Abstract
Clostridium cellulovorans has been one of promising microorganisms to use biomass efficiently; however the basic metabolic pathways have not been completely known. We carried out 13C-isotopomer-based target metabolome analysis, or carbohydrate conversion process analysis, for more profound understanding of metabolic pathways of the bacterium. Our findings that pyruvate + oxaloacetate, fumarate, and malate inside and outside cells exhibited 13C incorporation suggest that C. cellulovorans exactly fixed CO2 and partly operated the TCA cycle in a reductive manner. Accompanied with CO2 fixation, the microorganism was also found to produce and secrete lactate. Overall, our study demonstrates that a part of C. cellulovorans metabolic pathways related to glycolysis and the TCA cycle are involved in CO2 fixation.
Keywords: CO2 fixation, Clostridium cellulovorans, Target metabolomics
Introduction
C. cellulovorans, an anaerobic mesophilic bacterium, can degrade and assimilate not only various kinds of carbohydrates (including cellulose, xylan, pectin, cellobiose, glucose, fructose, galactose, and mannose) and but also actual biomass (rice straw and corn waste) (Tamaru et al. 2010b), and whose whole genome was recently sequenced for the first time by our group (Tamaru et al. 2010a). This wide spectrum of degradation depends on extracellular multi-protein complexes called cellulosomes in several cellulosome-producing Clostridium species reported; however, most of the researches focus on the cellulosome itself. In order to use Clostridium species for practical applications, it is important to elucidate the basic biology of these bacteria, especially their metabolic processes that are highly associated with the conversion of carbohydrates to final products.
C. cellulovorans has been suggested to have a CO2 fixation pathway, because of its ability to grow under a higher concentration of ‘100%’ CO2 compared to other Clostridium species (an atmosphere of 20% CO2 (C. cellulovorans); 5% CO2 (C. acetobutylicum and C. kluyveri); 10% CO2 (C. thermocellum and C. difficile) (Sleat et al. 1984; Amador-Noguez et al. 2010; Waller et al. 2013; Saujet et al. 2011; Thauer et al. 1968). Previously, a few studies have characterized the metabolic pathway of C. kluyveri and C. acetobutylicum (Jungermann et al. 1970; Amador-Noguez et al. 2010). In the genome analysis of C. cellulovorans (Tamaru et al. 2010a), the genes of 2 important CO2 fixation enzymes, namely pyruvate:ferredoxin oxidoreductase (PFOR) and phosphoenolpyruvic acid (PEP) carboxylase (PEPC) were annotated. Notably, PFOR of glycolysis and PEPC of the TCA cycle are both in the node of main metabolic pathways in C. cellulovorans. Therefore, the study of CO2 fixation by metabolome analysis would help to clarify the complete metabolic pathway of C. cellulovorans. In particular, 13C-labeling studies of metabolic products are useful for understanding the in vivo metabolism since 13-carbon isotope can distinguish fluxes through different pathways when these fluxes result in different positional isotopic enrichments in metabolic intermediates (Ratcliffe and Shachar-Hill 2006; McKinlay et al. 2007).
As illustrated in Figure 1, we carried out labeling experiments of metabolic intermediates by allowing C. cellulovorans to grow in medium with an atmosphere of ‘100%’ CO2 containing either NaH13CO3 or [U-13C]-glucose as a labeling reagent, followed by the GC/MS analysis. We demonstrated metabolic fluxes of C. cellulovorans and discussed the physiological meaning of CO2 fixation in the metabolic pathway of C. cellulovorans.
Figure 1.
Workflow for the investigation of CO 2 incorporation and the quantification of metabolites of interest.
