Abstract
The completion of the human genome reference sequence ushered in a new era for the study and discovery of human transposable elements. It now is undeniable that transposable elements, historically dismissed as junk DNA, have had an instrumental role in sculpting the structure and function of our genomes. In particular, long interspersed element-1 (LINE-1 or L1) and short interspersed elements (SINEs) continue to affect our genome, and their movement can lead to sporadic cases of disease. Here, we briefly review the types of transposable elements present in the human genome and their mechanisms of mobility. We next highlight how advances in DNA sequencing and genomic technologies have enabled the discovery of novel retrotransposons in individual genomes. Finally, we discuss how L1-mediated retrotransposition events impact human genomes.
Keywords: retrotransposon, polymorphism, transposable elements, genome diversity, mutation
INTRODUCTION
Approximately 45% of the human genome is derived from transposable elements (134). These include DNA transposons, long terminal repeat (LTR) retrotransposons, and non-LTR retrotransposons. Although most transposable elements have been rendered inactive through mutation, long interspersed element-1 (LINE-1 or L1) retrotransposition continues to diversify human genomes.
L1s comprise ~17% of human DNA (134). The L1-encoded proteins (ORF1p and ORF2p) can mobilize nonautonomous retrotransposons, other noncoding RNAs, and messenger RNAs, leading to the generation of processed pseudogenes (35, 62, 72, 81, 86, 102, 249). Thus, L1-mediated retrotransposition has generated a third of our genome.
In 1988, an examination of 240 unrelated males afflicted with hemophilia A revealed that independent mutagenic L1 insertions into exon 14 of the Factor VIII gene were responsible for the disease in two individuals (120). Heroic efforts to isolate an active progenitor L1 (67) and the development of a cultured cell retrotransposition assay (172) then helped elucidate the molecular mechanism of L1 retrotransposition. It now is apparent that L1s are alive and well in human populations, and that L1-mediated retrotransposition events account for approximately 1 of every 1,000 spontaneous, disease-producing insertions in man (40, 119).
Several reviews have discussed aspects of human retrotransposon biology and how the host genome defends itself from retrotransposon activity (6, 18, 51, 89, 182, 188). Here, we discuss recent progress in understanding the mechanism of L1 retrotransposition. We then highlight how new genomic technologies have illuminated the impact of L1-mediated retrotransposition events on human genetic variation and genome structure.
MOBILE ELEMENTS IN HUMAN GENOMES
Transposable elements are classified by whether they mobilize via a DNA (DNA transposons) or an RNA (retrotransposons) intermediate (Figure 1). They further are distinguished by whether they encode proteins to mediate their own mobility (autonomous elements) or rely upon proteins encoded by other elements (nonautonomous elements).
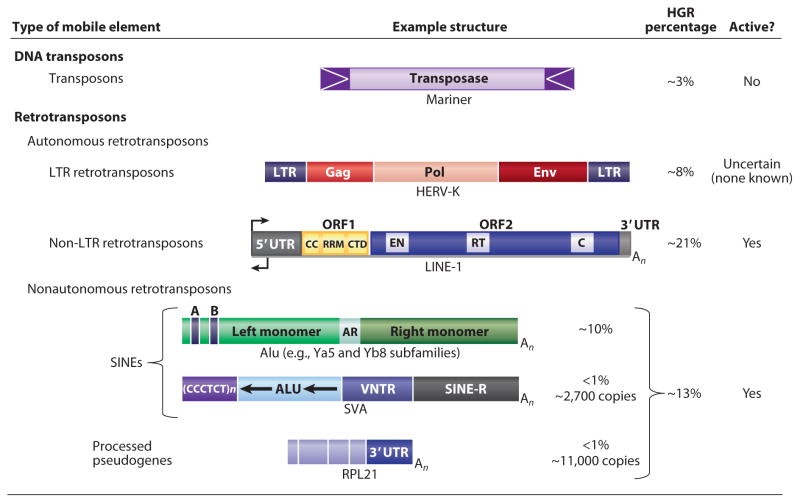
Figure 1.
The classes of mobile genetic elements in the human genome, showing the type of mobile element, the structure of representative elements, the percentage of each element in the human genome reference sequence (HGR), and whether each class of elements is currently active (134). Abbreviations for human endogenous retrovirus-K (HERV-K): LTR, long terminal repeat; Gag, group-specific antigen; Pol, polymerase; Env, envelope protein (dysfunctional). For LINE-1: UTR, untranslated region; CC, coiled coil; RRM, RNA recognition motif; CTD, carboxyl-terminal domain; EN, endonuclease; RT, reverse transcriptase; C, cysteine-rich domain. For Alu: A and B, component sequences of the RNA polymerase III promoter; AR, the adenosine-rich segment separating the 7SL monomers. For SINE-R/VNTR/Alu (SVA): VNTR, variable number of tandem repeats; SINE-R, domain derived from a HERV-K. An signifies a poly(A) tail.
DNA Transposons
DNA transposons generally move via a cut-and-paste mechanism. They tend to have a limited life span in higher eukaryotic genomes (134, 220), which likely is due to the accumulation of nonautonomous deletion derivatives that compete for the transposase encoded by autonomous elements. In most cases, transposase binds at DNA transposon inverted repeat sequences, “cuts” the transposon from its existing location, and then “pastes” it into a new genomic location (reviewed in 55).
DNA transposons comprise ~3% of the human genome reference sequence (HGR, NCBI 36/hg18). Sequence divergence among paralogous copies indicates that virtually all DNA transposons mobilized prior to the eutherian radiation, whereas a composite method indicates they have been extinct in the primate lineage for at least 37 million years (134, 192). Nevertheless, DNA transposons have had an enduring effect on the human genome. For example, the recombination activating genes, RAG1 and RAG2, which are critical for V(D)J recombination and immune system development, likely were domesticated from the Transib family of DNA transposons ~500 Mya (117, 262).
Engineered DNA transposons have practical applications, and can be exploited for useful purposes. For example, a reanimated salmon DNA transposon, Sleeping Beauty, has been used to discover genes implicated in cancer progression and shows promise as a vector in gene therapy studies (48, 96, 112, 113). Similarly, an insect DNA transposon, piggyBac, has been used to create gene-specific knockouts in mouse embryonic stem cells (65, 226). Finally, a zebrafish transposon, Tol2, shows promise as a mutagen in both the mouse and zebrafish germline (118, 121).
Retrotransposons
Retrotransposons mobilize via an RNA intermediate by a copy-and-paste mechanism and remain active in most mammalian genomes. They can be subdivided into two general classes, depending on whether they contain or lack LTRs (Figure 1).
Long terminal repeat retrotransposons: human endogenous retroviruses
LTR-containing elements, such as human endogenous retroviruses (HERVs), resemble retroviruses in both their structure and mobility mechanism. Most HERVs contain a nonfunctional envelope (ENV) gene, which relegates them to an intracellular existence (12).
HERVs and their nonautonomous derivatives comprise ~8% of the human genome (134). Virtually all HERVs are retrotransposition defective; however, a small number of HERV-K elements [where K denotes the host lysine transfer RNA (tRNA) that presumably initiates HERV-K (−) strand complementary DNA (cDNA) synthesis] are polymorphic with respect to presence/absence status in humans, indicating that they have retrotransposed since the human-chimpanzee divergence (22, 125, 148, 176). Moreover, some HERV-K elements contain intact open reading frames (ORFs) (160), and a reanimated HERV-K virus is infectious in cultured cell assays (63, 136). Thus, it is formally possible that rare HERV-K alleles retain the ability to move in modern humans.
Recent studies indicate that HERV-K retrotransposons are expressed in certain tumors, and chromosomal rearrangements involving HERV-K sequences have been implicated in prostate cancer (176, 234). HERV expression also has been implicated in the etiology of certain autoimmune and neurological diseases; however, these data remain controversial, and causal links between HERV expression and disease require additional experiments (176).
Although apparently immobile in humans, the remnants of endogenous retroviruses also can impact the function of mammalian genomes. A growing number of examples indicate that cis-acting sequences derived from endogenous retroviruses can play roles in the transcription, splicing, and/or epigenetic regulation of endogenous genes (47, 151, 165, 198). Moreover, sequences derived from endogenous retroviruses have been exapted by mammalian genomes, and now play important roles in placental development [e.g., Syncytin and peg10 (164, 186)].
Non–long terminal repeat retrotransposons: LINE-1 elements
L1s are the only known autonomously active human retrotransposons, and account for approximately one-sixth of our genome (134). Over 99.9% of L1s have been rendered inactive by 5′ truncations, inversions, and/or point mutations within the two L1-encoded ORFs (95, 134, 189). However, a consensus sequence derived from human genomic L1s suggested the existence of full-length, retrotransposition-competent L1s (RC-L1s) (208). Indeed, the subsequent isolation of the progenitors of mutagenic L1 insertions into the Factor VIII and dystrophin genes revealed that a cohort of L1s continue to mobilize in our genome (67, 106).
The structure and mobility mechanism of retrotransposition-competent LINE-1s
RC-L1s are ~6 kb in length and contain a 5′ untranslated region (UTR), two ORFs, and a 3′ UTR that is punctuated by a poly(A) tail (208) (Figure 1). The L1 5′ UTR houses an internal RNA polymerase II promoter that directs transcription from the 5′ end of the element (227); it also contains cis-acting binding sites for multiple transcription factors (5, 16, 133, 169, 232, 259).
Recent studies have demonstrated that the L1 5′ UTR contains a potent antisense promoter (L1 ASP). Transcription from the L1 ASP can lead to chimeric transcripts that contain a portion of the L1 5′ UTR and genomic sequences flanking the 5′ end of the L1 (181, 223). It is further speculated that these chimeric transcripts may either function in gene regulation or promote the formation of double-stranded L1 RNAs that regulate L1 retrotransposition by RNA interference-based mechanisms (159, 258). Interestingly, L1 ASP-derived chimeric transcripts have proven useful in the identification of expressed L1s from human embryonic stem cells, embryonic carcinoma cell lines, and somatic human tissues (77, 149, 223).
