SUMMARY
The shoot apical meristem (SAM) comprises a group of undifferentiated cells that divide to maintain the meristem and also give rise to all plant shoot organs. SAM fate is specified by HOMEODOMAIN-LEUCINE ZIPPER (HD-ZIP) transcription factors, which are targets of miR166/165. In Arabidopsis, AGO10 is a critical regulator of SAM maintenance, but the mechanism of regulation remains unknown. Here we demonstrate that AGO10 specifically recruits miR166/165. The AGO10-miR166/165 association is determined by the distinct structure of the miR166/165 duplex. Deficient loading of miR166 into AGO10 results in a defective SAM. AGO10 has a higher binding affinity for miR166 than does AGO1, a master repressor for miRNA targets. Notably, the miR166/165-binding ability of AGO10, but not its catalytic activity, is required for SAM development. We propose that AGO10 functions as a decoy for miR166/165 to maintain the SAM, preventing their incorporation into AGO1 complexes and the subsequent repression of HD-ZIP gene expression.
INTRODUCTION
Arabidopsis SAM contains several organized layers of stem cells located at the shoot tip. The SAM is maintained in a pluripotent state in its central region, and it also provides cells to the peripheral region to form differentiated organs. Among the factors that regulate whether or not cells in the SAM differentiate are class III HD-ZIP family genes, which include PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), and ATHB-8 and -15 (Barton, 2010).
The HD-ZIP genes are regulated by a group of small non-coding RNAs (sRNAs) in Arabidopsis (Mallory et al., 2004). sRNAs are processed by a Dicer-like enzyme from imperfectly self-folded hairpin precursors or double-strand RNAs to form sRNA duplexes (sRNA/*). Mature sRNAs are incorporated into AGO-centered RNA-induced silencing complexes (RISCs) to repress the expression of target genes at the transcriptional and posttranscriptional levels (Vaucheret, 2008). AGOs consist of a variable N-terminal domain and conserved C-terminal PAZ, MID and PIWI domains. The PAZ and MID domains recognize the 3′ and 5′ ends of sRNAs, respectively (Frank et al., 2010). The PIWI domain possesses an RNase H-like fold structure and carries out endonuclease activity directed by sRNAs against complementary RNA targets (Song et al., 2004).
The Arabidopsis genome contains 10 AGO genes whose functional diversity has been deduced from the nature of the bound sRNAs. AGO1 associates with most microRNAs (miRNAs) and a variety of small interfering RNAs (siRNAs) (Vaucheret, 2008), while AGO7 predominantly recruits miR390 to initiate trans-acting siRNA (ta-siRNA) production (Montgomery et al., 2008). AGO4, AGO6 and AGO9 all bind to endogenous 24 nt sRNAs to silence loci harboring repetitive DNA sequences, transposons and heterochromatin regions with partial redundancy (Havecker et al., 2010; Olmedo-Monfil et al., 2010). Given the large network of sRNAs in Arabidopsis, strict sorting mechanisms are required to channel different sRNAs into appropriate AGO complexes to assure the functional diversification and specification of individual RISC. Recent reports have shown that the 5′ terminal nucleotide of the sRNAs is a major determining factor for the selective association of these molecules with AGO proteins (Mi et al., 2008). sRNAs harboring a 5′ uridine are preferentially associated with AGO1. AGO2, AGO4, AGO6 and AGO9 favor sRNAs with an adenine in this position, whereas AGO5 prefers sRNAs with a 5′ cytosine. sRNA destination is also affected by the relative spatiotemporal expression patterns of AGOs and sRNA genes (Havecker et al., 2010).
AGO10 (originally identified as PNH or ZLL)plays a critical role in multiple developmental processes, such as the maintenance of undifferentiated stem cells in the SAM (Lynn et al., 1999; Moussian et al., 1998) and the establishment of leaf polarity (Liu et al., 2009). In the Arabidopsis ecotype Ler, ago 10 mutant seedlings (pnh/zll) display differentiated cells or complete organs in place of the SAM (denoted the pinhead phenotype), whereas these phenotypes are rarely seen in Col-0 background (Mallory et al., 2009). Recent studies indicate that AGO10 modulates these developmental processes by genetically repressing miR166/165, two related miRNAs that differ in sequence by only a single nucleotide (Liu et al., 2009). Both miRNAs target the same HD-ZIP family genes to regulate plant development (Jung and Park, 2007; Zhou et al., 2007).
