Abstract
BACKGROUND
Inflammation is commonly observed in radical prostatectomy specimens, and evidence suggests that inflammation may contribute to prostate carcinogenesis. Multiple microorganisms have been implicated in serving as a stimulus for prostatic inflammation. The pro-inflammatory anaerobe, Propionibacterium acnes, is ubiquitously found on human skin and is associated with the skin disease acne vulgaris. Recent studies have shown that P. acnes can be detected in prostatectomy specimens by bacterial culture or by culture-independent molecular techniques.
METHODS
Radical prostatectomy tissue samples were obtained from 30 prostate cancer patients and subject to both aerobic and anaerobic culture. Cultured species were identified by 16S rDNA gene sequencing. Propionibacterium acnes isolates were typed using multilocus sequence typing (MLST).
RESULTS
Our study confirmed that P. acnes can be readily cultured from prostatectomy tissues (7 of 30 cases, 23%). In some cases, multiple isolates of P. acnes were cultured as well as other Propionibacterium species, such as P. granulosum and P. avidum. Overall, 9 of 30 cases (30%) were positive for Propionibacterium spp. MLST analyses identified eight different sequence types (STs) among prostate-derived P. acnes isolates. These STs belong to two clonal complexes, namely CC36 (type I-2) and CC53/60 (type II), or are CC53/60-related singletons.
CONCLUSIONS
MLST typing results indicated that prostate-derived P. acnes isolates do not fall within the typical skin/acne STs, but rather are characteristic of STs associated with opportunistic infections and/or urethral flora. The MLST typing results argue against the likelihood that prostatectomy-derived P. acnes isolates represent contamination from skin flora.
Keywords: prostate cancer, inflammation, Propionibacterium acnes, MLST, infection
INTRODUCTION
Histologic specimens of prostate tissue from prostate cancer patients frequently exhibit unexplained acute and chronic inflammation and inflammation-associated lesions [1–3]. The development of prostatic inflammation may be related to microbial infection, as previous studies have demonstrated the presence of multiple microbial species in the prostates of prostate cancer patients [4,5]. Interestingly, many of the organisms identified are consistent with genera associated with inflammation-associated conditions including bacterial prostatitis and/or urinary tract infections [5]. Propionibacterium acnes (P. acnes) is a bacterium of particular interest in relation to prostate cancer. Propionibacterium acnes is a pro-inflammatory bacterium that is considered to be the etiological agent in the skin condition acne vulgaris and has also been reported in association with other inflammatory conditions including endocarditis, sarcoidosis, and post-surgical infections [6]. Propionibacterium acnes was first reported in association with prostate inflammation and cancer in 2005 [4]. Interestingly, this study reported that prostatectomy specimens from which P. acnes could be cultured were more likely to be inflamed, leading to the hypothesis that P. acnes-mediated inflammation may contribute to prostate carcinogenesis [4]. Several subsequent studies have also reported on the presence of P. acnes in prostate specimens [5,7,8]. Although not all studies have shown a positive association, the correlation between acne and/or plasma antibodies to P. acnes and prostate cancer incidence and outcomes has also been examined in multiple epidemiological studies [9–11]. In addition, in vitro studies have demonstrated that P. acnes is capable of inducing a strong inflammatory response in prostate cell lines [7,12,13].
Initially, sequencing of P. acnes tly and recA genes was used to categorize P. acnes strains into phylotypes I, II, and III [14,15]. A more recent strategy for typing bacterial strains is called multilocus sequence typing, or MLST, which has dissolved the population structure of the species P. acnes. MLST generates sequence types (STs) based on DNA sequencing and the determination of different alleles of internal fragments of housekeeping genes [16,17]. Related STs can form clonal complexes (CCs) based on their similarity to a central allelic profile. The MLST typing scheme for P. acnes again identified three divisions of P. acnes strains (I, II, and III) [16]. Division I was further subdivided into I–1a, I–1b, and I–2, and further into CCs. MLST analysis performed on 210 isolates of P. acnes from healthy individuals, patients with moderate to severe acne, and patients with various opportunistic infections (abscess, wounds, endocarditis, bursitis, hip prosthesis, etc.) demonstrated that severe acne isolates were predominantly classified into CCs belonging to group I–1a and I–1b strains, i.e., CC3, CC18, and CC31, whereas isolates associated with opportunistic infections were predominantly classified into CCs belonging to group I–2, II, and III strains, i.e., CC36, CC53/60, and CC43 [16].