Human ORF1 encodes a ~40-kDa protein (ORF1p) required for L1 retrotransposition (107, 172). ORF1p has an amino-terminal coiled-coil domain (107), a centrally located RNA recognition motif, and a basic carboxyl-terminal domain (123, 172). Biochemical experiments with mouse and human ORF1p demonstrate that the amino-terminal coiled-coil domain facilitates ORF1p trimer formation (123, 155). Structural and biochemical analyses also suggest that the RNA recognition motif and carboxyl-terminal domains play critical roles in ORF1p binding to nucleic acids (13, 114, 123, 128). Finally, ORF1p has nucleic acid chaperone activity, which may be important for L1 integration (156).
ORF2 encodes a ~150-kDa protein (ORF2p), which has endonuclease and reverse transcriptase activities that are critical for L1 retrotransposition (71, 78, 152, 158, 172). ORF2p also contains a cysteine-rich domain of unknown function near its carboxyl terminus that is required for retrotransposition (75, 172). Recent experiments suggest that human ORF2 translation occurs by an unconventional termination-reinitiation mechanism, and that the putative ORF2 AUG initiation codon is dispensable for translation, although it is conserved in all human L1s examined thus far (1, 66, 163).
The analysis of mutagenic L1 insertions in conjunction with biochemical and genetic assays has demonstrated that human ORF1p and ORF2p preferentially associate with their encoding messenger RNA (mRNA) (67, 72, 132, 249). This association, termed cis-preference, leads to the generation of an L1 ribonucleoprotein (RNP) particle (105, 131, 153). ORF1p is detectable in cytoplasmic RNPs from embryonic stem cells, embryonic carcinoma cell lines, and HeLa cells that overexpress engineered L1 elements (82, 83, 105, 131, 153). ORF1p and/or L1 RNA also have been detected in human oocytes, in some human somatic cells, and at select times during human and mouse germ cell development (21, 29, 54, 85, 236). In contrast, ORF2p appears to be much less abundant than ORF1p in L1 RNPs, and its detection has relied largely upon an assay to detect L1 reverse transcriptase activity in RNPs (132). However, epitope-tagging strategies recently have allowed reliable ORF2p detection in RNPs derived from cells transfected with L1 expression constructs by both Western blotting and immunofluorescence (68, 90). L1 RNPs also associate with stress granules, although determining whether this association is important for L1 retrotransposition requires further study (68, 93).
Experiments using adenovirus-based vectors suggest that L1 retrotransposition can occur in the absence of cell division (130). Thus, some components of L1 RNPs may enter the nucleus in the absence of nuclear envelope breakdown. L1 retrotransposition then likely occurs by target-site primed reverse transcription (TPRT) (53, 78, 144). During TPRT, the L1-encoded endonuclease generates a single-strand endonucleolytic nick in genomic DNA to expose a 3′-OH (78). The liberated 3′-OH is then used as a primer by the L1 reverse transcriptase to initiate cDNA synthesis using the L1 mRNA as a template (53, 78, 132). Molecular details regarding second-strand target-site cleavage and second-strand L1 cDNA synthesis require elucidation, but insights about both these steps in TPRT have arisen from studying a related retrotransposon, R2, from Bombyx mori (45). The process of TPRT generates a new L1 copy that generally is flanked by ~7–20-bp target-site duplications (TSDs) (119) (Figure 2).
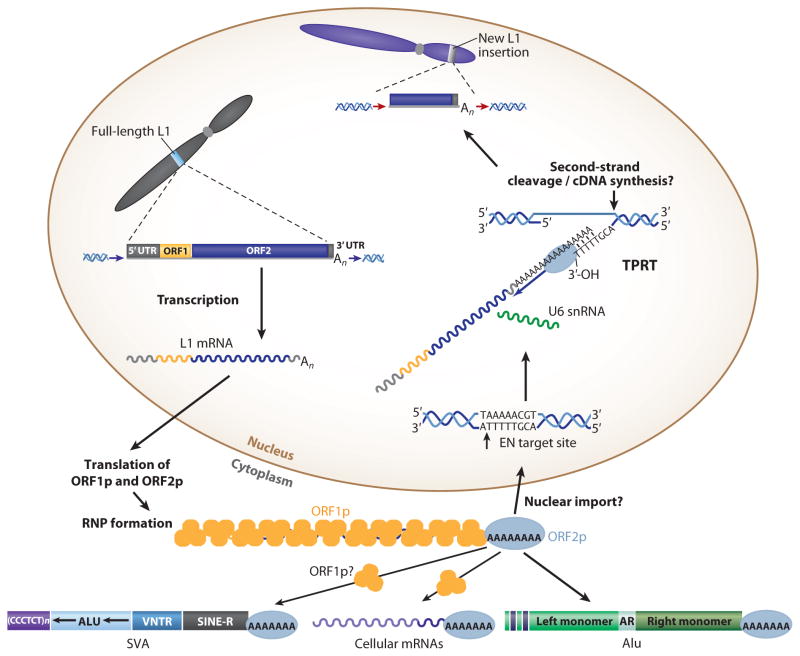
Figure 2.
A LINE-1 retrotransposition cycle. A full-length L1 (light blue bar on gray chromosome) is transcribed, the L1 messenger RNA (mRNA) is exported to the cytoplasm, and translation of ORF1p ( yellow circles) and ORF2p (blue oval ) leads to ribonucleoprotein (RNP) formation. Components of the L1 RNP are transported to the nucleus, and retrotransposition occurs by target-site primed reverse transcription (TPRT). During TPRT, the L1 endonuclease (EN) nicks genomic DNA, exposing a free 3′-OH that can serve as a primer for reverse transcription of the L1 RNA. The processes of second-strand cleavage, second-strand complementary DNA (cDNA) synthesis, and completion of L1 integration require elucidation. TPRT results in the insertion of a new, often 5′-truncated L1 copy at a new genomic location ( gray bar on purple chromosome) that generally is flanked by target-site duplications (red arrows). Alu, SINE-R/VNTR/Alu (SVA), and cellular mRNAs may hijack the L1-encoded protein(s) in the cytoplasm to mediate their trans mobilization. U6 small nuclear RNA (snRNA) may be integrated with L1 during TPRT. Question marks denote steps in the retrotransposition pathway of unknown mechanism.
Nonautonomous retrotransposons: Alu and SVA elements
The human genome also contains numerous nonautonomous retrotransposons that rely upon activity of L1-encoded proteins to mediate their mobility. These nonautonomous elements consist primarily of the small interspersed element (SINE) Alu, which accounts for ~10% of sequence in the HGR (134).
Alu elements arose in mammalian genomes ~65 Mya and contain two monomeric sequences derived from the signal recognition particle (SRP) 7SL RNA (14, 238). Active Alus are ~280 base pairs (bp) in length and end in an A-rich tail. The left monomer contains an internal RNA polymerase III promoter (14) and is separated from the right monomer by an adenosine-rich sequence (Figure 1). However, flanking genomic sequences also can influence Alu transcriptional initiation and termination (46, 49, 64, 91, 143, 239). Like L1s, Alus can be stratified into subfamilies (60). Recent computational analyses and studies in cultured cells suggest that there may be thousands of active Alu “core” elements in the HGR (24, 52). However, Alu Y elements, most notably Ya5 and Yb8 subfamily members, account for the vast majority of disease-producing insertions in humans (38).
Alu elements rely upon L1 ORF2p to facilitate their retrotransposition in trans (62). Mutations that either block Alu transcription or interfere with SRP9/14 protein binding adversely affect Alu retrotransposition in vitro (24). Indeed, the ability of the SRP9/14 proteins to interact with Alu RNA may be intimately tied to the evolution of active Alu subfamilies (24, 203).
Alu elements can also be co-opted to play roles in gene expression. For example, inverted Alu elements within the 3′ UTRs of some cellular mRNAs can result in adenosine-to-inosine RNA editing and preferential nuclear retention of the resultant edited mRNAs in differentiated cells (42). Additionally, Alu elements can be incorporated into existing transcription units by a process known as exonization, which may serve to enhance transcriptome diversification (139, 211, 217). Finally, the poly(A) tails flanking Alu elements may serve as a source for generating microsatellite sequences in human DNA (3).
SINE-R/VNTR/Alu (SVA) elements arose in primate lineages ~25 Mya and are present at ~2,700 copies in the human genome (244). They have a composite structure consisting of a variable-length hexameric repeat (CCCTCT)n that is sequentially followed by an inverted Alu-like sequence, a variable number of tandem repeats (VNTR) region, a sequence derived from the 3′ end of a HERV-K10 element (SINE-R), and a poly(A) tail (185, 187, 216) (Figure 1). Whether SVA elements contain functional promoter sequences remains an open question; however, it is likely that SVA elements are transcribed by RNA polymerase II and that the resultant SVA RNAs are mobilized to new genomic locations by the L1-encoded proteins (58, 101, 102, 187) (Figure 2). Indeed, the recent development of a cell-based assay for SVA mobilization should be instrumental in deciphering mechanistic details of SVA trans mobilization (102).
Nonautonomous retrotransposons: mobilization of cellular RNAs
Cellular messenger RNAs can occasionally use the L1-encoded proteins to mobilize to new genomic locations, thereby generating processed pseudogenes (72, 150, 249) (Figure 1). There are ~8,000–15,000 processed pseudogene copies in the HGR, and most are derived from genes that are highly expressed in the germline, such as housekeeping genes and ribosomal protein genes (235, 260, 261). Interestingly, some ribosomal protein processed pseudogenes (e.g., RPL21) occur at a relatively high copy number, suggesting that some property of these mRNAs allows them to recruit the L1-encoded proteins more effectively than other mRNAs (260).
Most processed pseudogenes are dead on arrival because they lack a functional promoter (242, 250). Thus, they can be used as molecular clocks to estimate mutation rates between species (94). However, some human processed pseudogenes are expressed, and a small number may encode functional genes or serve as sources of small interfering RNAs (siRNAs) with gene regulatory functions (103, 230, 242).
