mm.
100% sera used for each assay.
Effect of Sequence Length on PSA Activity
The substrate EHSSKLQ-AMC was chosen from among the others for additional characterization based on its PSA activity, its specificity for hydrolysis by PSA, and its relative stability in a variety of sera. On the basis of this starting sequence, shorter sequence lengths were then synthesized starting from the COOH terminus and analyzed for activity with PSA and other purified proteases (Table 1). In general, the shorter sequence length maintained specificity for proteolysis by PSA when compared to the panel of other purified extracellular proteases. The six-amino acid peptide, HSSKLQ, was even a better substrate for PSA without sacrificing any specificity. Next, Kms and kcats were determined for each sequence length (Table 2). From this group, the six-amino acid sequence HSSKLQ-AMC had the lowest Km. This peptide also maintained a high degree of specificity for PSA as well as good stability in sera (Table 3). The four-amino acid sequence SKLQ-AMC, although having a higher Km than HSSKLQ-AMC, had an equivalent kcat:Km ratio and serum stability. However, this shorter peptide was less water soluble than the histidine-containing HSSKLQ. Because of comparable activities (i.e., kcat:Km ratios) between the four- and six-amino acid sequences, the five-amino acid sequence was not synthesized. The three-amino acid sequence KLQ-AMC had detectable activity at 5 mm concentration, whereas the shorter one- and two-amino acid substrates had no detectable activity at 5 mm. Substrates were not evaluated at concentrations >5 mm because of poor solubility in aqueous buffer. On the basis of these combined data, the best substrate for PSA analysis is HSSKLQ-AMC.
PSA Activity in the PC-82 Human Xenograft
On the basis of its kcat:Km ratio, relative specificity, serum stability, and water solubility, the HSSKLQ-AMC substrate was used to analyze the enzymatic activity of PSA in the PC-82 xenograft model. PC-82 is an androgen-responsive tumor and is one of the few human prostate cancers that continues to secrete PSA during serial xenograft passage into nude mice (25). Sera from mice bearing 300–500-mg PC-82 tumors contain elevated levels of Tandem-R-detectable PSA (200–400 ng/ml).
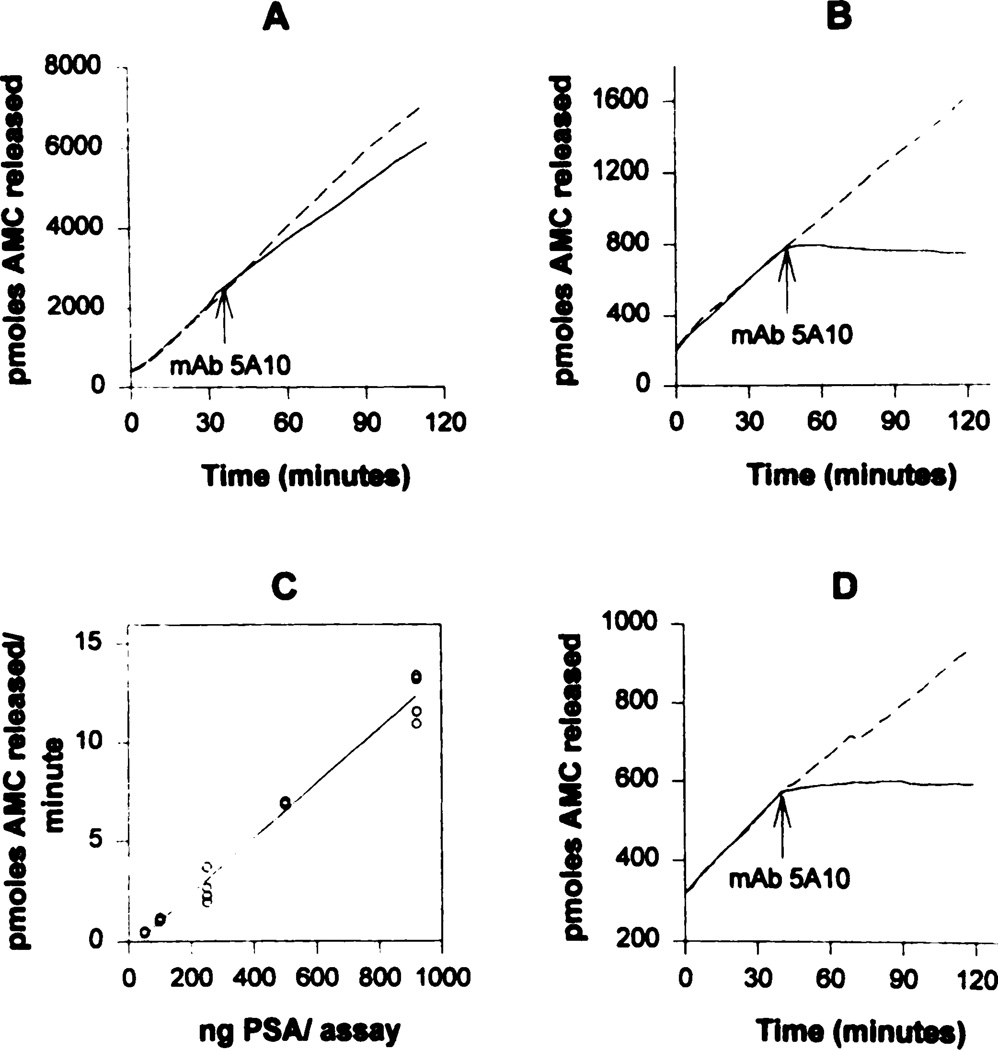