Propionibacterium acnes is an ubiquitous skin bacterium and is also reported to be a common culture contaminant. It is therefore often difficult to determine if the presence of P. acnes in surgical specimens (including radical prostatectomy specimens) has arisen from contamination from the skin of the patient and/or the medical staff or whether it represents a true infection of clinical significance [6,18–20]. The present study was undertaken to perform MLST analysis of P. acnes isolates from radical prostatectomy specimens in order to determine if the STs of these isolates are similar to the STs associated with healthy or diseased human skin or other anatomic locations and disease conditions.
MATERIALS AND METHODS
Prostate Tissue Samples
All specimens were collected under a Johns Hopkins Internal Review Board approved protocol. Post-prostatectomy tissue samples were obtained from 30 patients undergoing treatment for prostate cancer at the Johns Hopkins hospital. The clinical and pathological parameters of the patient samples are listed in Table I. Surgically resected prostates were placed in a sterile container and transported to the pathology suite. The prostate was then placed under a HEPA filtered laminar flow cabinet and a total of 10 tissue cores from peripheral prostate were collected into 2 ml of sterile PBS in a sterile 15 ml conical tube using a Bard Biopty gun and needles as previously described [5]. The prostate was maintained in a sterile field at all times during tissue collection.
TABLE I.
Clinical and Pathological Parameters of Patient Samples for Bacterial Culture
| Parameter | Value |
|---|---|
| Total number of patients | 30 |
| Mean age (range) | 57 (43–74) |
| Gleason score (number of patients) | |
| 6 | 8 |
| 3 + 4 = 7 | 11 |
| 4 + 3 = 7 | 6 |
| 8 | 2 |
| 9 | 3 |
| TNM stage | |
| T2 | 14 |
| T3 | 16 |
Bacterial Culture
The prostate tissues were first minced under a BSL-2 laminar flow hood using sterile razor blades and sterile petri dishes. Minced tissues were then equally divided into 5–8 ml of culture broth in polystyrene tubes for aerobic and anaerobic culture (BD Biosciences). For aerobic culture, minced tissues were cultured in Luria-Bertani (LB) broth (BD Biosciences) at 200 rpm at 37°C in a shaking incubator for a minimum of 1 week. Most positive aerobic cultures were positive for growth within 24–48 hr. For anaerobic culture, minced tissues were cultured in Brain Heart Infusion broth (BD Biosciences) in anaerobic pouches (GasPak EZ Anaerobe Gas System, BD Biosciences) at 37°C for at least 2 weeks. Most positive anaerobic cultures had visible growth within 1 week. For each sample collected, negative control cultures were performed for each type of broth and culture condition. Importantly, negative control cultures were never positive for bacterial growth.
Strain Identification
Bacteria from cultures positive for growth was harvested and gDNA was isolated using the modified protocol for Gram positive bacteria and the QIAamp DNA mini kit (Qiagen) or the MasterPure™ Gram Positive DNA Purification Kit (Epicentre). A universal primer set designed against the bacterial 16S rDNA gene, Ecoli9-F and Loop27-R was used for PCR as previously described [21]. Negative controls for 16S PCR were performed using sterile DNA-free water as template. The PCR cycling parameters were as follows: 94°C for 2 min, 35× cycles of 94°C for 30 sec, 53°C for 30 sec, and 72°C for 1 min and 72°C for 5 min. Purified PCR products were sent for Sanger Sequencing at the DNA Analysis Facility at Johns Hopkins. Sequencing results were analyzed by Standard Nucleotide BLAST search against reference bacterial genomic sequences (NCBI).