To date, there are no examples of de novo processed pseudogene retrotransposition events causing human disease. However, the expression of a processed pseudogene has been implicated in human facioscapulohumeral dystrophy patients (138, 222). Similarly, expression of an FGF4 processed pseudogene is associated with chondrodysplasia in 19 dog breeds, consistent with the idea that selective breeding can enrich for rare mutagenic L1-mediated insertion alleles (193).
The L1-encoded proteins also can mobilize other noncoding cellular RNAs, such as U6 small nuclear RNA (snRNA), to new genomic locations (35). Computational and experimental evidence suggests that the L1 reverse transcriptase can switch templates to the U6 snRNA during TPRT to generate U6/L1, and less frequently U6/processed pseudogene chimeras (33, 35, 81, 86) (Figure 2). Interestingly, some U6 pseudogenes, small uracil-rich RNAs (e.g., U1 snRNA), and small nucleolar RNAs (e.g., U3 snoRNA) end in poly(A) tails and are flanked by TSDs, suggesting that they also may have been mobilized by the L1-encoded proteins (23, 61, 81, 240, 248).
TECHNOLOGIES TO IDENTIFY HUMAN-SPECIFIC LINE-1s
Overview
The ability to discriminate human-specific L1-mediated retrotransposition events from the sheer mass of defective retrotransposons in the genome is akin to finding a needle in a haystack. However, an elegant combination of phylogenetic, molecular biological, computational, and modern genomic technologies has revolutionized our ability to identify human-specific L1-mediated retrotransposition events in both reference sequences and individual genomes (Figure 3). Some of the seminal findings allowing these advances are discussed below.
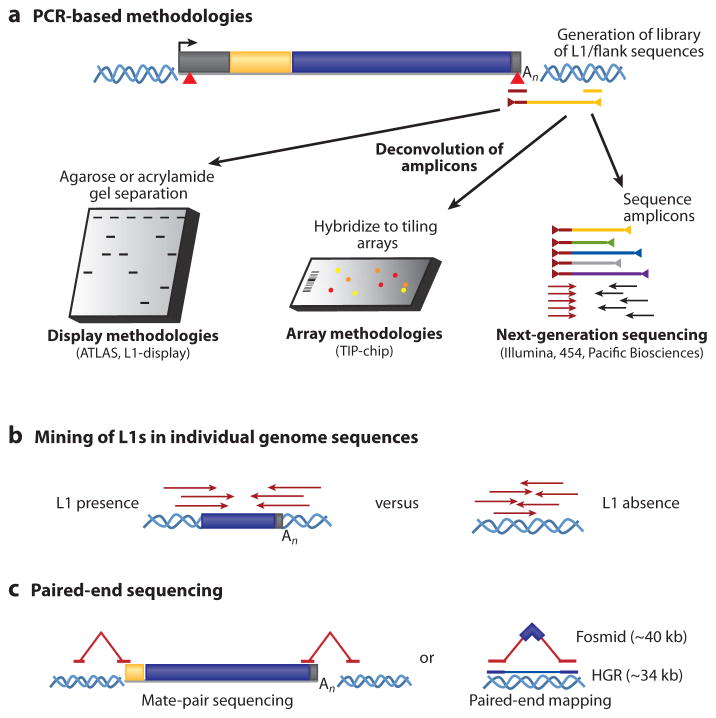
Figure 3.
Methods to detect LINE-1-mediated polymorphic human retrotransposition events in individual genomes. Modifications of these assays can also be used to identify other polymorphic retrotransposons in human DNA. (a) Polymerase chain reaction (PCR)–based methodologies. PCR using primers specific to diagnostic sequence variants in the L1 (red triangles and the corresponding maroon primer) and arbitrary oligonucleotides or primers complementary to ligated linkers ( yellow line) can be used to amplify human-specific L1s and their associated flanking sequences (maroon/yellow line flanked by triangles). The amplicon libraries are then resolved using electrophoresis, and individual products are cloned and sequenced (left). Alternatively, the amplicons can be hybridized to genome tiling microarrays (center), or directly characterized using high-throughput sequencing methodologies (right). Abbreviations: ATLAS, amplification typing of L1 active subfamilies; TIP-chip, transposon insertion profiling by microarray. (b) Mining of L1s in individual genome sequences. Whole-genome sequences, comparative genomics, or mining trace sequence databases can discover dimorphic L1s in individual genomes that are absent from reference genome assemblies. (c) Paired-end sequencing. Mate-pair reads containing one sequence from a uniquely mapping portion of genomic DNA and one sequence from an L1 can be used to identify novel retrotransposons (left) in individual genomes. Paired-end sequencing of fosmid inserts with restricted size distributions (~40 kb) allows the discovery of novel ~6-kb insertions (right) as well as deletions and inversions relative to a reference sequence. Fosmids containing insertions can then be screened for the presence of human-specific L1s. Abbreviation: HGR, human genome reference sequence. These methods are also described in another recent review (182).
A Brief Historical Perspective
L1s in the human genome have succeeded one another in a single lineage for the last ~40 million years (26–28, 122). Thus, new L1 subfamilies are continuously replacing older ones to dominate the expanding lineage of active elements (26, 28, 60, 122, 221). A majority of L1s are shared between human and chimp genomes, and likely represent retrotransposition-defective molecular fossils. However, studies in embryonic carcinoma cell lines revealed that a subset of expressed L1s contained diagnostic sequence variants within their 3′ UTR (e.g., an ACA instead of a GAG trinucleotide at positions 5930 to 5932 of L1.2; accession number M80343) (219). The subset of expressed L1s were designated the Ta (transcribed, subset a) L1 subfamily (219). Interestingly, all but one disease-producing L1 insertion identified to date are derived from the Ta subfamily (18, 89). The remaining mutagenic insertion was derived from the slightly older, human-specific pre-Ta subfamily of L1 (which contains an ACG trinucleotide at positions 5930 to 5932 relative to L1.2) (120).
The identification of sequence variants peculiar to human-specific L1s and the subsequent development of a cultured cell retrotransposition assay were instrumental in allowing the identification of RC-L1s in the HGR (Supplemental Figure 1; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org) (15, 31, 134, 172, 179). Comprehensive computational studies revealed that the HGR contains ~90 L1s with intact ORFs (31, 179). Polymerase chain reaction (PCR)–based cloning revealed that ~44 of these show a range of activity in cultured human HeLa cells (31). Unexpectedly, six highly active, or “hot,” L1s accounted for ~84% of the retrotransposition activity in the HGR. Extrapolation of the ~44 RC-L1s to the scale of a complete diploid genome suggested that the average human genome harbors ~80–100 active L1 elements (31, 205). Interestingly, limited genotyping analyses indicated that many active and retrotransposition-defective Ta subfamily L1s in the HGR are polymorphic with respect to presence or absence, indicating that many are recent insertions (9, 27, 31, 179).
Identification of Human-Specific LINE-1s by Mining Genome Sequences
Large-scale DNA sequencing projects have provided valuable resources to identify human L1-mediated retrotransposon polymorphisms. For example, comparative genomic analyses between the HGR and the draft chimpanzee genome allowed the identification of ~11,000 species-specific transposable elements, including 5,530 Alu, 1,174 L1, and 864 SVA elements specific to humans (167). Comparisons of DNA sequence trace files from 36 geographically diverse humans also enabled the identification of ~505 Alus, 65 L1s, 39 SVAs, 2 HERV-Ks, and 5 other polymorphic insertions (23). Similarly, comparison of a complete human diploid sequence (140) to the HGR allowed the identification of ~706 mobile element-associated structural variants, including the insertion of 584 Alus, 52 L1s, and 14 SVA elements (257). Mining 8 human genome sequences generated by next-generation sequencing yielded 4,342 Alu insertions absent from the HGR, 3,432 of which were additionally absent from a number of previous studies (108). Finally, a pilot analysis of sequence data obtained from ~200–300 individuals as part of the 1000 Genomes Project allowed the identification of over 1,000 L1 insertions that are absent from the HGR (69, 74, 168, 199). However, due to the low fold sequence coverage (2–4 fold for many genomes) and the composite nature of the HGR, it is likely that L1s are underrepresented in these analyses.
Clearly, the exploitation of human DNA sequence resources and nonhuman primate comparative genomics has revealed extensive human-specific transposable element diversity. A union of the above data has shown that these insertions are much more common in the population than once thought.
Experimental Approaches to Identify Novel Retrotransposon Insertions
The genomics revolution has revealed how structural variation contributes to interindividual genetic diversity. For example, representative oligonucleotide microarray analysis and array comparative genomic hybridization technologies have allowed the high-throughput identification of submicroscopic genetic differences of ~100 kb among individuals (110, 209). However, the size resolution of those techniques was not sufficient to permit identification of L1-mediated retrotransposition events.
More recently, a number of methods have been developed that are capable of detecting small- and intermediate-scale human structural variants (Figure 3). These methods include transposon display, array-based hybridization, second-generation DNA sequencing, and paired-end mapping of clone libraries (9, 25, 50, 69, 124, 129, 162, 168, 190, 191, 197, 215, 237, 246, 254). Together, these approaches have been instrumental in revealing transposon diversity in the genomes of geographically diverse individuals. A somewhat unexpected result is that mobile element dimorphisms account for a relatively large proportion of human genetic diversity (up to ~20%–25% of the interindividual genetic differences identified in some studies) (125, 129). The development of approaches to specifically identify these types of L1-mediated genetic variation is described below.
PCR-based display methods have allowed the identification of polymorphic L1 and Alu insertions in human genomes (9, 27, 34, 191, 200, 215). These methods exploit DNA sequence variants peculiar to human-specific retrotransposons (e.g., the ACA character present in Ta subfamily L1s) in conjunction with a pool of short, arbitrary oligonucleotides or sequences complementary to ligated linkers. PCR reactions utilizing this combination of retrotransposon-specific and degenerate or linker sequences then are used to generate complex amplicon libraries that contain the candidate retrotransposon and its immediate 5′ or 3′ flanking sequences. Sequencing of the flanking DNA and subsequent searches for sequence similarity then determine whether the candidate retrotransposon is present or absent in the HGR (9, 27). Together, these methods have identified numerous L1 and Alu insertion polymorphisms in individual genomes.