Initially, homogenates of PC-82 tumors were made, and the PSA concentration in these homogenates was determined using the Tandem-R assay. The enzymatic activity of these homogenates was then assayed using the substrate HSSKLQ-AMC, and the activity was compared to that of an equal amount of PSA purified from human seminal fluid (Fig. 1A). These PC-82 homogenates hydrolyzed the substrate at a 6-fold higher rate than the purified PSA, and most of this activity (i.e., >82 ± 2%) was not inhibited by the addition of anti-PSA antibody 5A10 (Fig. 1A). These studies indicated that the substrate HSSKLQ-AMC, although not a substrate for a variety of purified extracellular proteases, could be hydrolyzed by a combination of intracellular proteases.
Fig. 1.
Fluorometric assays using HSSKLQ-AMC substrate (0.4 mm) with neutralization of activity at indicated times by anti-PSA mAb 5A10 (10 µg/ml). A, PC-82 tumor homogenized in PSA assay buffer and assayed for PSA concentration by Tandem-R assay and aliquots containing 1 µg of total PSA assayed for activity. B, activity of PSA (1 µg) purified from human seminal plasma immunoprecipitated with mAb H117 (5 µg/ml) and protein A-Sepharose. C, linear range of enzymatic activity of immunoprecipitated PSA (human seminal plasma) using increasing amounts of PSA. D, activity of PSA (500 ng) immunoprecipitated from PC-82 tumor homogenates.  , activity without mAb 5A10;
, activity without mAb 5A10;  , activity with addition of mAb 5A10 at indicated times. All PSA assays were done in triplicate, and representative plots are shown.
, activity with addition of mAb 5A10 at indicated times. All PSA assays were done in triplicate, and representative plots are shown.
To further clarify this point, the family of peptide substrates based on the EHSSKLQ sequence was assayed for activity for the intracellular proteases cathepsin B, C, and D and esterase (Table 4). As the sequence length became incrementally shorter, the rate of hydrolysis by cathepsin B and D increased, whereas cathepsin C and esterase had no detectable activity with all of the sequences tested (Table 4). The single amino acid substrate, glutamine-AMC, was not hydrolyzed efficiently by any of these intracellular proteases.
Table 4.
PSA substrate hydrolysis by purified intracellular proteasesa
| pmol of substrate hydrolyzed/min/200 pmol of protease Cathepsins |
|||||
|---|---|---|---|---|---|
| Peptide sequenceb |
PSA | B | C | D | Esterasec |
| EHSSKLQ | 31.6 | 4.3 | UD | 2.2 | UD |
| HSSKLQ | 62.7 | 17.3 | UD | 3.8 | UD |
| SKLQ | 29.6 | 31.2 | UD | 6.4 | UD |
| KLQ | 0.4 | 87.0 | UD | ND | UD |
| LQ | UD | 190 | UD | 20.0 | UD |
| Q | UD | 1.0 | UD | 1.1 | 0.9 |
UD, undetectable (<0.1 pmol substrate of hydrolysis/min/200 pmol of protease); ND. Not done.
Substrates at 0.2 mm concentration in PSA assay buffer except SKLQ, KLQ, and LQ, in 1.4% acetonitrile/buffer, and Q-AMC. in 0.2% formic acid/buffer (pH 7.8).
Esterase from porcine liver.