Materials and methods
Cultivation conditions and growth rate analysis
C. cellulovorans 743B (ATCC 35296) was grown anaerobically at 37°C in an atmosphere of ‘100%’ CO2 unless otherwise noted. Liquid cultivation media contained the following reagents: 0.45 g/l KH2PO4 · H2O, 0.45 g/l K2HPO4, 0.9 g/l NaCl, 0.3675 g/l NH4Cl, 0.1575 g/l MgCl2 · 6H2O, 0.12 g/l CaCl2 · 2H2O, 5.2 mg/l Na2-EDTA, 1.5 mg/l FeCl2 · 4H2O, 0.942 mg/l CoCl2 · 6H2O, 0.85 mg/l MnCl2 · 4H2O, 0.07 mg/l ZnCl2 · 6H2O, 0.062 mg/l H3BO4, 0.036 mg/l Na2MoO4 · 2H2O, 0.024 mg/l NiCl2 · 6H2O, 0.017 mg/l CuCl2 · 6H2O, 5g/l NaHCO3, 4 g/l Bacto™ Yeast Extract (Becton and Dickinson Company), 3 g/l glucose, and 1 g/l L-cysteine. For labeling experiments, NaHCO3 and glucose were replaced by NaH13CO3 and [U-13C]-glucose, respectively (both 99% purity; Cambridge Isotope Laboratories, Andover, MA).
Quenching and extraction of intracellular metabolites
Quenching and metabolite extraction were carried out as previously described (Winder et al. 2008), with some modifications. In brief, culture broths were injected rapidly into 4 volumes of 60% aqueous methanol solution (−40°C) for quenching. Supernatants after centrifugation at 3000 × g at −9°C for 10 min for quenching were removed rapidly, and washed with 1 ml of 60% aqueous methanol (−40°C), followed by centrifugation at 3000 × g at −9°C for 10 min. Subsequently, supernatants were thoroughly removed, and cell pellets were frozen in liquid nitrogen and kept at −80°C until the following extraction procedures. Cell pellets were suspended in 500 μl of 100% methanol (−40°C), frozen in liquid nitrogen, and allowed to thaw on dry ice. After, the freeze-thaw cycle was performed 3 times in total, the suspensions were centrifuged at 16000 × g, at −9°C, for 5 min. Supernatants were retained and stored on dry ice, and another aliquot (500 μl) of 100% methanol (−40°C) was added to each pellet. The procedure was repeated twice, and the second aliquot of methanol was combined with the first one.
Metabolite derivatization
Extract aliquots and cultivation medium supernatants (20 μl each), as well as dilution series of standard mixtures of target metabolites (Table 1), were spiked with internal standards (ribitol, 10 or 1 μg for the extracellular or intracellular analysis, respectively) and lyophilized. Dried samples were subsequently derivatized in 2 stages, as previously described (Tsugawa et al. 2011). For oximation, 100 μl (50 μl for intracellular metabolites) of methoxyamine hydrochloride (Sigma-Aldrich, St. Louis, MO) in pyridine (20 mg/l) (Wako, Tokyo, Japan) was added and incubated at 30°C for 90 min. For trimethylsilylation, 50 μl (25 μl for intracellular metabolites) of N-methyl-N-(trimethylsilyl)trifluoroacetamide (GL Science, Tokyo, Japan) was added and incubated at 37°C for 30 min. Insoluble residues were removed by centrifugation at 12000 × g at 4°C for 5 min, and cultivation supernatants were transferred to clean vials.
Table 1.
Target metabolites detected by GC/MS
| Name | Retention time (min) | Formula | m/z Range |
|---|---|---|---|
| Pyruvate + OAA |
6.34 |
C6H12NO3Si |
174–177 |
| Lactate |
6.72 |
C8H19O3Si2 |
219–222 |
| Succinate |
14.45 |
C9H19O4Si2 |
247–251 |
| Fumarate |
15.31 |
C9H17O4Si2 |
245–249 |
| Malate |
17.42 |
C12H27O5Si3 |
335–339 |
| PEP |
18.39 |
C11H26O6PSi3 |
369–372 |
| Citrate |
19.79 |
C17H37O7Si4 |
465–471 |
| Ribitol (i.s.a) | 19.32 | C19H49O5Si5 | 219 |
(i.s.a), internal standard.