A derivation of classical display methods that employs suppression PCR methodology [amplification typing of L1 active subfamilies (ATLAS)] enabled the identification of nine full-length human-specific L1s from individual genomes (9). Interestingly, three out of seven tested sequences were “hot” in the cultured cell retrotransposition assay, and these three L1s were present at a minor allele frequency of less than 24% when genotyped in a panel of 90 geographically diverse individuals. Thus, these data provided additional evidence to support the hypothesis that young, human-specific L1s are underrepresented in the HGR.
Deconvolution of complex amplicon libraries generated by retrotransposon display approaches traditionally relied on electrophoretic fractionation using agarose or acrylamide gel systems and the subsequent cloning and characterization of individual molecules to map insertions to the HGR. The advent of new genomic technologies, including high-throughput DNA sequencing, now offers a means to revolutionize the discovery of polymorphic retrotransposon insertions (see Figure 3).
One method to identify dimorphic retrotransposon insertions, transposon insertion profiling by microarray (TIP-chip), employs the principles of transposon display to specifically amplify retrotransposons and their associated flanking sequences (80, 109, 253). The resultant amplicons are hybridized back to oligonucleotide arrays to identify sequences flanking the retrotransposons. TIP-chip allowed the discovery of numerous L1, Alu, and HERV-K insertion polymorphisms in genome-wide analyses. Application of TIP-chip to 69 unrelated individuals with X-linked intellectual disabilities also allowed the identification of L1 insertions within introns of the NHS and DACH2 genes, and mutations in these genes are implicated in intellectual disability (109). However, future studies are needed to determine whether the L1 insertions play a causal role in the observed cognitive phenotypes in these patients.
Another method to identify dimorphic L1 and Alu insertion polymorphisms employs traditional Sanger capillary sequencing and 454 or Illumina-based second-generation DNA sequencing technologies to deconvolute transposon-derived amplicon libraries (73, 111). Iskow et al. (111) identified 152 L1s from 38 ethnically diverse humans and 8 cell lines using capillary sequencing. They also identified 650 L1 and 403 Alu insertions in 30 lung or brain tumors and their matched nontumor controls via 454-based sequencing. These insertions were absent from both the HGR and a database of retrotransposon insertion polymorphisms (dbRIP) (134, 245), yielding 1,145 novel transposable elements. Ewing et al. (73) used an Illumina-based sequencing approach to identify 367 polymorphic L1s from a cohort of 25 humans that contained 15 unrelated individuals, individuals from 6 trios, and 3 pairs of monozygotic twins. Finally, Witherspoon et al. (255) used an Illumina-based sequencing approach to identify 487 Alu Yb8 and Yb9 insertions in 4 unrelated individuals that were absent from the HGR.
A third method to identify full-length or near-full-length L1 insertion polymorphisms involves paired-end DNA sequencing of fosmid libraries derived from individuals belonging to geographically diverse populations, which successfully identified intermediate-sized structural variants in human DNA (15, 124, 237). The screening of six individual libraries identified 68 L1s that were absent from the HGR. Remarkably, 37 of these 68 were “hot” L1s, 2 of which belonged to the pre-Ta L1 subfamily (15). Notably, unlike the methodologies described above, paired-end DNA sequencing of fosmid libraries does not involve PCR, allows identification of L1s in repetitive regions of the genome, and, though labor intensive, allows a comprehensive and relatively unbiased snapshot of L1 diversity. Together, the above methodologies have uncovered a virtual treasure trove of natural retrotransposon diversity in human genomes.
Allelic Heterogeneity in LINE-1 Activity
In addition to the polymorphic status of an L1, allelic heterogeneity may cause different L1 insertion alleles to exhibit different retrotransposition efficiencies in cultured cells. For example, L1.2A and L1.2B, which are likely progenitor alleles of a mutagenic insertion in the Factor VIII gene, exhibit a ~16-fold difference in their retrotransposition efficiencies because of amino acid substitutions near the ORF2p carboxyl terminus (67, 76, 120, 146). Similarly, the examination of three “hot” L1s from the HGR in a geographically diverse set of individuals revealed alleles with a wide range of retrotransposition activities (ranging from 0% to 390% of a reference L1) (212). Finally, a recent study identified an RC-L1 allele in an individual genome that apparently is defective in the HGR owing to a stop codon in ORF2p (15). Thus, both presence/absence of polymorphisms and allelic heterogeneity can influence L1 retrotransposition in an individual genome.
IMPACT OF MOBILE ELEMENTS ON MAMMALIAN GENOMES
LINE-1 as a Mutagen
A wealth of data is revealing the consequences of L1-mediated retrotransposition events in human genomes. Since their original discovery, approximately 65 disease-causing mutations in man have been attributed to L1-mediated retrotransposition events (18, 89). L1-mediated retrotransposition events can act as mutagens by directly disrupting exons (120). Similarly, insertions into introns can induce missplicing or exon skipping, thereby generating hypomorphic or null expression alleles (Figure 4) (19, reviewed in 89, 188). Finally, recent reports suggest the L1 endonuclease may cause double strand breaks, which in principle could lead to genomic instability (84, 142). Clearly, advances in genomics and the evolution of DNA sequencing technologies should allow the rapid identification of other disease-producing insertions in the coming years.
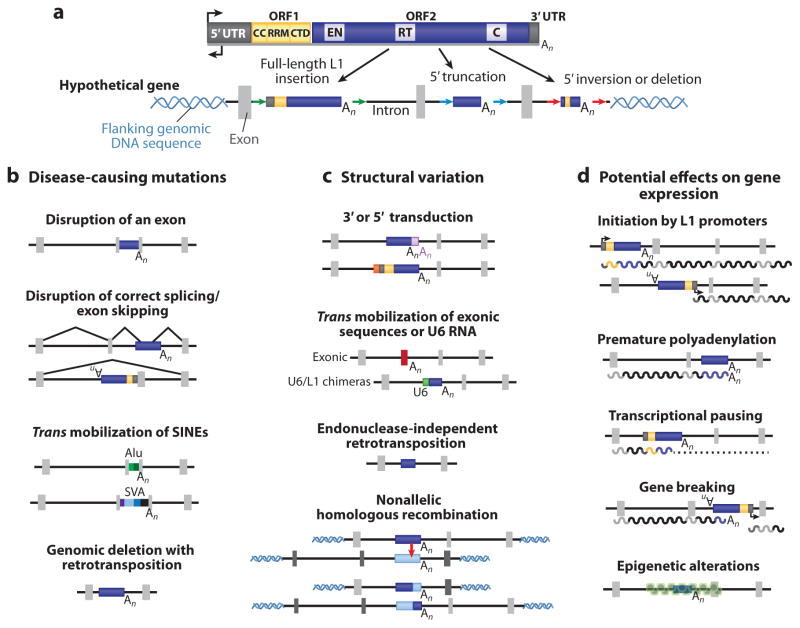
Figure 4.
Schematics highlighting the various ways that LINE-1-mediated retrotransposition events can impact the human genome. (a) A hypothetical wild-type gene locus. Light-gray rectangles represent exons, black lines represent introns, and helical lines represent flanking genomic DNA sequence. A full-length (left), 5′-truncated (center), and inverted or deleted L1 formed by twin priming (right) (189) are shown as intronic insertions. The arrows indicate target-site duplications (TSDs), and for simplicity are shown only in this panel. (b) Examples of L1-mediated processes that may result in disease. (c) Examples of structural variation caused by L1 insertions. The transduction figures show a 3′ transduction in light purple with its own poly(A) tail, and a 5′ transduction in orange. The nonallelic homologous recombination figure shows L1s at different loci (light and dark gray exons) acting as substrates for aberrant recombination (red arrow). (d ) Potential effects on gene expression caused by L1 insertion. Note that L1s in all the depicted events would generally contain TSDs, with the exception of endonuclease (EN)–independent retrotransposition events and some genomic deletions. Abbreviations: C, cysteine-rich domain; CC, coiled coil; CTD, carboxyl-terminal domain; RRM, RNA recognition motif; RT, reverse transcriptase; SVA, SINE-R/VNTR/Alu; UTR, untranslated region.
Effects on Gene Expression
L1 insertions can impact gene expression by a variety of mechanisms (Figure 4). For example, experimental studies have revealed that the adenosine-rich nature of the L1 transcript can introduce premature polyadenylation and/or RNA polymerase II transcriptional pause sites into genes, thereby attenuating their expression (97, 194). Interestingly, transcriptional pausing appears to depend on both the length of the L1 insertion and whether it is in the same transcriptional orientation as its resident gene (39, 97). The L1 ASP also can generate transcripts that, in principle, could affect gene expression (159, 181, 223, 258). Finally, in rare instances, L1 insertions may disrupt genes, leading to the generation of distinct transcription units through a phenomenon known as gene breaking (Figure 4) (252).
Approximate Rates of Heritable Retrotransposition Events
Determining the rate of germline retrotransposition in the human population remains an area of ongoing investigation. Estimates suggest that Alu elements are the most active retrotransposons in the human genome, with new insertions occurring in approximately 1 out of 20 live births (51, 257). L1 insertions follow, with estimates ranging between 1 out of 20 and 1 out of 200 births, depending upon the method used in the analysis (51, 73, 109, 141, 257). SVA insertions may be the least frequent retrotransposition events, occurring in approximately 1 out of 900 births (51, 257). In spite of disparate rates of retrotransposition, the different sizes and unique characteristics of L1, Alu, and SVA elements may pose distinctive challenges to the genome.