To allow the best substrate (i.e., HSSKLQ-AMC) to be useful for assaying the PSA enzymatic activity in homogenates of whole tumors, an immunoconcentration step was used to isolate enzymatically active PSA that was free of contamination by other intracellular proteases. PSA can be immunoprecipitated with mAb H117, a mAb that recognizes a shared epitope of PSA and hK2 outside their catalytic centers (21), and therefore, the immunoprecipitate does not inhibit the enzymatic activity of either enzyme. For example, when purified PSA from human seminal plasma is immunoprecipitated in this manner, it continues to have enzymatic activity when incubated with the PSA substrate HSSKLQ-AMC (Fig. 1B). Substrate hydrolysis is specific for PSA, because hK2 is unable to cleave the PSA substrate and because the proteolytic activity can be stopped immediately upon the addition of mAb 5A10, a mAb that only recognizes free PSA and not hK2 and inhibits the enzymatic activity of PSA (Ref. 22; Fig. 1B). These results validate that the immunoprecipitation protocol used can effectively capture enzymatically active PSA. Using this immunoprecipitation protocol and the HSSKLQ-AMC substrate at 0.4 mm concentration, PSA enzymatic activity was demonstrated to increase linearly with increasing amounts of purified seminal plasma PSA (i.e., linear range, 50–1000 ng of PSA; Fig. 1C). The lower limit of detection of PSA enzymatic activity with the HSSKLQ-AMC substrate (i.e., >0.1 pmol of AMC released/min) is 50 ng of total PSA per assay (Fig. 1C).
Using this validated immunoprecipitation protocol, PSA was immunoprecipitated with mAb H117 from homogenates made from PC-82 tumors and assayed for enzymatic activity using the HSSKLQ-AMC substrate (Fig. 1D). Like the enzymatic activity of the purified seminal plasma PSA immunoprecipitate, the enzymatic activity of the PSA immunoprecipitate from the PC-82 tumor homogenates was inhibited by >98% by the neutralizing mAb 5A10 (Fig. 1D). PSA immunoprecipitated from these PC-82 tumor homogenates possessed 70 ± 5% of the enzymatic activity of similar concentrations of immunoprecipitated PSA purified from seminal plasma.
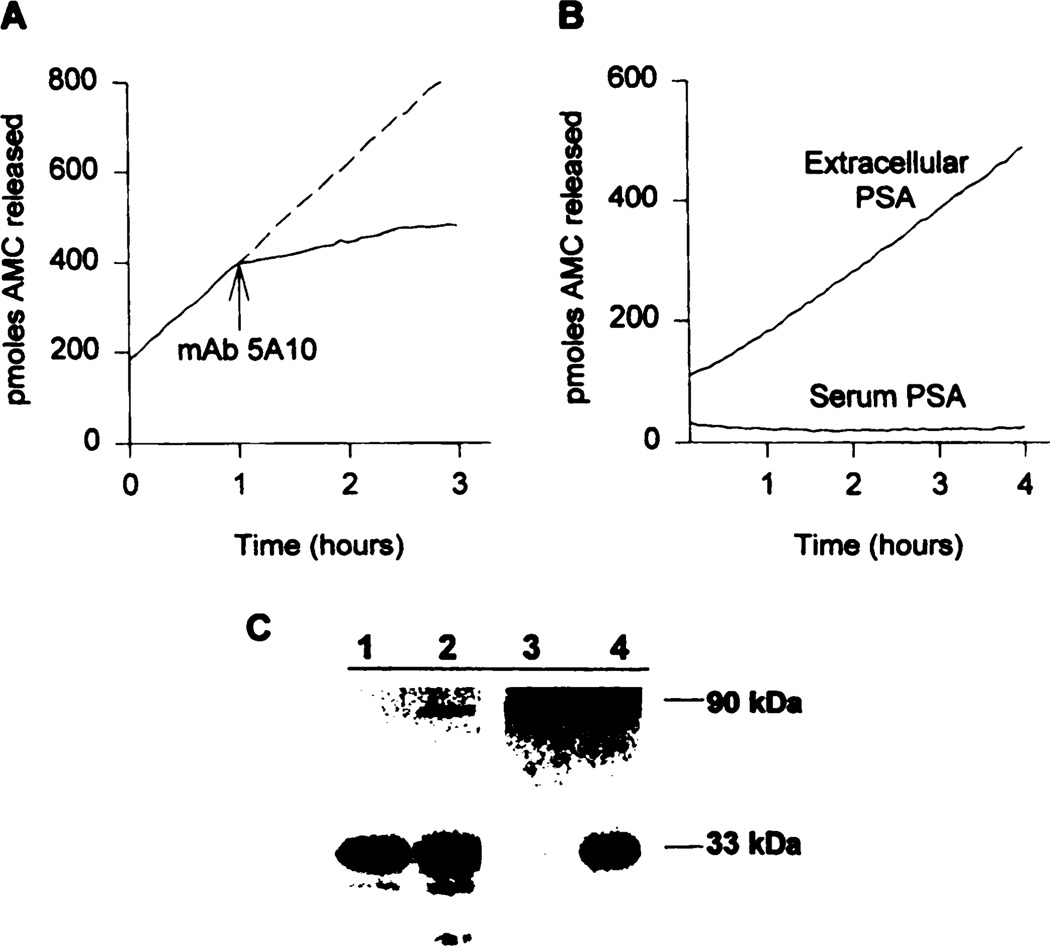
The PC-82 homogenate consists of PSA from both intracellular and extracellular compartments. To analyze the enzymatic activity of PSA only in the extracellular fluid, microcapillary tubes were used to wick fluid off the surface of freshly removed PC-82 tumors. The PSA concentration in this fluid was determined by Tandem-R assay, and after dilution to 1 µg/ml in PSA assay buffer, the PSA was immunoprecipitated according to the previously described protocol. The immunoprecipitates were then assayed for PSA enzymatic activity using the HSSKLQ-AMC substrate (0.4 mm; Fig. 2A). As expected, the enzymatic activity of the PSA immunoprecipitated from the PC-82 tumor extracellular fluid was also inhibited by mAb 5A10 (Fig. 2A). PSA immunoprecipitated from the PC-82 extracellular fluid possessed 66 ± 4% of the enzymatic activity of similar concentrations of immunoprecipitated PSA purified from seminal fluid.