GC/MS analysis and data processing
Derivatized metabolites were analyzed using GCMS-QP2010 Ultra (Shimadzu, Kyoto, Japan) equipped with a 30 m × 0.25 mm i.d. fused silica capillary column coated with 0.25-μm CP-SIL 8 CB low bleed (Agilent Technologies, Santa Clara, CA). Aliquots (1 μl) were injected in the split mode (25/1, supernatant analysis; 5/1, intracellular analysis) at 230°C, using helium as carrier gas at a flow rate of 1.12 ml/min. The column temperature was held at 80°C for 2 min isothermally, raised to 130°C (4°C/min) and then to 330°C (25°C/min), and maintained for 6 min isothermally. The interface and MS source temperatures were 250°C and 200°C, respectively, and the ion voltage was 1 kV. Data were collected by GCMS solution (Shimadzu), and identified metabolites are shown in Table 1. Mass isotopomer distributions were corrected for natural isotope abundance as previously described (Nanchen et al. 2007). The GC/MS analysis was performed on 3 biological replicates of each sample.
Results
CO2 incorporation into C. cellulovorans metabolites
According to previous reports that CO2 was required for culturing some Clostridium species, we speculate that C. cellulovorans also has the activity of CO2 fixation. Our speculation is further supported by the fact that C. cellulovorans, whose genes related to CO2 fixation were also annotated in the genome of C. cellulovorans (Tamaru et al. 2010a), can be cultivated in media containing higher CO2 concentrations, even at ‘100%’, compared to other Clostridium species. Therefore, in this study, we cultivated C. cellulovorans in media containing NaH13CO3 instead of NaHCO3 and then examined a massive number of metabolites derived from C. cellulovorans and cultivation supernatants using GC/MS. Figure 2 shows the ratios of each metabolite in media containing either NaHCO3 or NaH13CO3. Higher values of relative fractions when C. cellulovorans was cultivated in media containing NaH13CO3 indicate that 13C atoms derived from NaH13CO3 were incorporated into specific metabolites. Our results demonstrate that when C. cellulovorans was cultivated in media containing NaH13CO3, the relative fractions of pyruvate + oxaloacetate (OAA), lactate, fumarate, and malate inside (Figure 2a) and outside (Figure 2b) cells were significantly higher than those from C. cellulovorans cultivated in media containing NaHCO3. Based on these findings, C. cellulovorans evidently is able to incorporate 13C atoms into abovementioned metabolites, and therefore has the ability to fix CO2.
Figure 2.
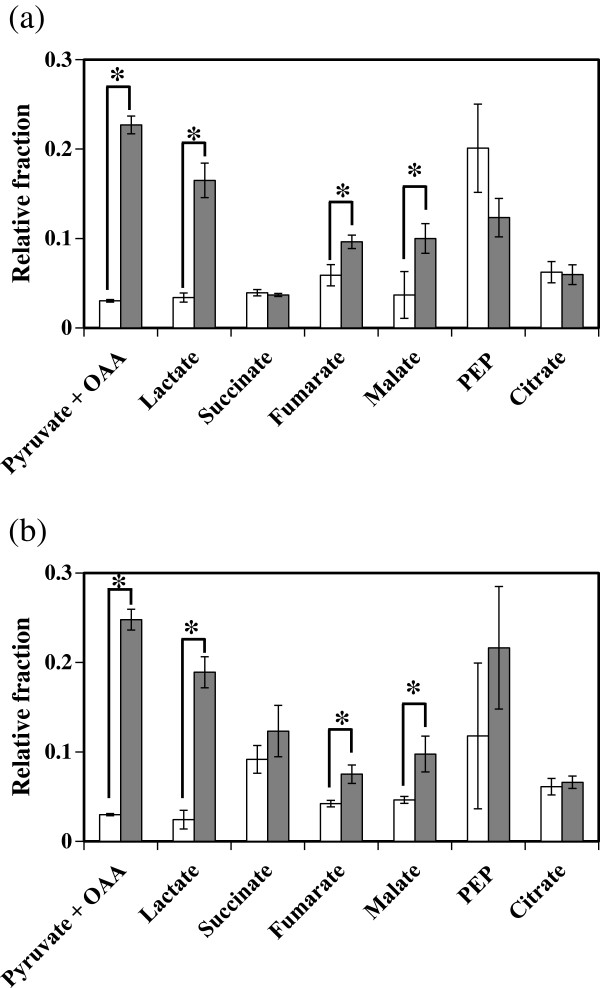
Incorporation of CO2 into several metabolites inside and outside of the cells. The vertical axis represents the relative fraction of 13C-labeled metabolites, which was calculated as follow as: m1/ , where m1 and mi are the corrected intensities and n is the number of carbon atoms in metabolites (n = 3 in the case of lactate, for example). (a) Intracellular metabolites. (b) Extracellular metabolites. White and gray bars represent the ratios of each metabolite in media containing NaHCO3, and NaH13CO3, respectively. Error bars and asterisks indicate standard deviations and significant difference (*P < 0.01), respectively.