The above estimates are subject to ascertainment biases, and may represent only minimal estimates of the actual de novo retrotransposition frequency. For example, because some studies relied on comparisons with the HGR, they may underestimate the contribution of low-allele-frequency insertions to human genetic diversity (257). In fact, Iskow et al. (111) suggested that low-allele-frequency L1-mediated retrotransposition events, representing either rare or perhaps private L1 and/or Alu alleles, may be present in “virtually all personal genomes in the human populations.” Consistently, fosmid-based paired-end DNA sequencing, TIP-chip profiling, and the preliminary examination of the 1000 Genomes Project data have readily allowed the detection of rare L1 retrotransposition events in geographically diverse individuals (15, 74, 109, 168). Clearly, the exhaustive DNA sequencing of parent/offspring trios at high coverage should provide more accurate estimates of L1-mediated germline retrotransposition events in the near future.
Somatic LINE-1 Retrotransposition: Insights from Cancer Studies
In addition to acting as a germline mutagen, studies have revealed that L1 retrotransposition also occurs in certain somatic cells. Historically, the identification of a mutagenic L1 insertion into the adenomatous polyposis coli (APC) gene in a colorectal tumor that was absent from adjacent nontumor tissue established that L1 could retrotranspose in somatic cells (166).
Recent data generated using L1 display approaches combined with 454-based DNA sequencing led to the discovery of 9 de novo somatic L1 retrotransposition events in 6 of 20 non small-cell lung cancers that were absent from matched adjacent normal tissue samples (111). Interestingly, tumors containing new L1 retrotransposition events also exhibited global patterns of hypomethylation, providing a correlative link between epigenetic changes and increased L1 retrotransposition in tumors. Hypomethylation of the L1 5′ UTR also has been observed in malignant cells and cancer tissues, and is correlated with an increase in L1 mRNA and/or ORF1p expression (2, 4, 20, 89). Similarly, 5-azacytidine treatment leads to an elevation of L1-ASP-driven chimeric transcripts in nonmalignant breast epithelial cells (56). Thus, the above data suggest that the rate of L1 retrotransposition, and by proxy Alu and SVA retrotransposition, may be elevated in some cancers. Further studies should allow a greater understanding of whether and/or how often L1-mediated retrotransposition occurs in cancer and whether these events play a causal role in tumorigenesis.
Somatic LINE-1 Retrotransposition During Normal Development
Human genetic approaches, experiments conducted in transgenic animals, and cell culture models further suggest that L1 retrotransposition can occur in normal cells at discrete times during development. For example, genetic analyses have conclusively demonstrated both germline and somatic mosaicism of a mutagenic L1 retrotransposition event in the mother of a male patient afflicted with X-linked choroideremia (241). Thus, the mutagenic L1 insertion must have occurred during early embryonic development of the mother prior to partitioning of the germline. Consistent with this hypothesis, engineered L1s can retrotranspose in human embryonic stem cells (82).
Engineered human L1s can also retrotranspose during the early stages of mouse and rat embryonic development; however, most of the resultant insertions are not heritable (116). These findings are remarkable and suggest that L1 mRNA may be transferred from the gametes to the zygote to undergo retrotransposition at a later time in development, generating somatic mosaicism in the resultant offspring.
Unexpectedly, recent studies also revealed that engineered human L1s can retrotranspose at discrete times during neuronal development (54, 177, 178). These retrotransposition events could, in principle, generate somatic mosaicism in the nervous system and have the potential to affect intraindividual neuronal variability (54, 177, 178). In fact, sensitive TaqMan PCR-based approaches suggest that certain regions of the brain have an increase in human-specific L1 content relative to heart and/or liver tissues isolated from the same individual (54, 178). The observed increase in L1 DNA copy number in the brain suggests elevated levels of endogenous L1 retrotransposition in brain. However, other non-mutually-exclusive mechanisms must be considered when accounting for these L1 copy number differences (195, 251). Indeed, formal proof of endogenous L1 retrotransposition in brain will require the characterization of new L1 insertions from individual neurons. Although this remains a daunting task, advances in DNA sequencing technologies—including the ability to comprehensively characterize genome sequences from single or small pools of cells—should allow rigorous testing of these interesting observations.
Together, the above findings overturned the long-held dogma that L1-mediated retrotransposition could occur only in germ cells, and has led to speculations about how L1-meditated retrotransposition events may affect intraindividual genetic variation (116, 154, 177, 178, 218). Time and rigorous experimentation will tell whether somatic L1 retrotransposition events represent stochastic genetic noise that is tolerated by the host genome or whether these events have functional consequences in disease pathogenesis and/or neuronal development.
LINE-1 AS AN AGENT OF GENOME DIVERSIFICATION
LINE-1 Retrotransposition by Target-Site Primed Reverse Transcription
Canonical TPRT generally results in the insertion of an L1 or nonautonomous retrotransposition event at a new genomic location flanked by short TSDs (Figures 2 and 4). On occasion, TPRT also can lead to small deletions of target-site DNA that vary from approximately 2 to 50 bp in length, and/or to the addition of nontemplated or filler nucleotides at the 5′ genomic DNA/L1 junction sequence (5, 135, 180). Similar target-site alterations were observed upon examining engineered L1 retrotransposition events in transformed cell lines, human embryonic stem cells, and neuronal progenitor cells (54, 82, 86, 87, 177, 228).
Approximately 35% of Ta subfamily L1 retrotransposition events represent full-length insertions (26). The remaining L1s are 5′ truncated (95) and often contain short micro-homologies at the 5′ genomic DNA/L1 junction sequence (8, 86, 87, 157, 228, 263). Additionally, approximately 25% of L1s contain L1 inversion or deletion events that likely are generated by a process termed twin priming (189) (Figure 4). The high frequency of 5′ truncation associated with new L1 retrotransposition events remains enigmatic and may reflect host defense or DNA repair processes that act to either dissociate the L1 reverse transcriptase from the nascent L1 cDNA or degrade the L1 mRNA template prior to the completion of reverse transcription.
LINE-1 Retrotransposition-Mediated Deletion Events
In addition to acting as an insertional mutagen, L1-mediated retrotransposition events can lead to various forms of human structural variation. For example, studies conducted in HeLa and HCT116 cells revealed that ~10% of retrotransposition events derived from engineered human L1s are associated with the formation of chimeric L1s accompanied by intrachromosomal deletions, intrachromosomal duplication or inversions, and perhaps interchromosomal translocations (86, 87, 228). Comparisons of the pre- and postintegration sites of chimeric retrotransposition events suggested that DNA recombination processes such as single-strand annealing, synthesis-dependent strand annealing, and perhaps nonhomologous end joining are involved in the formation of these aberrant structures (86, 87, 228).
L1 retrotransposition-mediated genomic deletions are not peculiar to transformed human cells. For example, comparative biological approaches between the human and chimpanzee reference genomes enabled the discovery of 30 L1 and 19 Alu retrotransposition-mediated deletion events, which together account for a loss of ~26 kb of human genomic DNA in the past six million years (36, 100, 202).
L1 retrotransposition-mediated deletion events have also been observed in human genetic diseases (41). For example, a full-length L1 insertion that was accompanied by a deletion of ~46 kb, including 7 exons of the PDHX gene, led to a sporadic case of pyruvate dehydrogenase complex deficiency (170). Similarly, a ~4-kb L1 insertion was accompanied by a ~17-kb deletion that disrupted 3 exons of the EYA1 gene, leading to a sporadic case of branchio-oto-renal syndrome (173); however, the structure of this event suggests that it may have occurred by an endonuclease-independent (ENi) L1 retrotransposition mechanism (see below).
Alu and SVA retrotransposition-mediated deletion events also have impacted the human genome. For example, an Alu retrotransposition-mediated deletion approximately 1 Mya resulted in the loss of a 92-bp exon of the CMP-Neu5Ac hydroxylase gene, leading to a loss-of-function frameshift mutation in humans (104). Similarly, an SVA retrotransposition-mediated deletion apparently resulted in the loss of a 14-kb region of genomic DNA encompassing the entire HLA-A gene in three Japanese families containing patients afflicted with leukemia (229). Thus, although relatively rare, retrotransposition-mediated deletion continues to impact the human genome and represents a mechanism that can lead to both human structural variation and disease.
Postintegration LINE-1- and Alu-Mediated Recombination Processes
The abundance of L1 and Alu retrotransposons in the human genome provides numerous potential substrates for postintegration recombination events that can lead to disease and/or structural variation in human genomes (reviewed in 51, 89). For example, nonallelic homologous recombination (NAHR) events between two inverted Alu elements originally were observed in a ~5-kb deletion that included exons of the Low-Density Lipoprotein Receptor gene, resulting in a case of familial hypercholesterolemia (137). Similarly, NAHR between genomic L1s has been implicated in sporadic cases of phosphorylase kinase deficiency, Alport syndrome, and Ellis–van Creveld syndrome (32, 210, 233). Finally, comparative biological and genomic studies have revealed that NAHR events between L1s or Alus have generated structural variants in the chimpanzee and human genomes (98, 99, 125, 145, 213). Notably, young Alu elements are significantly enriched in regions flanking segmental duplications, suggesting that Alu-mediated recombination events may be involved in some segmental duplication expansions observed in humans (11). Personal genomics and advances in DNA sequencing likely will continue to reveal that inter-retrotransposon recombination events represent a significant portion of structural variation in human genomes.
Endonuclease-Independent Insertions
Experiments in XRCC4-deficient and DNA protein kinase catalytic subunit (DNA-PKcs)–deficient Chinese hamster ovary cell lines, which are defective in the nonhomologous end-joining pathway of DNA repair, revealed an alternative mechanism of ENi L1 retrotransposition distinct from conventional TPRT (174, 175). Characterization of ENi insertions revealed that they generally integrated at noncanonical L1 endonuclease cleavage sites, lacked TSDs, and frequently were truncated at both their 5′ and 3′ ends. In addition, some ENi retrotransposition events were associated with genomic DNA deletions and/or contained cDNA fragments derived from non-L1 cellular RNAs that likely were reverse transcribed during the integration process. L1 insertions bearing the hallmarks of ENi retrotransposition events have been identified in the human genome, again showing how cultured cell models can predict events that occur in nature (214, 224). Together, these findings suggested that ENi retrotransposition might bypass the requirement for endonuclease activity by initiating reverse transcription at endogenous lesions in genomic DNA, thereby acting as a molecular bandage (175, 243). Consistent with the above hypothesis, some ENi retrotransposition events in DNA-PKcs-deficient Chinese hamster ovary cells integrate at dysfunctional telomeres (174). Thus, ENi retrotransposition shows curious similarities to the action of telomerase and may represent a form of RNA-mediated DNA repair utilized by retrotransposons that lack an endonuclease domain (57, 88, 174).