Although the genetic functions of AGO10 have been described, the molecular mechanism by which it regulates SAM development remains unknown. Here, using an unbiased biochemical approach, we show that AGO10 specifically binds to miR166/165. This association is determined by distinct structural features of miR166/165 duplex; specifically by a combination of a mismatch (12U/*8U) and several adjacent pairings in the duplex. We further show that deficient loading of miR166/165 into AGO10 resulted in the pinhead phenotype. The defective SAM in an ago10 mutant was rescued by simply sequestering miR166/165 in the expression niche of AGO10, but not AGO1. Notably, AGO10 has a higher affinity for miR166 than does AGO1, leading to the preferential loading of miR166 into AGO10. Moreover, the binding capability of AGO10 to miR166/165, but not its catalytic activity, is both necessary and sufficient for the proper SAM development. We propose that AGO10 controls SAM development by specifically sequestering miR166/165 and preventing their loading into AGO1, allowing miR166/165 activity to be antagonized, and therefore the HD-ZIP genes to be upregulated.
RESULTS
Identification of AGO10-associated sRNAs
To identify AGO10-bound sRNAs, we purified AGO10 complexes from flowers of ago10-3; PAGO10-8His-Flag (HF)-AGO10 transgenic plants (Supplemental Information, Fig. S1A–E) using a two-step affinity purification. As shown in Figure 1A, the first step of the purification, using a Ni-NTA column, efficiently enriched the sample for dual-tagged AGO10 protein. An additional immunoprecipitation with an anti-Flag monoclonal antibody removed the remaining non-specifically bound proteins and yielded HF-AGO10 complexes of high purity. The identity of the isolated HF-AGO10 was confirmed by western blot with an anti-Flag antibody (Fig. 1A).
Figure 1.
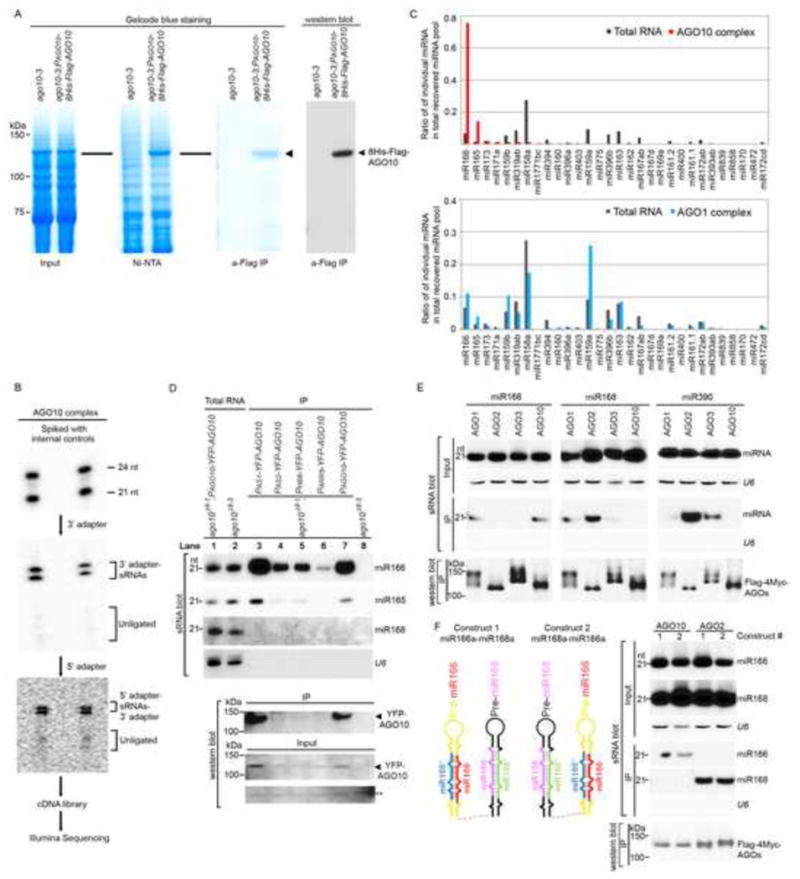
AGO10 predominantly recruits miR166/165. (A) Two-step affinity purification of epitope-tagged AGO10-containing RISCs. (B) Cloning and sequencing of AGO10-associated sRNAs. The sRNAs recovered from AGO10 complexes were spiked with 32P-labeled internal 21 and 24 nt sRNA controls and traced throughout the entire cloning process. (C) Approximately 90% of AGO10-bound miRNAs were miR166/165. (D–F) The specific AGO10-miR166/165 interaction was confirmed in Arabidopsis (D) and in N. bentha (E and F). sRNA blots were conducted with total RNA and sRNAs recovered from immunoprecipitated AGO complexes (IP). Western blot analyses were done with the crude extract and aliquots of the IP products using anti-YFP or -Myc antibodies. A cross-reacting band (**) served as a loading control. See also Fig. S1.