Glucose metabolism into metabolic pathway intermediates
Next, to understand the whole strategy of glucose metabolism in C. cellulovorans, we examined a massive number of metabolites inside bacterial cells that were cultivated in media containing [U-13C]-glucose. In this way, there is a report how metabolites flow in metabolic pathway of C. acetobutylicum have been analyzed (Amador-Noguez et al. 2010). To more understand metabolites flow in C. cellulovorans, we observed how 13C atoms were incorporated into some metabolites. The results shown in Figure 3 indicate that 13C atoms derived from [U-13C]-glucose were incorporated into pyruvate + OAA, lactate, fumarate, and malate inside the cells. These results also demonstrate the following 4 points. First, both PFOR and PEPC fixed CO2. It is because that pyruvate had only two 13C atoms of three carbons (Figure 3). The results indicated that pyruvate was converted from acetyl-CoA associated with CO2 fixation once pyruvate became acetyl-CoA, which is constructed two carbons in acetyl group. In the same way, as malate and fumarate had three 13C atoms, they could be prepared from PEP by PEPC associated with CO2 fixation. Second, PFOR initiated the reversible conversion of pyruvate to acetyl-CoA. Third, the amount of PEP flowing into the TCA cycle could be much less than that flowing into pyruvate, acetyl-CoA, and lactate. Fourth, under this condition, 13C atoms were not incorporated into succinate and citrate.
Figure 3.
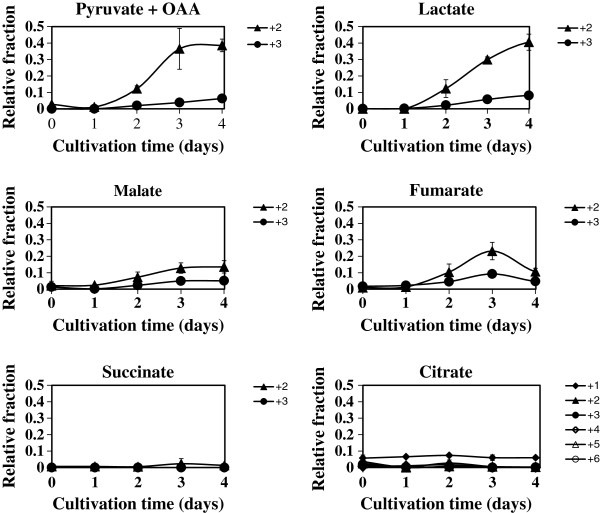
Dynamic incorporation of [U-13C]-glucose into target metabolites. The abscissa and vertical axes represent the cultivation time of C. cellulovorans and the relative fraction of indicated labeled compounds. ' + 1', ' + 2', ' + 3', ' + 4', ' + 5', and ' + 6' mean the number of 13-carbon isotope in each metabolite incorporated from [U-13C]-glucose. Error bars represent standard deviations.
Lactate secretion accompanied with CO2 fixation
As shown in Figures 2 and 3, a flux of lactate was observed in C. cellulovorans, in agreement with the previous report (Sleat et al. 1984). Therefore, we checked the amount of secreted lactate by C. cellulovorans cultivated in media containing NaH13CO3 (Figure 4a). We further calculated the percentage of 13C incorporation into secreted lactate. The results show that, accompanied with CO2 fixation, C. cellulovorans produced lactate at a constant rate after 2 days (Figure 4b).