LINE-1-Mediated Transduction
The examination of disease-producing L1 insertions and experiments using the cultured cell retrotransposition assay revealed that L1s are able to mobilize genomic DNA sequences flanking their 3′ (and less commonly their 5′) ends by a process termed L1-mediated transduction (Figure 4). The transduction of 3′ sequences is relatively common, and suggests that RNA polymerase II frequently bypasses the natural L1 polyadenylation site and instead uses a site in flanking genomic DNA (106, 171, 172). Computational analyses and the examination of L1 structural variants in individual genomes have revealed that 3′ transductions flank ~20% of human-specific L1s in the HGR and that some 3′ transductions are over 1 kb in length (15, 92, 125, 196). Extrapolations based on these data suggest that L1-mediated 3′ transductions may comprise as much as ~19–30 Mb of the human genome (92, 196).
L1 3′ transductions create recognizable sequence tags, which can be used to infer parent/offspring relationships between full-length L1s and their progeny. For example, a ~489-bp region of genomic DNA flanking a mutagenic L1 insertion into the dystrophin gene was instrumental in identifying an active L1 progenitor allele (106). Similarly, shared 3′ transduction sequences were recently used to identify minifamilies of L1s that contain active, rare alleles in the human population (15, 125). L1s from two of these 3′ transduction families, LRE3 and L1RP, are responsible for disease-producing insertions in sporadic cases of chronic granulomatous disease and X-linked retinitis pigmentosa, respectively (30, 127, 207). Thus, the examination of transduction families that include highly active elements may be used as a way to discover novel, rare, and highly active L1s in human populations.
The cultured cell L1 retrotransposition assay revealed that 3′ transduction could, in principle, lead to the mobilization of exons and/or regulatory DNA sequences to new genomic locations, and may represent a mechanism of exon shuffling (171, 172). To date, there are no in vivo examples of L1 3′ transduction leading to the formation of a new gene. However, SVA elements are also frequently associated with 3′ transductions (187), and SVA-mediated 3′ transduction events prior to the divergence of humans and great apes led to the dispersion of the AMAC gene to three distinct places in the human genome (256). Whether these copies of the AMAC gene are functional requires validation, but the potential for genomic diversification through mobile element-mediated dispersion is clear.
The ability of the L1-encoded proteins to mobilize non-L1 RNAs to new genomic locations in trans provides another potential mechanism for exon dispersal and/or the creation of new genes. For example, retrotransposition of a cyclophilin A cDNA into the TRIM5-α locus of New World monkeys led to the creation of a chimeric protein that confers HIV resistance to owl monkeys (206). Similarly, a novel testes-specific hominoid gene, PIPSL, is a chimeric ubiquitin-binding protein derived from a fusion of the PIP5K1A and 26S proteasome subunit RNAs that underwent retrotransposition (7, 184). The PIPSL gene appears to have been subject to positive selection and is conserved among humans, suggesting a functional role in modern genomes. Finally, it appears that exons from the CFTR and ATM genes have been mobilized in trans to new genomic locations by the L1 retrotransposition machinery (70, 201). However, it remains possible that some of these examples represent L1-mediated 3′ transductions that were so severely 5′ truncated that they lack L1 sequences (70, 171).
In principle, L1 5′ transduction can occur if a transcript initiating upstream of an L1 undergoes retrotransposition. Because 5′ transduction can only be found by examining full-length L1s, they appear to be much less common than 3′ transductions. However, potential examples of L1 5′ transduction have been identified in the HGR, in cultured cell experiments, and in a mutagenic mouse insertion (39, 134, 228, 249). In contrast, 5′ transduction events frequently accompany SVA retrotransposition events, which is consistent with the idea that some SVA elements rely on host promoter sequences for their transcription (58, 101). Indeed, a 5′ transduction derived from the MAST2 gene was used to identify a family of SVA elements (SVAF) that likely are amplifying in the human genome (58, 101).
Epigenetic Phenomena Related to LINE-1s
L1s may also play critical roles in the epigenetic regulation of host genes (Figure 4). For example, recent research suggests that indicator cassettes delivered into the genome by engineered L1 retrotransposons can be epigenetically silenced either during or immediately after their integration (54, 83, 177). It will be interesting to determine whether silencing is specific for the retrotransposed L1 sequence or whether it can affect the epigenetic status of adjacent genes.
The ability of L1s to affect the epigenetic regulation of genes may not be restricted to new retrotransposition events. For example, the ability of L1s to accumulate on sex chromosomes over evolutionary time led Mary Lyon (147) to speculate that L1s might function as cis-acting booster elements to aid in the spreading of heterochromatin formation observed during X-inactivation. Consistent with this notion, some genes that escape X-inactivation are in relatively L1-poor regions of the X-chromosome (10, 37), and L1 density appears to correlate positively with heterochromatin spread in X/autosomal translocations (231). Indeed, recent experiments suggest that L1s might play an active role in nucleating heterochromatin formation on the inactive X chromosome (44). Clearly, although these findings are still in their early stages, further research should allow even more discoveries about how L1-associated epigenetic changes impact gene expression.
CLOSING REMARKS
It is now clear that transposable elements are an integral part of our genomes. In the coming years, technological breakthroughs in DNA sequencing and genome annotation will undoubtedly uncover many more examples of how transposable elements impact biological processes. For example, high-coverage, longer-read sequencing of trios and monozygotic twins and/or the ability to sequence the genomes of single or small populations of cells will yield more information about the actual rate of L1-mediated retrotransposition events in both germline and somatic cells, and should clarify the role of L1-mediated retrotransposition events in certain types of human cancers. It will be interesting to determine whether genetically distinct populations or particular individuals are more prone to L1-mediated retrotransposition events.
Because transposable elements can be considered intracellular genomic parasites, they also provide an ideal means to study host-parasite interactions. The availability of new experimental reagents, such as epitope-tagged engineered human L1s, combined with cell-culture-based retrotransposition assays now allows a way to identify host factors that act to combat the constant assault of transposable elements on the genome. We probably will discover even more examples where genetic differences in host factors correlate with variation in L1 retrotransposition rates (e.g., 43, 89, 126, 183, 225).
Finally, future studies will enlighten us about how transposable element-derived sequences serve as seeds for evolutionary change. There is an ever-growing number of examples of regulatory elements derived from transposable elements that are required for proper gene expression (e.g., 17, 115, 204, 247; reviewed in 79), and we anticipate this list will expand. Indeed, transposable elements may provide a mechanism to amplify and distribute cis-acting DNA sequences to new genomic locations, which after selection may function in gene regulation—a potentially prescient hypothesis put forth by Davidson & Britten (59) over 40 years ago.
We have come a long way since Barbara McClintock’s (161) discovery of mobile genetic elements in maize over half a century ago. In the past decade alone, we have witnessed an exponential growth in knowledge about how transposable elements have impacted the human as well as other mammalian genomes. Though much progress has been made, our work has just begun. The coming years likely will identify how transposable elements contribute to phenotypic variation and human-specific traits. Clearly, the “junk” in our genomes is stepping into the limelight. It is an exciting time for transposable element research; the human genetics community should take greater notice of these dynamic elements.
Supplementary Material
Acknowledgments
We thank Drs. Haig Kazazian and Mark Batzer for their critical and insightful review of the manuscript. We thank Aurélien Doucet, Billy Giblin, Nancy Leff, John Moldovan, Sandra Richardson, and other members of the Moran lab for helpful comments and discussions. C.R.B. was supported in part by NIH training grants T32GM7544 and T32000040. J.V.M. is supported by NIH grants GM060518 and GM082970, and is also an investigator of the Howard Hughes Medical Institute. J.L.G.-P. is supported by an ISCIII-CSJA (FEDER/EMER07/056), a Marie Curie IRG action (FP7-PEOPLE-2007-4-3-IRG), CICE (P09-CTS-4980), Proyectos en Salud PI-002 from Junta de Andalucía (Spain), and the Spanish Ministry of Health (FEDER/FIS PI08171). R.M.B. was supported by a Wellcome Trust project grant (075163/Z/04/Z) to R.M.B. and Prof. Sir Alec Jeffreys, FRS. Finally, we thank our spouses for their support and patience while writing this review. Henry Levin and J.V.M. have recently written a review that also discusses various aspects of L1 biology.
Footnotes
DISCLOSURE STATEMENT
J.V.M. is an inventor on a patent, “Compositions and methods of use of human retrotransposons,” application no. 60/006,831, issued November 2000. He has not received any money from the patent, and its issuance does not influence the opinions in this review.
Contributor Information
Christine R. Beck, Email: cregina@umich.edu.
John V. Moran, Email: moranj@umich.edu.