Multilocus Sequence Typing
For bacterial isolates that were identified as P. acnes, MLST was performed per the typing scheme described by Lomholt and Kilian [16]. Nine housekeeping genes were amplified by PCR and used for sequence analysis (cel, coa, fba, gms, lac, oxc, pak, recA, and zno) [16]. Negative PCR controls were performed for each primer set using sterile DNA-free water as template. In two P. acnes strains (from patients no. 20 and no. 22), we were unable to amplify the oxc allele and the zno allele. This was presumably due to mismatches in the primer sets for these strains using the Lomholt and Kilian MLST scheme. In these cases, the following replacement primers were used: oxc2-F 5′-AGGCGTGCTGCCGGAAAAG-3′, oxc2-R 5′-CAC-CACCGGCGTCAGGATT-3′, and zno2-R 5′-TCA-TATGCCGCGTCGACCTC-3′. The PCR cycling parameters for all housekeeping genes except for recA were as follows: 96°C for 40 sec, 35× cycles of 94°C for 35 sec, 55°C for 40 sec and 72°C for 40 sec and 72°C for 7 min. The PCR cycling parameters for recA were as follows: 95°C for 3 min, 35× cycles of 95°C for 1 min, 55°C for 30 sec and 72°C for 90 sec and 72°C for 10 min. Purified PCR products were sent for Sanger Sequencing at the DNA Analysis Facility at Johns Hopkins or Genomic Services at Beckman Coulter Genomics. ST of each isolate was determined using a publically available MLST database (http://pacnes.mlst.net) [16]. Allele sequences can be found at http://pacnes.mlst.net.
The identification of CCs and their founders based on allele profiles was achieved by eBURST analysis at http://eburst.mlst.net/using the eBURST version 2 clustering algorithm, which was developed and is hosted by Imperial College London and is based on principles originally described by Feil et al. [22].
RESULTS
Bacterial Culture
The results of bacterial culture from prostatectomy tissues are shown in Table II. Half of the patient samples were negative for bacterial growth. As determined by 16S rDNA sequence analysis, in the remaining cases, P. acnes was the most frequently cultured species, isolated from 23% of patient samples. Other species cultured from prostatectomy tissues included P. avidum (7%), P. granulosum (3%), Staphylococcus epidermidis (17%), and Corynebacterium glucuronolyticum (7%). As shown in Table III, in several cases either more than one species of bacteria or more than one strain of P. acnes was cultured. Interestingly, in two cases, P. acnes was isolated from an aerobic culture.
TABLE II.
Bacteria Isolated From Prostatic Tissue of 30 Unselected Patients With Prostate Cancer
| Organism | No. of patientsa (%) |
|---|---|
| No bacterial growth | 15 (50%) |
| Corynebacterium glucuronolyticum | 2 (7%) |
| Propionibacterium acnes | 7 (23%) |
| Propionibacterium avidum | 2 (7%) |
| Propionibacterium granulosum | 1 (3%) |
| Staphylococcus epidermidis | 5 (17%) |
More than one species was cultured from two cases.
There were no significant correlations between P. acnes culture status and patient age, Gleason score, or tumor stage. Interestingly, there was a borderline significant correlation between tumor stage (pT2 vs. pT3) and cases that were positive for culture of S. epidermidis (Fisher’s exact test, P = 0.045, Table IV).
TABLE IV.
Association Between Prostate Cancer Pathological Stage and Bacterial Culture Status
| Stage | No. of cases (P. acnes, +) | No. of cases (P. acnes, −) | P valuea | No. of cases (S. epidermidis, +) | No. of cases (S. epidermidis, −) | P valuea |
|---|---|---|---|---|---|---|
| pT2 | 4 | 10 | 0 | 14 | ||
| pT3 | 3 | 13 | 0.6746 | 5 | 11 | 0.0447 |
As determined by Fisher’s exact test.
MLST Analysis of Prostatectomy-Derived P. acnes Isolates
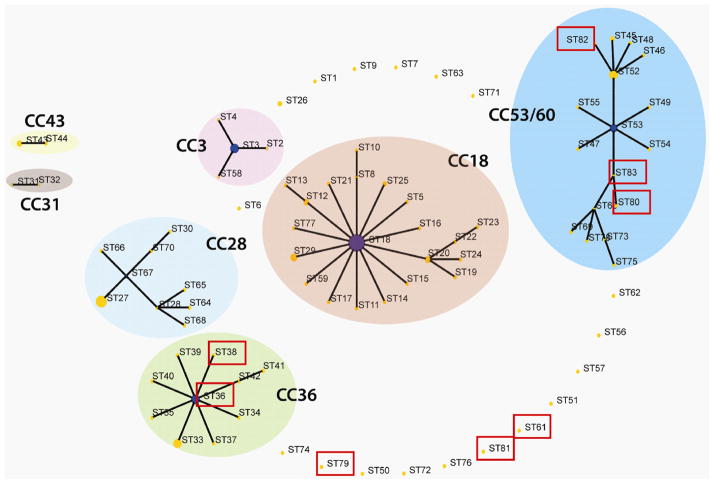
In all, nine different strains of P. acnes from the present study were cultured and subject to MLST analyses (Table III). In addition to these isolates, we also performed MLST typing on a prostatectomy-derived P. acnes isolate from a previous study [5]. The results of MLST analysis are shown in Table V. We observed eight different STs among the prostatectomy-derived P. acnes isolates (Table V). In most cases (six of the eight STs), the allelic profile of the P. acnes strains could not be matched completely with the known STs defined in the MLST database (differing at one to two alleles) [16]. We therefore assigned new STs (ST61 and ST79-83). An eBURST analysis revealed that prostatectomy tissue-derived P. acnes strains belong to two CCs: CC36, the representative CC of group I–2 strains, and CC53/60, a major CC of group II strains (Fig. 1). The newly assigned STs (ST61 and ST79-83) are all type II strains; they are either part of or closely related to CC53/60.