Fig. 2.
A, enzymatic activity of PSA (400 ng) immunoprecipitated from PC-82 tumor extracellular fluid using HSSKLQ-AMC substrate (0.4 mm) with and without the addition of mAb 5A10 (10 µg/ml) at indicated times. B, activity of PSA (200 ng) immunoprecipitated from pooled sera of PC-82 tumor-bearing animals compared to similar amounts of PSA immunoprecipitated from PC-82 tumor extracellular fluid. C, Western blot analysis of immunoprecipitated PSA (25 ng PSA/lane). Lane 1, purified human seminal plasma PSA; Lane 2; purified PSA incubated for 24 h with 10-fold molar excess ACT; Lane 3, PSA from pooled serum of PC-82 tumor-bearing animals; Lane 4, PSA from extracellular fluid from PC-82 tumor. All PSA assays were done in triplicate, and representative plots are shown.
PSA was also immunoprecipitated from the pooled sera of animals bearing PC-82 tumors and assayed for enzymatic activity using the HSSKLQ-AMC substrate (Fig. 2B). The lack of PSA enzymatic activity in serum (Fig. 2B) is consistent with previous studies showing that PSA in human serum is enzymatically inactivated by interaction with the major extracellular protease inhibitors ACT and α2-macroglobulin (21). These inhibitors are present in a 104–105-fold molar excess to PSA in serum (15, 22, 23). PSA forms a 1:1 molar ratio complex with ACT by splitting the reactive center of the inhibitor, thus forming a Mr 90,000 stable PSA-ACT covalent complex (Ref. 26; Fig. 2C, Lane 2).
Western blot analysis of the PSA in the sera of PC-82-bearing animals demonstrated that the majority of the immunoprecipitable PSA is in the Mr 90,000 PSA-ACT complex (Fig. 2C, Lane 3), which is enzymatically inactive, as shown in Fig. 2B. In the immunoprecipitated extracellular fluid obtained from the PC-82 tumors, PSA can be found in two forms on Western blot, ≈25% being the Mr 90,000 PSA-ACT complex and the remaining 75% in the Mr 33,000 uncomplexed, free form of PSA (Fig. 2C, Lane 4). The Mr 33,000 free PSA is the enzymatically active form that is capable of hydrolyzing the HSSKLQ-AMC substrate Fig. 2A.
Previously, it has been demonstrated that the complex formed between PSA and α2-macroglobulin does not result in the complete inactivation of PSA, because smaller substrates can still be hydrolyzed by the PSA-α2-macroglobulin complex (21). The PSA-α2-macroglobulin complex is not immunoreactive with PSA antibodies, and therefore, this complex would not be captured during the immunoprecipitation of PSA from the sera of PC-82 tumor bearing animals. To determine whether PSA can be inhibited completely by the full complement of serum protease inhibitors present in mouse blood, plasma was obtained from nude mice, and purified, enzymatically active human seminal plasma PSA was added to a concentration of 5 µg/ml. Samples were incubated at 37°C for 24 h and then assayed directly for PSA activity using the HSSKLQ-AMC substrate. In this experiment, there was no difference in the rate of hydrolysis of the HSSKLQ-AMC substrate in the PSA-spiked mouse plasma versus control plasma (n = 6, data not shown), demonstrating that the PSA was inhibited completely by the mouse plasma. The addition of the anti-PSA mAb 5A10 resulted in no further inhibition of hydrolysis in both PSA-containing and control plasma.