Figure 4.
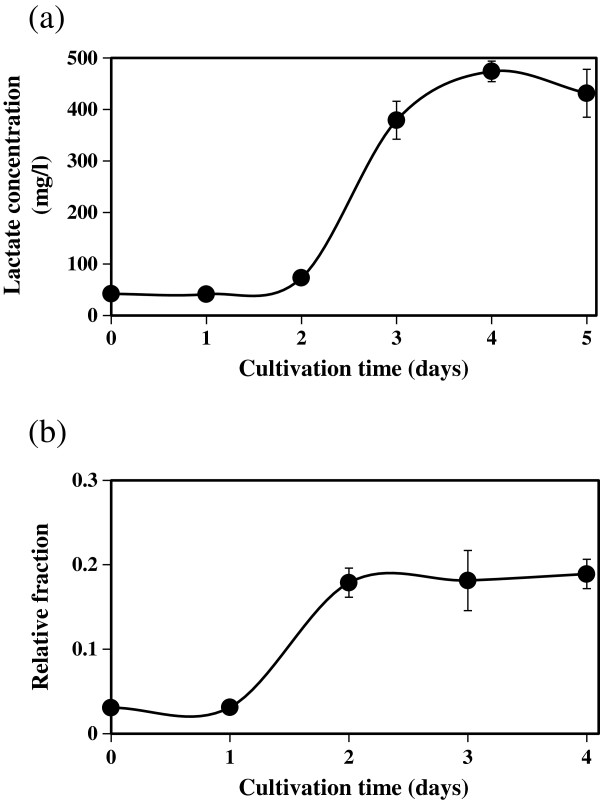
Lactate secretion accompanied with CO2 fixation. (a) The time course of lactate concentration in cultivation supernatants. (b) The time course of relative fraction of lactate in cultivation supernatants. Relative fractions were calculated as described in Figure 2. Error bars represent standard deviations.
Discussion
Using target metabolomics, we demonstrate here that C. cellulovorans produces lactate, malate, and fumarate. As illustrated in the metabolic map, including the TCA cycle of C. cellulovorans (Figure 5), we propose that C. cellulovorans produces lactate accompanied with CO2 fixation and generates fumarate by partly operating the TCA cycle in a reductive manner (Figure 5a). The reason why C. cellulovorans operates these metabolic pathways (lactate and fumarate production and CO2 fixation, except for the PEPC reaction) could be the preservation of redox balance in the cell. That is, the reactions of lactate and malate production (operated by lactate dehydrogenase and malate dehydrogenase, respectively) might be accompanied with the regeneration of 1 molecule of NAD(P)+. In addition, the reaction of CO2 fixation by PFOR produces oxidized ferredoxin, and a molecule of oxidized ferredoxin subsequently produces 2 molecules of NAD+. These reactions may help oxidizing agents to be used in glycolysis. We also examined the existence of CO2 fixation enzymes (PFOR and PEPC) by the proteome analysis (data not shown). Compared to the flux from glycolysis to the TCA cycle, the flux to lactate could be dominant, since our results show that higher amounts of 13C atoms were incorporated into lactate, but not malate and fumarate (Figure 3).
Figure 5.
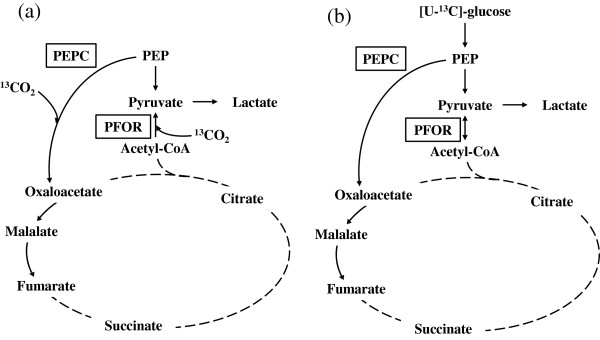
Proposed pathway for CO2 incorporation in C. cellulovorans. Possible metabolic maps of C. cellulovorans in media containing NaH13CO3(a) and [U-13C]-glucose (b). Solid lines, possible pathways with directions indicated by arrows; dashed lines, impossible pathways in our studies. Lactate is produced from pyruvate, whereas fumarate is generated from oxaloacetate and malate. CO2 is fixed by PFOR and PEPC. In contrast, no citrate is produced from oxaloacetate.