sRNAs associated with the isolated AGO10 complexes were recovered, cloned, and sequenced using Illumina technology (Fig. 1B). After removing internal controls, adapter sequences and reads with lengths <19 nt or >28 nt, the remaining sRNA sequences were mapped to the Arabidopsis genome. In total, 3,574,215 genome-matched sRNA reads were obtained, representing 395,448 unique sequences. Approximately 85% of AGO10-associated sRNAs were 21 nt in length, and 97% contained a 5′ uridine. Further analysis revealed that 80% of the AGO10-bound sRNAs were annotated miRNAs (Table S1), indicating that AGO10 is mainly engaged in the miRNA pathway. The remaining AGO10-bound sRNAs were derived from ta-siRNAs (7.6%), repeat-associated sequences (1.4%), natural antisense siRNAs (Sunkar et al., 2007) (1.1%) and annotated coding regions (9%).
The sRNAs that matched previously defined miRNA families were analyzed further. As a control, miRNAs from crude extracts and AGO1 complexes that were purified in the same manner as AGO10 were also cloned. Absolute read counts of individual miRNA species as a reflection of AGO-associated sRNAs in the isolated AGO complexes, however, are not directly comparable due to differences in the total number of sRNA reads obtained from each sample (Table S1). Therefore, sRNA association with AGO complexes was assessed by calculating the ratios of individual miRNA family reads relative to total miRNA reads in the AGO complexes. These ratios were further compared with those obtained from the crude extracts. Surprisingly, 90% of the AGO10-bound miRNAs were miR166/165, whereas these miRNAs made up only 8% of the total miRNAs present in the crude extract (Fig. 1C). Thus, miR166/165 was enriched more than 11-fold in the AGO10 complex compared to the crude extract. This pattern was not observed for the miRNAs associated with either AGO1 or other AGO complexes (Table S1 and Fig. 1C; Mi et al., 2008).
AGO10 interacts specifically with miR166/165
To investigate if AGO10 prefers miR166/165, we conducted sRNA blot analysis with sRNAs isolated from purified YFP-AGO10 complexes. Consistent with the sequencing results, miR166/165 were overrepresented in AGO10 immunoprecipitates, whereas other miRNAs were below detectable limits (Fig. 1D, lane 7). AGO10 displays a spatiotemporally dynamic expression pattern emanating from the SAM, the adaxial side of the cotyledons to the vasculature (Tucker et al., 2008). To investigate whether AGO10 recruits miR166/165 throughout its dynamic expression, we isolated YFP-AGO10 expressed from the promoter sequences of a few marker genes that are expressed at specific embryonic stages and in specific embryonic regions, such as Homeobox Gene 8 (HB8), Arabidopsis response factor 5 (ARR5), Asymmetric leaves 1 (AS1) or Asymmetric leaves 2 (AS2) (Tucker et al., 2008). sRNA blot analysis showed that AGO10 was consistently accompanied by miR166/165 (Fig. 1D).