PSA Activity in Human Prostate Tissue and Serum
Extracellular fluid from normal and malignant prostate tissue was assayed for its PSA concentration using the Tandem-R assay. The extracellular PSA concentrations ranged from 30 to 110 µg/g for the normal prostate tissue (n = 8; mean = 13.9 ± 6.6 µg/ml) and 20–110 µg/g for the prostate cancers (n = 7; mean = 9.9 ± 6.3 µg/ml). Individual samples were diluted to a final concentration of 4 µg/ml and assayed directly for PSA enzymatic activity using the HSSKLQ-AMC substrate. To determine the percentage of PSA enzymatic activity in these samples, the anti-PSA mAb 5A10, which inhibits >99% of the enzymatic activity of PSA from purified human seminal plasma, was added to a final concentration of 50 µg/ml. The percentage decrease in slope upon addition of mAb 5A10 versus control samples was then calculated. For normal prostate tissue, the mAb 5A10 inhibited 78.0 ± 2.4% (n = 16) of the activity in the extracellular fluid, whereas in the fluid from prostate cancers, the mAb 5A10 inhibited 89.1 ± 2.0% (n = 13) of the activity. These results demonstrate that PSA is enzymatically active in the extracellular fluid obtained from both normal and malignant prostate tissues and that PSA is responsible for the majority of HSSKLQ-AMC substrate hydrolysis in the extracellular fluid from these tissues.
In additional studies, the HSSKLQ-AMC substrate was used to assay without immunoprecipitation the enzymatic activity of PSA in the sera of four men with prostate cancer and elevated Tandem-R-detectable serum PSA levels ranging from 165 to 689 ng/ml. Using these serum samples, no hydrolysis of the HSSKLQ-AMC substrate was detected (i.e., <0.1 pmol AMC released/min).
DISCUSSION
The aim of the present study was to identify a PSA-specific peptide substrate that can be used as a carrier for targeted delivery of cytotoxic agents to sites of metastatic prostate cancer. Substrates for PSA include Sg I and II (16, 19), the extracellular matrix components fibronectin (16) and laminin (27), the insulin-like growth factor binding proteins (28), the single-chain form of urokinase-type plasminogen activator (29) and parathyroid hormone-related protein (30). PSA, through its ability to degrade extracellular matrix components, may also play a role in tumor cell invasion and metastases (27). From a map of PSA cleavage sites within Sg I and II (16, 19) we have generated a series of PSA substrates. On the basis of assays comparing the proteolysis of these substrates by a panel of purified proteases, determination of kinetic parameters, and measurements of stability in sera, one substrate, HSSKLQ-AMC, was selected for further analysis. This substrate has high specificity for PSA, and it is stable in human sera, including sera from men with elevated PSA levels and human plasma. The Km for this substrate is 470 µm, which is a 3–5-fold improvement over the Kms for previously used PSA substrates, which have been primarily chymotrypsin substrates with Kms for PSA in the 1.3–2 mm range (15, 17, 20).
Besides having an improved Km and kcat, the HSSKLQ substrate is not measurably hydrolyzed by a panel of purified extracellular proteases, including chymotrypsin and trypsin. The only other protease with any significant ability to hydrolyze this substrate is the intracellular lysosomal cysteine protease, cathepsin B. In normal cells, lysosomal proteases are secreted extracellularly as inactive zymogens that are then taken up by the cell and delivered to its lysosomal compartment via the mannose-6-phosphate receptor pathway (31). Several laboratories have reported an association between increased plasma membrane association and/or activity of cathepsin B and either malignant conversion or enhanced metastatic capability (32, 33). Cathepsin B is not present in significant amounts in the extracellular fluid of normal cells but appears to be substantially elevated in the extracellular fluid surrounding neoplastic cells, including prostate cancer cells (34, 35). The proteolysis of HSSKLQ-AMC by cathepsin B demonstrates that this substrate is not entirely PSA specific. However, proteolysis by active cathepsin B secreted by cancer cells may result in additional, selective activation of the prodrug within sites of metastatic prostate cancer.
Besides cathepsin B, HSSKLQ-AMC is not a substrate for a large panel of purified extracellular proteases. The HSSKLQ-AMC substrate, however, is hydrolyzed readily by tumor homogenates, indicating that intracellular proteases, presumably from lysosomes, are capable of cleaving this substrate. The nonspecific hydrolysis of the HSSKLQ-AMC substrate by intracellular proteases requires that an immunoconcentration step be used prior to assaying the enzymatic activity of PSA in tumor homogenates. This also requires that an incubation of the immunoconcentrated proteins with a specific PSA-neutralizing antibody be performed to ensure that the enzymatic activity seen is due to the PSA and not due to hydrolysis by other intracellular or extracellular proteases.