Our findings also indicate that little citrate and succinate was produced from glucose (Figure 3). Isocitrate dehydrogenase, which operates downstream of citrate in the TCA cycle and operates in an oxidative manner with NAD(P)+, could not be used. It is known that citrate is produced from glutamate in some organisms. The metabolic information of C. acetobutylicum (Amador-Noguez et al. 2010) also suggested that C. cellulovorans could use amino acids (glutamate/glutamine) to make other metabolites. If C. cellulovorans produces citrate from glutamate, redox balance would be better maintained because glutamate dehydrogenase or glutamate synthase uses NADP+ and isocitrate dehydrogenase uses NADPH. In particular, when C. cellulovorans lives under a reductive condition, such a pathway is more reasonable than the pathway of citrate production from acetyl-CoA in an oxidative manner. To examine this hypothesis in the future investigation, it will be a promising approach to study how 13C atoms are incorporated into metabolites when using media containing 13C-labeled glutamate. Some other amino acids may be needed to maintain the metabolic pathway in C. cellulovorans, because the bacterium cannot be cultivated in media without yeast extract, which has glutamate (Sleat et al. 1984).
As mentioned above, we speculate that C. cellulovorans could use the mechanism to maintain redox balance, because the oxidizability (the ability to oxygenate other metabolites) is valuable for the condition which was absent from O2. C. cellulovorans lives under anaerobic conditions because photosynthesis is not operated under the natural growth condition of C. cellulovorans (wood chip). It has been reported that CO2 fixation is useful to maintain redox balance in microorganisms that have the Calvin cycle or TCA cycle (McKinlay and Harwood 2010); therefore CO2 fixation could be a common mechanism to regulate redox balance.
Notably, lactate production in cultivation supernatants after 4 days was 474 mg/l (= 31.6 mol per 100 mol glucose), which is about twice of that reported in the previous study (Sleat et al. 1984), where cellobiose was used as a carbon source.
Here, we demonstrated that, accompanied with CO2 fixation, C. cellulovorans produced several kinds of organic acids and that the TCA cycle was partly operated in a reductive manner at the metabolite level. These presented results would provide important information for the application of C. cellulovorans as industrial cellulosome-producing bacteria.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MS, HM, HM, YT, EF, KK and MU conceived and designed the study. MS, HS, HM and EF analyzed the data. MS, HS, HM, KK and MU drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Masahiro Shinohara, Email: shinohara.masahiro.23m@st.kyoto-u.ac.jp.
Hiroshi Sakuragi, Email: hsaku@kais.kyoto-u.ac.jp.
Hironobu Morisaka, Email: morisaka@kais.kyoto-u.ac.jp.
Hideo Miyake, Email: miyake@bio.mie-u.ac.jp.
Yutaka Tamaru, Email: ytamaru@bio.mie-u.ac.jp.
Eiichiro Fukusaki, Email: fukusaki@bio.eng.osaka-u.ac.jp.
Kouichi Kuroda, Email: k_kuro@kais.kyoto-u.ac.jp.
Mitsuyoshi Ueda, Email: miueda@kais.kyoto-u.ac.jp.
Acknowledgements
This research was supported by CREST and the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN).