The predominant association of miR166/165 with AGO10 could be explained by the coincident overlapping expression patterns of the AGO10 and miR166/165 genes. To rule out this possibility, we conducted competitive immunoprecipitation (IP) assays in an N. benthamiana (bentha) expression system. The Arabidopsis genome has seven copies of miR166 and two copies of miR165 genes. We chose to use only miR166a for further experiments because miR166/165 derived from all precursors were effectively loaded into AGO10, although the absolute amounts of miR166/165 in the AGO10 immunoprecipitates were proportional to the expression levels of miR166/165 and the amounts of recovered AGO10 protein (Fig. S1 F and G). We next co-expressed different 35S-Flag-4 Myc-AGO constructs with precursors of miR166a, 168a and 390b in N. bentha (Fig. 1E). We immunoaffinity-purified AGO1, 2, 3 and 10 complexes and assayed their binding to particular miRNAs. Figure 1E shows that AGO10 indeed only recruited miR166 but not miR168 or 390, while other AGOs did not demonstrate this preference. Interestingly, AGO2 was reported to favor sRNAs with a 5′ A (Mi et al., 2008), while in our experiments it also bound to miR168 (Fig. 1E). To more precisely control the spatiotemporal co-expression of the tested miRNAs in the transient system, we constructed clusters of miRNA precursors, represented by two contiguous miRNA precursors in the same vectors driven by the 35S promoter (i.e., 35S-miR166a-miR168a or 35S-miR168a-miR166a) (Fig. 1F, left schematic). Both miR166 and miR168 were efficiently processed regardless of the order in which the precursors were placed (Fig. 1F, right panel). When co-expressed with the AGO genes, only miR166, but not miR168, was sorted into AGO10. Our co-IP competition experiments clearly indicated that there is a specific interaction between AGO10 and miR166/165 in planta.
The miR166 sequence is not the major determinant for its specific association with AGO10
We hypothesized that miR166/165 possesses some unique sequences that are recognized by AGO10. To test this, we generated a series of point mutations in the miR166 sequence by swapping nucleotides between miR166 and its complementary strand in the precursor (Fig. 2A). Nucleotide swapping was employed to maintain major/minor groove structures in the miRNA/* duplex. The only two exceptions were nucleotides 12 and 13 in the miR166 strand, which share the same sequence (U) with the star strand (* nucleotides 8 and 7). In these cases, mutations of U to A were created in both strands. Most miR166 mutants expressed from the miR166a precursor accumulated to levels comparable with the wild type miR166, except for the miR166 C11G *G9C mutation, the expression of which was barely detectable (Fig. 2B). An additional mutation (miR166 C11U) also resulted in poor yield (Fig. S2A), suggesting that C11 is critical for the effective processing of miR166. When co-expressed with AGO10, most miR166 mutants, except for miR166 U1A *A19U, were efficiently loaded into AGO10 (Fig. 2B–E). The 5′ nucleotide mutation precluded its loading into AGO10, suggesting that the previously reported rule of 5′ nucleotide discrimination (Mi et al., 2008) could now extend to AGO10. Two other mutations (miR166 C7G * G13C and G10U *U10G) impaired the loading of the miRNAs into AGO10 less severely (Fig. 2E).
Figure 2.