TABLE III.
Prostate Tissue Samples Positive for Bacteria Growth and Species Identification by 16S rDNA Sequence Analysis
| Patient no. | Gleason grade | Stage | Aerobic bacteriaa | Anaerobic bacteriaa |
|---|---|---|---|---|
| 5 | 3 + 3 = 6 | T2 | P. acnes (99%, NC_017535) | P. acnes (99%, NC_017535) |
| 8 | 3 + 4 = 7 | T3A | – | S. epidermidis (100%, NC_004461) |
| 9 | 3 + 4 = 7 | T2 | – | P. acnes (99%, NC_017535) |
| 10 | 3 + 4 = 7 | T3B | P. acnes (99%, NC_017535) | (1) P. acnes (100%, NC_017535) (2) P. avidum (99%, NZ_JH165055) |
| 11 | 3 + 4 = 7 | T2 | – | P. granulosum (99%, NR_025276) |
| 15 | 4 + 4 = 8 | T3A | S. epidermidis (100%, NC_004461) | P. acnes (99%, NC_017535) |
| 16 | 3 + 4 = 7 | T3A | – | P. avidum (99%, NZ_JH165055) |
| 19 | 3 + 4 = 7 | T2 | – | P. acnes (99%, NC_017535) |
| 20 | 3 + 3 = 6 | T2 | – | P. acnes (100%, NC_017535) |
| 22 | 4 + 3 = 7 | T3A | – | P. acnes (99%, NC_017535) |
| 23 | 4 + 3 = 7 | T3A | – | C. glucuronolyticum (97%, NZ_GG667131) |
| 24 | 5 + 4 = 9 | T3B, N1 | S. epidermidis (99%, NZ_GG696777) | |
| 25 | 4 + 5 = 9 | T3A | S. epidermidis (100%, NC_004461) | |
| 26 | 4 + 3 = 7 | T2 | C. glucuronolyticum (100%, NZ_GG667131) | |
| 27 | 4 + 3 = 7 | T3A | S. epidermidis (100%, NC_004461) |
Closest match to GenBank reference genomic sequence (% similarity, Accession no.).
TABLE V.
MLST Profiles of Prostate-Derived P. acnes Isolates
| Patient no. | MLST profile (cel-coa-fba-gms-lac-oxc-pac-recA-zno) | STa | CCb | Divisiona |
|---|---|---|---|---|
| 5 (Aerobic) | 3-9-7-11-7-3-5-6-9 | 61 | Singleton | II |
| 5 | 3-13-8-11-7-3-11d-6-9 | 79 | Singleton | II |
| 9 | 5-9-3-3-4-3-5-2-9 | 36 | 36 | I–2 |
| 10 (Aerobic) | 3-13-7-11-7-7-5-6-9 | 80 | 53 | II |
| 10-1 | 3-13-11d-11-7-3-5-6-9 | 81 | Singleton | II |
| 15 | 3-9-7-11-7-3-5-6-9 | 61 | Singleton | II |
| 19 | 5-9-3-3-2-3-5-2-9 | 38 | 36 | I–2 |
| 20 | 3-13-8-11-7-7-5-6-14 | 82 | 53 | II |
| 22 | 3-13-7-11-7-7-5-6-14 | 83 | 53 | II |
| PA-2c | 5-9-3-3-4-3-5-2-9 | 36 | 36 | I–2 |
Fig. 1.
Population snapshots of P. acnes generated by eBURST analysis based on the MLST allele profiles.