LITERATURE CITED
- 1.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–24. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176:39–44. doi: 10.1016/0378-1119(96)00205-3. [DOI] [PubMed] [Google Scholar]
- 3.Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: a source for the genesis of primate microsatellites. Genomics. 1995;29:136–44. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- 4.Asch HL, Eliacin E, Fanning TG, Connolly JL, Bratthauer G, Asch BB. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol Res. 1996;8:239–47. [PubMed] [Google Scholar]
- 5.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–55. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28:527–39. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 7.Babushok DV, Ohshima K, Ostertag EM, Chen X, Wang Y, et al. A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res. 2007;17:1129–38. doi: 10.1101/gr.6252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–50. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badge RM, Alisch RS, Moran JV. ATLAS: a system to selectively identify human-specific L1 insertions. Am J Hum Genet. 2003;72:823–38. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA. 2000;97:6634–39. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–34. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet. 2006;7:149–73. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 13.Basame S, Wai-Lun Li P, Howard G, Branciforte D, Keller D, Martin SL. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J Mol Biol. 2006;357:351–57. doi: 10.1016/j.jmb.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 14.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–79. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 15.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–70. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2:1697–702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 17.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 18.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–58. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 19.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–10. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010;38:3909–22. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol. 2005;79:12507–14. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–51. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, et al. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–83. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–28. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 27.Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14:1221–31. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. 2001;18:2186–94. doi: 10.1093/oxfordjournals.molbev.a003765. [DOI] [PubMed] [Google Scholar]
- 29.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol. 1994;14:2584–92. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, et al. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet. 2002;71:327–36. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–85. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burwinkel B, Kilimann MW. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J Mol Biol. 1998;277:513–17. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- 33.Buzdin A, Gogvadze E, Kovalskaya E, Volchkov P, Ustyugova S, et al. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 2003;31:4385–90. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buzdin A, Ustyugova S, Gogvadze E, Lebedev Y, Hunsmann G, Sverdlov E. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum Genet. 2003;112:527–33. doi: 10.1007/s00439-002-0904-2. [DOI] [PubMed] [Google Scholar]
- 35.Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of L1. Genomics. 2002;80:402–6. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 36.Callinan PA, Wang J, Herke SW, Garber RK, Liang P, Batzer MA. Alu retrotransposition-mediated deletion. J Mol Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- 37.Carrel L, Park C, Tyekucheva S, Dunn J, Chiaromonte F, Makova KD. Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet. 2006;2:e151. doi: 10.1371/journal.pgen.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll ML, Roy-Engel AM, Nguyen SV, Salem AH, Vogel E, et al. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J Mol Biol. 2001;311:17–40. doi: 10.1006/jmbi.2001.4847. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum Mol Genet. 2006;15:2146–56. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- 40.Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat. 2005;25:207–21. doi: 10.1002/humu.20133. [DOI] [PubMed] [Google Scholar]
- 41.Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. 2005;117:411–27. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 42.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–78. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–53. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 44.Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–69. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 45.Christensen SM, Eickbush TH. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol Cell Biol. 2005;25:6617–28. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu WM, Liu WM, Schmid CW. RNA polymerase III promoter and terminator elements affect Alu RNA expression. Nucleic Acids Res. 1995;23:1750–57. doi: 10.1093/nar/23.10.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–14. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 48.Collier LS, Largaespada DA. Hopping around the tumor genome: transposons for cancer gene discovery. Cancer Res. 2005;65:9607–10. doi: 10.1158/0008-5472.CAN-05-3085. [DOI] [PubMed] [Google Scholar]
- 49.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Res. 2009;19:545–55. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 51.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cordaux R, Hedges DJ, Batzer MA. Retrotransposition of Alu elements: How many sources? Trends Genet. 2004;20:464–67. doi: 10.1016/j.tig.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Craig N, Craigie R, Gellert M, Lambowitz A. Mobile DNA II. Washington, DC: ASM; 2002. [Google Scholar]
- 56.Cruickshanks HA, Tufarelli C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics. 2009;94:397–406. doi: 10.1016/j.ygeno.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 57.Curcio MJ, Belfort M. The beginning of the end: links between ancient retroelements and modern telomerases. Proc Natl Acad Sci USA. 2007;104:9107–8. doi: 10.1073/pnas.0703224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Damert A, Raiz J, Horn AV, Lower J, Wang H, et al. 5′-transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davidson EH, Britten RJ. Regulation of gene expression: possible role of repetitive sequences. Science. 1979;204:1052–59. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- 60.Deininger PL, Batzer MA, Hutchison CA, III, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–11. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- 61.Denison RA, Van Arsdell SW, Bernstein LB, Weiner AM. Abundant pseudogenes for small nuclear RNAs are dispersed in the human genome. Proc Natl Acad Sci USA. 1981;78:810–14. doi: 10.1073/pnas.78.2.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 63.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–56. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dewannieux M, Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86:378–81. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–83. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 66.Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, et al. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol Cell Biol. 2007;27:4685–97. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–8. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 68.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ejima Y, Yang L. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum Mol Genet. 2003;12:1321–28. doi: 10.1093/hmg/ddg138. [DOI] [PubMed] [Google Scholar]
- 71.Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem. 2004;279:27753–63. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 72.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–67. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 73.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–70. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ewing AD, Kazazian HH., Jr Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–90. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fanning T, Singer M. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 1987;15:2251–60. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farley AH, Luning Prak ET, Kazazian HH., Jr More active human L1 retrotransposons produce longer insertions. Nucleic Acids Res. 2004;32:502–10. doi: 10.1093/nar/gkh202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 78.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–16. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 79.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gabriel A, Dapprich J, Kunkel M, Gresham D, Pratt SC, Dunham MJ. Global mapping of transposon location. PLoS Genet. 2006;2:e212. doi: 10.1371/journal.pgen.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–11. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–77. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 83.Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–73. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–93. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Georgiou I, Noutsopoulos D, Dimitriadou E, Markopoulos G, Apergi A, et al. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum Mol Genet. 2009;18:1221–28. doi: 10.1093/hmg/ddp022. [DOI] [PubMed] [Google Scholar]
- 86.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–95. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–25. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 88.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci USA. 2007;104:9352–57. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 90.Goodier JL, Mandal PK, Zhang L, Kazazian HH., Jr Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet. 2010;19:1712–25. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goodier JL, Maraia RJ. Terminator-specific recycling of a B1-Alu transcription complex by RNA polymerase III is mediated by the RNA terminus-binding protein La. J Biol Chem. 1998;273:26110–16. doi: 10.1074/jbc.273.40.26110. [DOI] [PubMed] [Google Scholar]
- 92.Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–57. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 93.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–83. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Graur D, Shuali Y, Li WH. Deletions in processed pseudogenes accumulate faster in rodents than in humans. J Mol Evol. 1989;28:279–85. doi: 10.1007/BF02103423. [DOI] [PubMed] [Google Scholar]
- 95.Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. EMBO J. 1984;3:1753–59. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–83. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 98.Han K, Lee J, Meyer TJ, Remedios P, Goodwin L, Batzer MA. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci USA. 2008;105:19366–71. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han K, Lee J, Meyer TJ, Wang J, Sen SK, et al. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 2007;3:1939–49. doi: 10.1371/journal.pgen.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han K, Sen SK, Wang J, Callinan PA, Lee J, et al. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–52. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH., Jr Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009;19:1983–91. doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr245. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005;33:2374–83. doi: 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N. Alu-mediated inactivation of the human CMP-N-acetylneuraminic acid hydroxylase gene. Proc Natl Acad Sci USA. 2001;98:11399–404. doi: 10.1073/pnas.191268198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–39. [PMC free article] [PubMed] [Google Scholar]
- 106.Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH., Jr A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet. 1994;7:143–48. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 107.Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem. 1992;267:19765–68. [PubMed] [Google Scholar]
- 108.Hormozdiari F, Alkan C, Ventura M, Hajirasouliha I, Malig M, et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2011;21:840–49. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–82. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 111.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–10. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 113.Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, et al. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–22. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Januszyk K, Li PW, Villareal V, Branciforte D, Wu H, et al. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J Biol Chem. 2007;282:24893–904. doi: 10.1074/jbc.M702023200. [DOI] [PubMed] [Google Scholar]
- 115.Jurka J. Conserved eukaryotic transposable elements and the evolution of gene regulation. Cell Mol Life Sci. 2008;65:201–4. doi: 10.1007/s00018-007-7369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–54. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 119.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 120.Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–66. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 121.Keng VW, Ryan BJ, Wangensteen KJ, Balciunas D, Schmedt C, et al. Efficient transposition of Tol2 in the mouse germline. Genetics. 2009;183:1565–73. doi: 10.1534/genetics.109.100768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci USA. 2009;106:731–36. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–47. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet. 1999;8:1557–60. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 128.Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci USA. 1997;94:10155–60. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–26. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kubo S, del Seleme MC, Soifer HS, Perez JL, Moran JV, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci USA. 2006;103:8036–41. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. 2005;14:3237–48. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 132.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–60. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 133.Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12:1097–105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 135.Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 2004;14:2253–60. doi: 10.1101/gr.2745804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–46. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–53. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–91. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 140.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li X, Scaringe WA, Hill KA, Roberts S, Mengos A, et al. Frequency of recent retrotransposition events in the human factor IX gene. Hum Mutat. 2001;17:511–19. doi: 10.1002/humu.1134. [DOI] [PubMed] [Google Scholar]
- 142.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu WM, Schmid CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21:1351–59. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 145.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lutz SM, Vincent BJ, Kazazian HH, Jr, Batzer MA, Moran JV. Allelic heterogeneity in LINE-1 retrotransposition activity. Am J Hum Genet. 2003;73:1431–37. doi: 10.1086/379744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–37. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 148.Macfarlane C, Simmonds P. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J Mol Evol. 2004;59:642–56. doi: 10.1007/s00239-004-2656-1. [DOI] [PubMed] [Google Scholar]