References
- Amador-Noguez D, Feng XJ, Fan J, Roquet N, Rabitz H, Rabinowitz JD. Systems-level metabolic flux profiling elucidates a complete, bifurcated tricarboxylic acid cycle in Clostridium acetobutylicum. J Bacteriol. 2010;3:4452–4461. doi: 10.1128/JB.00490-10. doi:10.1128/JB.00490-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungermann KA, Schmidt W, Kirchniawy FH, Rupprecht EH, Thauer RK. Glycine formation via threonine and serine aldolase. Its interrelation with the pyruvate formate lyase pathway of one-carbon unit synthesis in Clostridium kluyveri. Eur J Biochem. 1970;3:424–429. doi: 10.1111/j.1432-1033.1970.tb01097.x. doi:10.1111/j.1432-1033.1970.tb01097.x. [DOI] [PubMed] [Google Scholar]
- McKinlay JB, Harwood CS. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc Natl Acad Sci USA. 2010;3:11669–11675. doi: 10.1073/pnas.1006175107. doi:10.1073/pnas.1006175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinlay JB, Shachar-Hill Y, Zeikus JG, Vieille C. Determining Actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metabolic Eng. 2007;3:177–192. doi: 10.1016/j.ymben.2006.10.006. doi:10.1016/j.ymben.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Nanchen A, Fuhrer T, Sauer U. Determination of metabolic flux ratios from 13C-experiments and gas chromatography–mass spectrometry data: protocol and principles. Meth Mol Biol. 2007;3:177–197. doi: 10.1007/978-1-59745-244-1_11. doi:10.1007/978-1-59745-244-1_11. [DOI] [PubMed] [Google Scholar]
- Ratcliffe RG, Shachar-Hill Y. Measuring multiple fluxes through plant metabolic networks. Plant J Cell mol Biol. 2006;3:490–511. doi: 10.1111/j.1365-313X.2005.02649.x. doi:10.1111/j.1365-313X.2005.02649.x. [DOI] [PubMed] [Google Scholar]
- Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol. 2011;3:3186–3196. doi: 10.1128/JB.00272-11. doi:10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat R, Mah RA, Robinson R. Isolation and characterization of an anaerobic, cellulolytic bacterium. Clostridium cellulovorans sp. nov. Appl Environ Microbiol. 1984;3:88–93. doi: 10.1128/aem.48.1.88-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru Y, Miyake H, Kuroda K, Nakanishi A, Kawade Y, Yamamoto K, Uemura M, Fujita Y, Doi RH, Ueda M. Genome sequence of the cellulosome-producing mesophilic organism Clostridium cellulovorans 743B. J Bacteriol. 2010;3:901–902. doi: 10.1128/JB.01450-09. doi:10.1128/JB.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru Y, Miyake H, Kuroda K, Ueda M, Doi RH. Comparative genomics of the mesophilic cellulosome-producing Clostridium cellulovorans and its application to biofuel production via consolidated bioprocessing. Environ Technol. 2010;3:889–903. doi: 10.1080/09593330.2010.490856. doi:10.1080/09593330.2010.490856. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Henninger H, Wenning J, Decker K. The energy metabolism of Clostridium kluyveri. Eur J Biochem. 1968;3:173–180. doi: 10.1111/j.1432-1033.1968.tb00189.x. doi:10.1111/j.1432-1033.1968.tb00189.x. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Bamba T, Shinohara M, Nishiumi S, Yoshida M, Fukusaki E. Practical non-targeted gas chromatography/mass spectrometry-based metabolomics platform for metabolic phenotype analysis. J Biosci Bioeng. 2011;3:292–298. doi: 10.1016/j.jbiosc.2011.05.001. doi:10.1016/j.jbiosc.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Waller BH, Olson DG, Currie DH, Guss AM, Lynd LR. Exchange of type II dockerin-containing subunits of the Clostridium thermocellum cellulosome as revealed by SNAP-tags. FEMS Microbiol Letters. 2013;3:46–53. doi: 10.1111/1574-6968.12029. doi:10.1111/1574-6968.12029. [DOI] [PubMed] [Google Scholar]
- Winder CL, Dunn WB, Schuler S, Broadhurst D, Jarvis R, Stephens GM, Goodacre R. Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Analytical Chem. 2008;3:2939–2948. doi: 10.1021/ac7023409. doi:10.1021/ac7023409. [DOI] [PubMed] [Google Scholar]