 Prostatectomy-derived P. acnes strains are associated with these STs.
Prostatectomy-derived P. acnes strains are associated with these STs.
Comparison of Prostatectomy-Derived P. acnes CCs and STs to Previously Characterized Strains
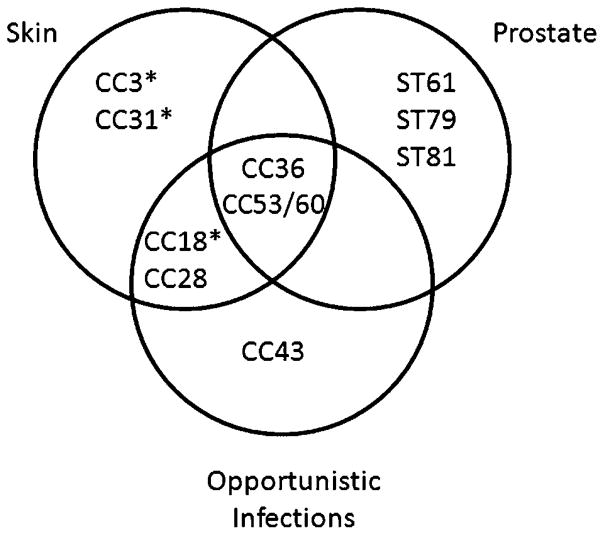
We compared the results of MLST analysis of prostatectomy tissue-derived P. acnes strains to previous MLST studies that have been conducted on 210 P. acnes isolates from human skin, severe acne, and opportunistic infections [16] and 75 human skin and acne-associated isolates from a cohort in the Unites States included as part of the Human Microbiome Project (HMP; Fig. 2) [23]. MLST analysis was previously performed on these strains [24]. Human skin isolates are distributed across the spectrum of CCs and are most predominantly strains of group I–1a (CC3, CC18, and CC28). Strains isolated from opportunistic infections [16] most often belong to CC36 and CC53/60. Likewise, prostatectomy-derived P. acnes isolates were identified as CC36 and CC53/60 strains as well as CC53/60-related singletons. Interestingly, as shown in Figure 3, prostatectomy-derived P. acnes isolates do not overlap with CCs determined to be associated with isolates from severe acne [16].
Fig. 2.
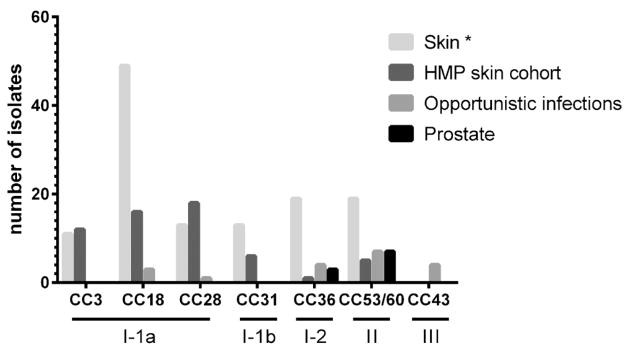
Comparison of prostatectomy-associated P. acnes strains to previously described strain collections of isolates from skin and opportunistic infections [16] and skin isolates from a HMP skin cohort. Singletons ST61, ST79, and ST81 are related to CC53/60 and are grouped in this CC for the purpose of this figure. *From Ref.16.
Fig. 3.
Venn diagram of the association between CCs of P. acnes strains from prostatectomy tissues in the present study compared with isolates from opportunistic infections and skin from a previous study [16].* Severe acne isolates are associated with these CCs. ST61, ST79, and ST81 are singletons and related to CC53/60.