In men with prostate cancer, PSA levels in the blood can exceed 1000 ng/ml. This serum PSA is derived from the PSA in the extracellular fluid. Upon escape into the extracellular compartment, PSA is initially enzymatically active, as demonstrated by the PC-82 and human prostate cancer results presented. Extracellular prostate cancer fluid can hydrolyze the HSSKLQ-AMC substrate and can form complexes with the extracellular protease inhibitor, ACT. Once in the serum, however, PSA has no assayable enzymatic activity due to its complexing with ACT and α2-macroglobulin (15, 23). A small portion of PSA occurs in a free, noncomplexed (Mr 33,000) form that is apparently enzymatically inactive due to its inability to form complexes with the excess protease inhibitors (23). The enzymatic inactivity of this Mr 33,000 form of PSA in the blood is supported by our data showing that little to no hydrolysis of the PSA substrate HSSKLQ-AMC occurred in the blood of men with elevated PSA levels. PSA immunoprecipitated from the blood serum of PC-82-bearing animals also had no enzymatic activity, and Western blot analysis demonstrated that the majority of the serum PSA is found in the Mr 90,000 PSA-ACT complex.
The small amount of free PSA in the serum most likely represents either the PSA proenzyme form or PSA that has been cleaved internally. PSA is produced as a preproenzyme. Prior to secretion, a 17-amino acid hydrophobic signal sequence is removed from the NH2 terminus to produce an enzymatically inactive proenzyme (36). The inactive proenzyme has a seven-amino acid activation peptide that must be released through cleavage at the ultimate arginine preceding the NH2-terminal isoleucine in the mature form of PSA to gain enzymatic activity (36). This cleavage site predicts that a trypsin-like protease converts the zymogen into enzymatically active PSA, and indeed, trypsin has been shown to activate recombinant pro-PSA produced in Escherichia coli (37). PSA secreted by PC-82 into the extracellular fluid is processed correctly, because the PSA isolated from this fluid is enzymatically active. These results demonstrate that PSA is activated before entering the serum and not as a result of processing by a protease present within the blood or contributed by any of the other accessory sex glands in the male genital tract. The enzymatic activity in the extracellular fluid indicates that the processing enzyme must be present either within the prostate cancer cells themselves or in the extracellular fluid. If it is in the extracellular fluid, this processing enzyme is produced either by the PC-82 cells themselves or by the surrounding tissue. To resolve this issue, conditioned media from primary cultures of PC-82 prostate cancer cells established in serum-free defined media to prevent stromal cell growth are being assayed using the AMC-tagged seven-amino acid PSA propeptide sequence as a substrate for this PSA-processing enzyme.
In conclusion, we have identified a peptide with the amino acid sequence HSSKLQ that can be used both as a substrate to measure PSA enzymatic activity in extracellular fluids and as a carrier to target prodrugs for activation within sites of metastatic prostate cancer producing enzymatically active PSA. We have documented that the PC-82 human prostatic cancer cells secrete enzymatically active PSA into their extracellular fluid. Thus, the PC-82 xenograft model can be used for testing whether therapeutic prodrug activation is possible in vivo. We have also demonstrated that PSA is enzymatically active in the extracellular fluid obtained from both normal prostate tissue and prostate cancer. We are continuing this work by coupling specific amino-TG analogues to the peptide HSSKLQ to evaluate their efficacy against PC-82 tumors in vivo.
ACKNOWLEDGMENTS
The expert assistance of John C. Lamb and Sue Dalrymple in obtaining animal tissue and processing human prostate tissues is greatly appreciated. We also thank Dr. Daniel Chan (Johns Hopkins Department of Pathology, Johns Hopkins School of Medicine) for supplying the PSA-containing human serum.
Footnotes
The work reported was supported by a CapCure Award (to J. T. I.).
The abbreviations used are: TG, thapsigargin; PSA, prostate-specific antigen; Sg, semenogelin; AMC, amino-4-methyl-coumarin; mAb, monoclonal antibody; ACT, α1-antichymotrypsin.