- 149.Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, et al. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol. 2011;31:300–16. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Maestre J, Tchenio T, Dhellin O, Heidmann T. mRNA retroposition in human cells: processed pseudogene formation. EMBO J. 1995;14:6333–38. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Martin F, Maranon C, Olivares M, Alonso C, Lopez MC. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J Mol Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- 153.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991;11:4804–7. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Martin SL. Developmental biology: jumping-gene roulette. Nature. 2009;460:1087–88. doi: 10.1038/4601087a. [DOI] [PubMed] [Google Scholar]
- 155.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc Natl Acad Sci USA. 2003;100:13815–20. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. 2001;21:467–75. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Martin SL, Li WL, Furano AV, Boissinot S. The structures of mouse and human L1 elements reflect their insertion mechanism. Cytogenet Genome Res. 2005;110:223–28. doi: 10.1159/000084956. [DOI] [PubMed] [Google Scholar]
- 158.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–10. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 159.Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006:71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mayer J, Meese E, Mueller-Lantzsch N. Multiple human endogenous retrovirus (HERV-K) loci with gag open reading frames in the human genome. Cytogenet Cell Genet. 1997;78:1–5. doi: 10.1159/000134614. [DOI] [PubMed] [Google Scholar]
- 161.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36:344–55. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–41. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McMillan JP, Singer MF. Translation of the human LINE-1 element, L1Hs. Proc Natl Acad Sci USA. 1993;90:11533–37. doi: 10.1073/pnas.90.24.11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mi S, Lee X, Li X, Veldman GM, Finnerty H, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–89. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 165.Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 1994;8:1463–72. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 166.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–45. [PubMed] [Google Scholar]
- 167.Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–79. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Minakami R, Kurose K, Etoh K, Furuhata Y, Hattori M, Sakaki Y. Identification of an internal cis-element essential for the human L1 transcription and a nuclear factor(s) binding to the element. Nucleic Acids Res. 1992;20:3139–45. doi: 10.1093/nar/20.12.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mine M, Chen JM, Brivet M, Desguerre I, Marchant D, et al. A large genomic deletion in the PDHX gene caused by the retrotranspositional insertion of a full-length LINE-1 element. Hum Mutat. 2007;28:137–42. doi: 10.1002/humu.20449. [DOI] [PubMed] [Google Scholar]
- 171.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–34. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 172.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–27. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 173.Morisada N, Rendtorff ND, Nozu K, Morishita T, Miyakawa T, et al. Branchio-oto-renal syndrome caused by partial EYA1 deletion due to LINE-1 insertion. Pediatr Nephrol. 2010;25:1343–48. doi: 10.1007/s00467-010-1445-x. [DOI] [PubMed] [Google Scholar]
- 174.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–12. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 175.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–65. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 176.Moyes D, Griffiths DJ, Venables PJ. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 2007;23:326–33. doi: 10.1016/j.tig.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 177.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 178.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–46. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet. 2002;71:312–26. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Narita N, Nishio H, Kitoh Y, Ishikawa Y, Minami R, et al. Insertion of a 5′ truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Investig. 1993;91:1862–67. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nigumann P, Redik K, Matlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–34. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 182.O’Donnell KA, Burns KH. Mobilizing diversity: transposable element insertions in genetic variation and disease. Mob DNA. 2010;1:21. doi: 10.1186/1759-8753-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–59. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ohshima K, Igarashi K. Inference for the initial stage of domain shuffling: tracing the evolutionary fate of the PIPSL retrogene in hominoids. Mol Biol Evol. 2010;27:2522–33. doi: 10.1093/molbev/msq138. [DOI] [PubMed] [Google Scholar]
- 185.Ono M, Kawakami M, Takezawa T. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res. 1987;15:8725–37. doi: 10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ono R, Nakamura K, Inoue K, Naruse M, Usami T, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–6. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- 187.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–51. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–38. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 189.Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–65. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ovchinnikov I, Rubin A, Swergold GD. Tracing the LINEs of human evolution. Proc Natl Acad Sci USA. 2002;99:10522–27. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ovchinnikov I, Troxel AB, Swergold GD. Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res. 2001;11:2050–58. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pace JK, II, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–32. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–98. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–66. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 195.Peterson SE, Westra JW, Paczkowski CM, Chun J. Chromosomal mosaicism in neural stem cells. Methods Mol Biol. 2008;438:197–204. doi: 10.1007/978-1-59745-133-8_16. [DOI] [PubMed] [Google Scholar]
- 196.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–15. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Robins DM, Samuelson LC. Retrotransposons and the evolution of mammalian gene expression. Genetica. 1992;86:191–201. doi: 10.1007/BF00133720. [DOI] [PubMed] [Google Scholar]
- 199.Rouchka E, Montoya-Durango DE, Stribinskis V, Ramos K, Kalbfleisch T. Assessment of genetic variation for the LINE-1 retrotransposon from next generation sequence data. BMC Bioinforma. 2010;11(Suppl 9):S12. doi: 10.1186/1471-2105-11-S9-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Roy AM, Carroll ML, Kass DH, Nguyen SV, Salem AH, et al. Recently integrated human Alu repeats: finding needles in the haystack. Genetica. 1999;107:149–61. [PubMed] [Google Scholar]
- 201.Rozmahel R, Heng HH, Duncan AM, Shi XM, Rommens JM, Tsui LC. Amplification of CFTR exon 9 sequences to multiple locations in the human genome. Genomics. 1997;45:554–61. doi: 10.1006/geno.1997.4968. [DOI] [PubMed] [Google Scholar]
- 202.Salem AH, Kilroy GE, Watkins WS, Jorde LB, Batzer MA. Recently integrated Alu elements and human genomic diversity. Mol Biol Evol. 2003;20:1349–61. doi: 10.1093/molbev/msg150. [DOI] [PubMed] [Google Scholar]
- 203.Sarrowa J, Chang DY, Maraia RJ. The decline in human Alu retroposition was accompanied by an asymmetric decrease in SRP9/14 binding to dimeric Alu RNA and increased expression of small cytoplasmic Alu RNA. Mol Cell Biol. 1997;17:1144–51. doi: 10.1128/mcb.17.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sasaki T, Nishihara H, Hirakawa M, Fujimura K, Tanaka M, et al. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci USA. 2008;105:4220–25. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 206.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–73. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 207.Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet. 1998;19:327–32. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- 208.Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O’Hara B, et al. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–25. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–28. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 210.Segal Y, Peissel B, Renieri A, de Marchi M, Ballabio A, et al. LINE-1 elements at the sites of molecular rearrangements in Alport syndrome-diffuse leiomyomatosis. Am J Hum Genet. 1999;64:62–69. doi: 10.1086/302213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sela N, Mersch B, Hotz-Wagenblatt A, Ast G. Characteristics of transposable element exonization within human and mouse. PLoS One. 2010;5:e10907. doi: 10.1371/journal.pone.0010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Seleme MC, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH., Jr Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci USA. 2006;103:6611–16. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sen SK, Han K, Wang J, Lee J, Wang H, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sen SK, Huang CT, Han K, Batzer MA. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007;35:3741–51. doi: 10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sheen FM, Sherry ST, Risch GM, Robichaux M, Nasidze I, et al. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, et al. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region: molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–76. [PubMed] [Google Scholar]
- 217.Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci USA. 2011;108:2837–42. doi: 10.1073/pnas.1012834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–54. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–97. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–48. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 221.Smit AF, Toth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–17. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 222.Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–85. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Srikanta D, Sen SK, Huang CT, Conlin EM, Rhodes RM, Batzer MA. An alternative pathway for Alu retrotransposition suggests a role in DNA double-strand break repair. Genomics. 2009;93:205–12. doi: 10.1016/j.ygeno.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–98. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sun LV, Jin K, Liu Y, Yang W, Xie X, et al. PBmice: an integrated database system of piggyBac (PB) insertional mutations and their characterizations in mice. Nucleic Acids Res. 2008;36:D729–34. doi: 10.1093/nar/gkm790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–29. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, et al. Human L1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–38. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 229.Takasu M, Hayashi R, Maruya E, Ota M, Imura K, et al. Deletion of entire HLA-A gene accompanied by an insertion of a retrotransposon. Tissue Antigens. 2007;70:144–50. doi: 10.1111/j.1399-0039.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- 230.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–38. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tang YA, Huntley D, Montana G, Cerase A, Nesterova TB, Brockdorff N. Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenetics Chromatin. 2010;3:10. doi: 10.1186/1756-8935-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–15. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Temtamy SA, Aglan MS, Valencia M, Cocchi G, Pacheco M, et al. Long interspersed nuclear element-1 (LINE1)-mediated deletion of EVC, EVC2, C4orf6, and STK32B in Ellisvan Creveld syndrome with borderline intelligence. Hum Mutat. 2008;29:931–38. doi: 10.1002/humu.20778. [DOI] [PubMed] [Google Scholar]
- 234.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–99. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 235.Torrents D, Suyama M, Zdobnov E, Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–67. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92:1520–24. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 238.Ullu E, Esposito V, Melli M. Evolutionary conservation of the human 7 S RNA sequences. J Mol Biol. 1982;161:195–201. doi: 10.1016/0022-2836(82)90286-8. [DOI] [PubMed] [Google Scholar]
- 239.Ullu E, Weiner AM. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. Nature. 1985;318:371–74. doi: 10.1038/318371a0. [DOI] [PubMed] [Google Scholar]
- 240.Van Arsdell SW, Denison RA, Bernstein LB, Weiner AM, Manser T, Gesteland RF. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell. 1981;26:11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- 241.Van den Hurk JA, Meij IC, Seleme MC, Kano H, Nikopoulos K, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–92. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 242.Vanin EF. Processed pseudogenes: characteristics and evolution. Annu Rev Genet. 1985;19:253–72. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 243.Voliva CF, Martin SL, Hutchison CA, III, Edgell MH. Dispersal process associated with the L1 family of interspersed repetitive DNA sequences. J Mol Biol. 1984;178:795–813. doi: 10.1016/0022-2836(84)90312-7. [DOI] [PubMed] [Google Scholar]
- 244.Wang H, Xing J, Grover D, Hedges DJ, Han K, et al. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 245.Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat. 2006;27:323–29. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wang J, Wang W, Li R, Li Y, Tian G, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 2007;104:18613–18. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Weber MJ. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–61. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 251.Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, et al. Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol. 2010;518:3981–4000. doi: 10.1002/cne.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–78. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Wheelan SJ, Scheifele LZ, Martinez-Murillo F, Irizarry RA, Boeke JD. Transposon insertion site profiling chip (TIP-chip) Proc Natl Acad Sci USA. 2006;103:17632–37. doi: 10.1073/pnas.0605450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–76. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 255.Witherspoon DJ, Xing J, Zhang Y, Watkins WS, Batzer MA, Jorde LB. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci USA. 2006;103:17608–13. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Xing J, Zhang Y, Han K, Salem AH, Sen SK, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–26. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 259.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–40. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhang Z, Harrison P, Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002;12:1466–82. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–58. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 263.Zingler N, Willhoeft U, Brose HP, Schoder V, Jahns T, et al. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15:780–89. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.