DISCUSSION
The association between P. acnes and disease conditions has been difficult to confirm largely because P. acnes is the most predominant species found on human skin and is reported to be a common culture contaminant. Even the strongest association between P. acnes and a disease condition—as a causative agent in acne vulgaris—remains controversial [25]. Although multiple studies have now reported on the ability to culture P. acnes from prostate cancer tissues [4,5], the question still remains as to whether the presence of this species represents a true prostatic infection or contamination from patient skin, the medical team, or the surgical environment. In the present study, we aimed to begin to address this question by employing a newly established MLST scheme to compare the STs/CCs of prostatectomy tissue-derived P. acnes isolates to previous collections of P. acnes isolates from healthy skin, severe acne, and opportunistic infections. The results of these analyses indicated that the prostatectomy-derived P. acnes isolates included in this study do not overlap with STs/CCs associated with severe acne, but instead overlap with CCs associated with opportunistic infections. Healthy skin-associated isolates are somewhat uniformly distributed among CCs (with the exception of ST18 strains that are thought to represent an “epidemic clone” of P. acnes that is frequently associated with severe acne and prevalent on human skin [16]), and prostatectomy tissue-derived P. acnes isolates did fall within the same CCs as some isolates from healthy skin. However, if the prostatectomy tissue-derived P. acnes isolates were simply reflective of normal skin flora, they would be expected to fall within a broad spectrum of CCs (and especially within the I–1a group) and not confined to distinct CCs in the I–2 and II groups. Instead, none of the prostatectomy tissue-derived P. acnes isolates belong to group I–1a. Moreover, some prostatectomy tissue-derived P. acnes isolates are unique and represent new STs within group II.
There are few studies that have been performed to characterize the normal microbial constituents of the adult male urethral flora. Many of these studies have relied on urine culture and the most commonly recognized species thought to inhabit the male urethra include Staphylococcus spp., Corynebacterium spp., Enterococcus spp., and streptococci [26,27]. Interestingly, in addition to Propionibacterium spp., two of these urethra flora-associated species were cultured from prostatectomy tissues in the present study (Staphylococcus spp. and Corynebacterium spp.). Studies that have utilized PCR-based molecular techniques have also identified P. acnes in the urine of adult males [28,29]. In the study by Shannon et al. [28], urethral P. acnes isolates were found to be associated with phylogenetic clusters IB and II (analogous to I–2 and II in [16]). In all, the results of the present study indicate that the bacterial isolates obtained from prostatectomy specimens may reflect urethral flora as opposed to skin flora. This would support the theory that these bacterial strains may infect the prostate, as the proposed route that bacteria may infect the prostate is via the urethra. On the other hand, the presence of these species in prostatectomy tissues could also represent contamination of the prostatectomy specimen from urethral flora, perhaps due to catheterization of the patient prior to surgery, and this remains a topic of future studies.
We discovered an interesting significant, but borderline (P = 0.045), correlation between the ability to culture S. epidermidis from prostatectomy tissues and advanced stage (T3) prostate cancer (Table IV). Staphylococcus epidermidis has been previously associated with chronic bacterial prostatitis and is therefore implicated in the pathogenesis of prostatic inflammation [30,31]. The isolation of S. epidermidis from prostatectomy tissues has also been previously reported [4,5]. On the other hand, it is known that necrotic tumors can become infected with bacteria from endogenous sources, especially when they occur next to a site where bacteria flora resides (such as the urethra) [32]. Additional studies must be conducted to determine if this association holds up in a larger sample size and whether the relationship is causal or consequent in regard to tumorigenesis.
CONCLUSIONS
MLST typing results indicated that prostate-derived P. acnes isolates do not fall within the typical skin/acne STs, but rather are characteristic of STs associated with opportunistic infections and phylogenetic clusters associated with urethral flora. The MLST analysis results argue against the likelihood that prostatectomy-derived P. acnes isolates represent contamination from skin flora. The question of whether P. acnes truly establishes prostatic infections that arose from urethral flora or whether its presence in prostatectomy tissues represents contamination from urethral flora remains a topic of future studies.
Acknowledgments
Grant sponsor: Department of Defense Prostate Cancer Research Program Award; Grant number: W81XWH-11-1-0521; Grant sponsor: The Johns Hopkins University Prostate Cancer SPORE; Grant number: 5P50CA058236; Grant sponsor: Prostate Cancer Foundation (PCF); Grant sponsor: Prevent Cancer Foundation; Grant sponsor: International Max Planck Research School for Infectious Diseases and Immunology.
This work was supported by Department of Defense Prostate Cancer Research Program Award W81XWH-11-1-0521 and The Johns Hopkins University Prostate Cancer SPORE Grant (5P50CA058236; K.S.S). K.S.S. is also supported as the Chris and Felicia Evensen Prostate Cancer Foundation (PCF) Young Investigator and was supported by a fellowship from the Prevent Cancer Foundation. T.N.M. was funded by the International Max Planck Research School for Infectious Diseases and Immunology. We would like to thank Dr. Mogens Kilian for assistance with eBURST analyses.