REFERENCES
- 1.Yagoda A, Petrylak D. Cytotoxic chemotherapy for advanced hormone-resistant prostate cancer. Cancer (Phila.) 1993;71:1098–1109. doi: 10.1002/1097-0142(19930201)71:3+<1098::aid-cncr2820711432>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 2.Parker SL, Tong T, Bolden S, Wingo PA. Cancer statistics, 1996. CA Cancer J. Clin. 1996;65:5–27. doi: 10.3322/canjclin.46.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Schultz H, Isaacs JT, Coffey DS. A critical review of the concept of total androgen ablation in the treatment of prostatic cancer. Prog. Clin. Biol. Res. 1987;243A:1–19. [PubMed] [Google Scholar]
- 4.Berges RS, Vukanovic J, Epstein JI, CarMichel M, Cisek L, Johnson DE, Veltri RW, Walsh PC, Isaacs JT. Implication of cell kinetic changes during the progression of human prostatic cancer. Clin. Cancer Res. 1995;1:473–480. [PMC free article] [PubMed] [Google Scholar]
- 5.Isaacs JT, Lundmo P. Chemotherapeutic induction of programmed cell death in nonproliferating prostate cancer cells. Proc. Am. Assoc. Cancer Res. 1992;33:588–589. [Google Scholar]
- 6.Furuya Y, Lundmo P, Short AD, Gill DL, Isaacs JT. The role of calcium, pH, and cell proliferation in the programmed (apoptotic) death of androgen-independent prostatic cancer cells induced by thapsigargin. Cancer Res. 1994;54:6167–6175. [PubMed] [Google Scholar]
- 7.Furuya Y, Isaacs JT. Proliferation dependent versus independent programmed cell death of prostatic cancer cells involves distinct gene regulation. Prostate. 1994;25:301–309. doi: 10.1002/pros.2990250604. [DOI] [PubMed] [Google Scholar]
- 8.Lin XS, Denmeade SR, Cisek L, Isaacs J. The mechanism and role of growth arrest in programmed (apoptotic) death of prostate cancer cells induced by thapsigargin. Prostate. 1997 doi: 10.1002/(sici)1097-0045(19971101)33:3<201::aid-pros9>3.0.co;2-l. in press, [DOI] [PubMed] [Google Scholar]
- 9.Denmeade SR, Isaacs JT. Activation of programmed (apoptotic) cell death for the treatment of prostate cancer. Adv. Pharmacol. 1996;35:281–306. doi: 10.1016/s1054-3589(08)60278-1. [DOI] [PubMed] [Google Scholar]
- 10.Wang MC, Valenzuela LA, Murphy GP, Chu TM. Purification of a human prostate specific antigen. Invest. Urol. 1979;17:159–163. [PubMed] [Google Scholar]
- 11.Lilja H. A kallikrein-like serine protease in prostatic fluid cleaves the predominant seminal vesicle protein. J. Clin. Invest. 1985;76:1899–1903. doi: 10.1172/JCI112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watt KWK, Lee P-J, M’Timkulu T, Chan W-P, Loor R. Human prostate-specific antigen: structural and functional similarity with serine proteases. Proc. Natl. Acad. Sci. USA. 1986;83:3166–3170. doi: 10.1073/pnas.83.10.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Diamandis EP, Levesque M, Giai M, Roagna R, Ponzone R, Sismondi P, Monne M, Croce CM. Prostate specific antigen in breast cancer, benign breast disease and normal breast tissue. Breast Cancer Res. Treat. 1996;40:171–178. doi: 10.1007/BF01806212. [DOI] [PubMed] [Google Scholar]
- 14.Giai M, Yu H, Roagna R, Ponzone R, Katsaros D, Levesque MA, Diamandis EP. Prostate-specific antigen in serum of women with breast cancer. Br. J. Cancer. 1995;72:728–731. doi: 10.1038/bjc.1995.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensson A, Laurell C-B, Lilja Hans. Enzymatic activity of prostate-specific antigen and its reactions with extracellular serine protease inhibitors. Eur. J. Biochem. 1990;194:755–765. doi: 10.1111/j.1432-1033.1990.tb19466.x. [DOI] [PubMed] [Google Scholar]
- 16.Lilja H, Abrahamsson P-A, Lundwall A. Semenogelin, the predominant protein in human semen. J. Biol. Chem. 1989;264:1894–1900. [PubMed] [Google Scholar]
- 17.Akiyama K, Nakamura T, Iwanaga S, Hara H. The chymotrypsin-like activity of human prostate-specific antigen γ-seminoprotein. FEBS Lett. 1987;225:1894–1900. doi: 10.1016/0014-5793(87)81151-1. [DOI] [PubMed] [Google Scholar]
- 18.Lilja H, Oldbring J, Rannevik G, Laurell C-B. Seminal vesicle-secreted proteins and their reactions during gelation and liquefication of human semen. >J. Clin. Invest. 1987;80:281–285. doi: 10.1172/JCI113070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malm J, Hellman J, Lilja H. Mapping of the unique enzyme characteristics of PSA. J. Urol. 1997;157(Suppl.):345. [Google Scholar]
- 20.Kurkela R, Herrala A, Henttu P, Nal H, Vihko P. Expression of active, secreted human prostate-specific antigen by recombinant baculovirus-infected insect cells on a pilot scale. Biotechnology. 1995;13:1230–1234. doi: 10.1038/nbt1195-1230. [DOI] [PubMed] [Google Scholar]