Footnotes
Disclosure statement: A.M.D. is currently an employee of Predictive Biosciences Inc., who also holds a part-time adjunct appointment at the Johns Hopkins University School of Medicine. However, no funding or other support was provided by the company for any of the work in this manuscript. The terms of the relationship between A.M.D. and Predictive Biosciences are managed by the Johns Hopkins University in accordance with its conflict of interest policies.
References
- 1.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7(4):256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sfanos KS, Wilson BA, De Marzo AM, Isaacs WB. Acute inflammatory proteins constitute the organic matrix of prostatic corpora amylacea and calculi in men with prostate cancer. Proc Natl Acad Sci USA. 2009;106(9):3443–3448. doi: 10.1073/pnas.0810473106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sfanos KS, De Marzo AM. Prostate cancer and inflammation: the evidence. Histopathology. 2012;60(1):199–215. doi: 10.1111/j.1365-2559.2011.04033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen RJ, Shannon BA, McNeal JE, Shannon T, Garrett KL. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? J Urol. 2005;173(6):1969–1974. doi: 10.1097/01.ju.0000158161.15277.78. [DOI] [PubMed] [Google Scholar]
- 5.Sfanos KS, Sauvageot J, Fedor HL, Dick JD, De Marzo AM, Isaacs WB. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate. 2008;68(3):306–320. doi: 10.1002/pros.20680. [DOI] [PubMed] [Google Scholar]
- 6.Jakab E, Zbinden R, Gubler J, Ruef C, von Graevenitz A, Krause M. Severe infections caused by Propionibacterium acnes: an underestimated pathogen in late postoperative infections. Yale J Biol Med. 1996;69(6):477–482. [PMC free article] [PubMed] [Google Scholar]
- 7.Fassi Fehri L, Mak TN, Laube B, Brinkmann V, Ogilvie LA, Mollenkopf H, Lein M, Schmidt T, Meyer TF, Brüggemann H. Prevalence of Propionibacterium acnes in diseased prostates and its inflammatory and transforming activity on prostate epithelial cells. Int J Med Microbiol. 2011;301(1):69–78. doi: 10.1016/j.ijmm.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Alexeyev O, Bergh J, Marklund I, Thellenberg-Karls C, Wiklund F, Gronberg H, Bergh A, Elgh F. Association between the presence of bacterial 16S RNA in prostate specimens taken during transurethral resection of prostate and subsequent risk of prostate cancer (Sweden) Cancer Causes Control. 2006;17(9):1127–1133. doi: 10.1007/s10552-006-0054-2. [DOI] [PubMed] [Google Scholar]
- 9.Severi G, Shannon BA, Hoang HN, Baglietto L, English DR, Hopper JL, Pedersen J, Southey MC, Sinclair R, Cohen RJ, Giles GG. Plasma concentration of Propionibacterium acnes antibodies and prostate cancer risk: results from an Australian population-based case-control study. Br J Cancer. 2010;103(3):411–415. doi: 10.1038/sj.bjc.6605757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutcliffe S, Giovannucci E, Isaacs WB, Willett WC, Platz EA. Acne and risk of prostate cancer. Int J Cancer. 2007;121(12):2688–2692. doi: 10.1002/ijc.23032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galobardes B, Smith GD, Jeffreys M, Kinra S, McCarron P. Acne in adolescence and cause-specific mortality: lower coronary heart disease but higher prostate cancer mortality. Am J Epidemiol. 2005;161(12):1094–1101. doi: 10.1093/aje/kwi147. [DOI] [PubMed] [Google Scholar]
- 12.Drott J, Alexeyev O, Bergstrom P, Elgh F, Olsson J. Propionibacterium acnes infection induces upregulation of inflammatory genes and cytokine secretion in prostate epithelial cells. BMC Microbiol. 2010;10(1):126. doi: 10.1186/1471-2180-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mak TN, Fischer N, Laube B, Brinkmann V, Metruccio MM, Sfanos KS, Mollenkopf HJ, Meyer TF, Brüggemann H. Propionibacterium acnes host cell tropism contributes to vimentin-mediated invasion and induction of inflammation. Cell Microbiol. 2012;14(11):1720–1733. doi: 10.1111/j.1462-5822.2012.01833.x. [DOI] [PubMed] [Google Scholar]