- 21.Lovgren J, Piironen T, Overmo C, Dowell B, Karp M, Pettersson K, Lilja H, Lundwall A. Production of recombinant PSA and hk2 and analysis of their immunologic cross-reactivity. Biochem. Biophys. Res. Commun. 1995;213:888–895. doi: 10.1006/bbrc.1995.2212. [DOI] [PubMed] [Google Scholar]
- 22.Pettersson K, Piironen T, Seppälä M, Liukkonon L, Christensson A, Matikainen M-T, Suonpää M, Lövgren T, Lilja H. Free and complexed prostate-specific antigen (PSA): in vitro stability, epitope map, and development of immunofluoremetric assays for specific and sensitive detection of free PSA and PSA-α1-anti-chymotrypsin complex. Clin. Chem. 1995;41:1480–1488. [PubMed] [Google Scholar]
- 23.Lilja H, Christensson A, Dahlén U, Matikainen MT, Nilsson O, Pettersson K, Lövgren T. Prostate specific antigen in human serum occurs predominantly in complex with α1-anti-chymotrypsin. Clin. Chem. 1991;37:1618–1625. [PubMed] [Google Scholar]
- 24.Hoehn W, Schroeder FH, Riemann JF, Joebsis AC, Hermanek P. Human prostatic adenocarcinoma: some characteristics of a serially transplantable line in nude mice (PC-82) Prostate. 1980;1:94–104. doi: 10.1002/pros.2990010113. [DOI] [PubMed] [Google Scholar]
- 25.Csapo Z, Brand K, Wlather R, Fokas K. Comparative experimental study of the serum prostate specific antigen and prostatic acid phosphatase in serially transplantable human prostatic carcinoma lines in nude mice. J. Urol. 1988;140:1032–1038. doi: 10.1016/s0022-5347(17)41921-5. [DOI] [PubMed] [Google Scholar]
- 26.Kyprianou N, English HF, Isaacs JT. Programmed cell death during regression of PC-82 human prostate cancer following androgen ablation. Cancer Res. 1990;50:3748–3753. [PubMed] [Google Scholar]
- 27.Webber MM, Waghray A, Bello D. Prostate specific antigen, a serine protease, facilitates human prostatic cancer cell invasion. Clin. Cancer Res. 1995;1:1089–1094. [PubMed] [Google Scholar]
- 28.Cohen P, Graves HCB, Peehl DM, Kamarei M, Giudice LC, Rosenfeld RG. Prostate-specific antigen (PSA) is an insulin-like growth factor binding protein-3 protease found in seminal plasma. J. Clin. Endocrinol. Metab. 1992;75:1046–1053. doi: 10.1210/jcem.75.4.1383255. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida E, Ohmura S, Sugiki M, Maruyama M, Mihara H. Prostate-specific antigen activates single-chain urokinase-type plasminogen activator. Int. J. Cancer. 1995;63:863–865. doi: 10.1002/ijc.2910630618. [DOI] [PubMed] [Google Scholar]
- 30.Iwamura M, Hellman J, Cockett AT, Lilja H, Gershagen S. Alteration of the hormonal bioactivity of parathyroid hormone-related protein (PTHrP) as a result of limited proteolysis by prostate-specific antigen. Urology. 1996;48:317–325. doi: 10.1016/S0090-4295(96)00182-3. [DOI] [PubMed] [Google Scholar]
- 31.Sloane BF, Moin K, Krepela E, Rozhin J. Cathepsin B and its endogenous inhibitors: role in tumor malignancy. Cancer Metast. Rev. 1990;9:333–352. doi: 10.1007/BF00049523. [DOI] [PubMed] [Google Scholar]
- 32.Sloane BF, Rozhin J, Taylor H, Crissman JD, Honn KV. Cathepsin B: association with plasma membrane in metastatic tumors. Proc. Natl. Acad. Sci. USA. 1986;83:2483–2487. doi: 10.1073/pnas.83.8.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krepela E, Bartek J, Skalova D, Vicar J, Rasnick D, Taylor-Papadimitriou J, Hallowes RC. Cytochemical and biochemical evidence of cathepsin B in malignant, transformed and normal breast epithelial cells. J. Cell Sci. 1987;87:145–154. doi: 10.1242/jcs.87.1.145. [DOI] [PubMed] [Google Scholar]
- 34.Sinha AA, Wilson MJ, Gleason DF, Reddy PK, Sameni M, Sloane BF. Immunohistochemical localization of cathepsin B in neoplastic human prostate. Prostate. 1995;26:171–178. doi: 10.1002/pros.2990260402. [DOI] [PubMed] [Google Scholar]
- 35.Sinha AA, Gleason DF, Staley NA, Wilson MJ, Sameni M, Sloane BF. Cathepsin B in angiogenesis of human prostate: an immunohistochemical and immunoelectron microscopic analysis. Anat. Rec. 1995;241:353–362. doi: 10.1002/ar.1092410309. [DOI] [PubMed] [Google Scholar]
- 36.Lundwall A, Lilja H. Molecular cloning of human prostate-specific antigen cDNA. FEBS Lett. 1987;214:317–322. doi: 10.1016/0014-5793(87)80078-9. [DOI] [PubMed] [Google Scholar]
- 37.Takayama TK, Fujikawa K, Davie EW, Lange PH. Comparisons of recombinant with seminal plasma-purified human prostate specific antigen. Proc. Am. Urol. Assoc. 1996;155:338A. [Google Scholar]