- 14.McDowell A, Valanne S, Ramage G, Tunney MM, Glenn JV, McLorinan GC, Bhatia A, Maisonneuve J-F, Lodes M, Persing DH, Patrick S. Propionibacterium acnes types I and II represent phylogenetically distinct groups. J Clin Microbiol. 2005;43(1):326–334. doi: 10.1128/JCM.43.1.326-334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDowell A, Perry AL, Lambert PA, Patrick S. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol. 2008;57(2):218–224. doi: 10.1099/jmm.0.47489-0. [DOI] [PubMed] [Google Scholar]
- 16.Lomholt HB, Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS ONE. 2010;5(8):e12277. doi: 10.1371/journal.pone.0012277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDowell A, Gao A, Barnard E, Fink C, Murray PI, Dowson CG, Nagy I, Lambert PA, Patrick S. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology. 2011;157(7):1990–2003. doi: 10.1099/mic.0.049676-0. [DOI] [PubMed] [Google Scholar]
- 18.Pan SC, Wang JT, Hsueh PR, Chang SC. Endocarditis caused by Propionibacterium acnes: an easily ignored pathogen. J Infect. 2005;51(4):e229–e231. doi: 10.1016/j.jinf.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Nisbet M, Briggs S, Ellis-Pegler R, Thomas M, Holland D. Propionibacterium acnes: an under-appreciated cause of post-neurosurgical infection. J Antimicrob Chemother. 2007;60(5):1097–1103. doi: 10.1093/jac/dkm351. [DOI] [PubMed] [Google Scholar]
- 20.Levy PY, Fenollar F, Stein A, Borrione F, Cohen E, Lebail B, Raoult D. Propionibacterium acnes postoperative shoulder arthritis: an emerging clinical entity. Clin Infect Dis. 2008;46(12):1884–1886. doi: 10.1086/588477. [DOI] [PubMed] [Google Scholar]
- 21.Sfanos K, Harmody D, Dang P, Ledger A, Pomponi S, McCarthy P, Lopez J. A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Syst Appl Microbiol. 2005;28(3):242–264. doi: 10.1016/j.syapm.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186(5):1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huiying L. Metagenomic study of the human skin microbiome associated with acne. Nat Precedings. 2010 http://dx.doi.org/10.1038/npre.2010.5305.1.
- 24.Kilian M, Scholz CFP, Lomholt HB. Multilocus sequence typing and phylogenetic analysis of Propionibacterium acnes. J Clin Microbiol. 2012;50(4):1158–1165. doi: 10.1128/JCM.r06129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapadin AN, Fleischmajer R. Tetracyclines nonantibiotic properties and their clinical implications. Clin Infect Dis. 2006;54(2):258–265. doi: 10.1016/j.jaad.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Montagnini Spaine D, Mamizuka EM, Pereira Cedenho A, Srougi M. Microbiologic aerobic studies on normal male urethra. Urology. 2000;56(2):207–210. doi: 10.1016/s0090-4295(00)00615-4. [DOI] [PubMed] [Google Scholar]
- 27.Willen M, Holst E, Myhre EB, Olsson AM. The bacterial flora of the genitourinary tract in healthy fertile men. Scand J Urol Nephrol. 1996;30(5):387–393. doi: 10.3109/00365599609181315. [DOI] [PubMed] [Google Scholar]
- 28.Nelson DE, Van Der Pol B, Dong Q, Revanna KV, Fan B, Easwaran S, Sodergren E, Weinstock GM, Diao L, Fortenberry JD. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE. 2010;5(11):e14116. doi: 10.1371/journal.pone.0014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon BA, Cohen RJ, Garrett KL. Polymerase chain reaction-based identification of Propionibacterium acnes types isolated from the male urinary tract: evaluation of adolescents, normal adults and men with prostatic pathology. BJU Int. 2006;98(2):388–392. doi: 10.1111/j.1464-410X.2006.06273.x. [DOI] [PubMed] [Google Scholar]
- 30.Kloos WE, Bannerman TL. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7(1):117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickel CJ, Costerton JW. Coagulase-negative Staphylococcus in chronic prostatitis. J Urol. 1992;147:389–401. doi: 10.1016/s0022-5347(17)37247-6. [DOI] [PubMed] [Google Scholar]
- 32.Brook I. Bacteria from, solid tumours. J Med Microbiol. 1990;32(3):207–210. doi: 10.1099/00222615-32-3-207. [DOI] [PubMed] [Google Scholar]