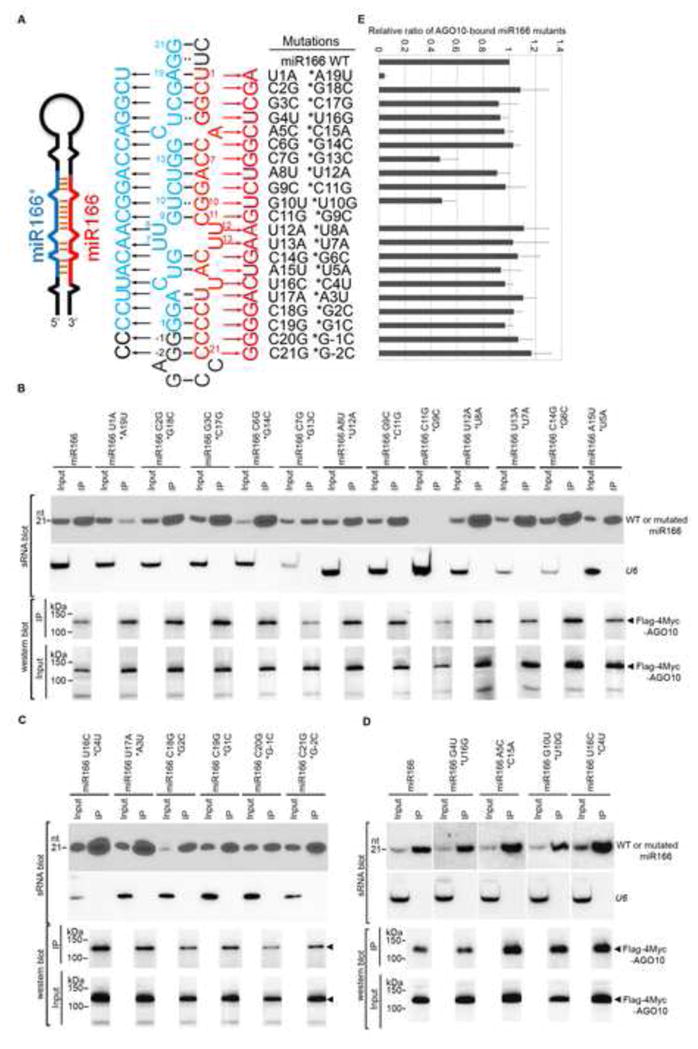
Few mutations in the miR166 sequence compromised miRNA loading into AGO10. (A) Schematics of point mutations in miR166 and its * strand. Predicted foldback of miR166a (Left panel). Paired single mutations in miR166 (red) and its * strand (blue) (Middle panel). The outside region of the miR166/166* duplex is shown in black. Numbers (i.e. 1–21) were given next to miR166/166* to show its orientation. −1 and −2 indicate their relative positions to the start of the miR166* strand. (B–D) Loading of miR166 mutants into AGO10 in N. bentha. Analyses of sRNA and western blots were conducted as in Figure 1E. (E) The relative mean signals of miR166 mutants/AGO10 were normalized to that of miR166/AGO10 with ±SD from seven experiments. Note: miR166 C11G *G9C was not detected in the input. See also Fig. S2.
The simple 5′ nucleotide rule, however, does not explain AGO10 specificity for miR166/165 because most Arabidopsis miRNAs contain a 5′ U but do not extensively bind to AGO10. miR166/165 contain four or five adjacent cytosines at their 3′ ends. This unique feature raises the possibility that AGO10 might contain a special pocket in its PAZ domain to recognize the cytosine tails. To test this, double mutations were introduced at the 3′ end of miR166 (C18G C19G, *G2C G1C; C20G C21G, *G-1C G-2C). Similarly, a miR166 mutation (C2G G3C, *G18C C17G) close to the 5′ end was introduced as a control. Figure S2B shows that none of the double mutations had a significant effect on their loading into AGO10. To further exclude the possibility that the unique C-tail is a major determining factor for AGO10-miR166 binding, we engineered a mutant form of miR168 with “CCCC” replacing the four nucleotides at its 3′ end. When co-expressed with AGO1 and AGO10, miR168-CCCC was loaded into AGO1 as efficiently as miR168; however, it was not detected in AGO10 complexes (Fig. S2 C and D). Taken together, these data indicate that a unique 3′ end of four Cs is neither necessary nor sufficient for specific AGO10-miR166/165 association.
The miR166/166* structure determines the predominant AGO10-miR166 association
Because most single mutations and tested double mutations in miR166 did not significantly affect its association with AGO10, we next hypothesized that the internal structure of the miR166/166* duplex might be important for its interaction with AGO10. To test this possibility, we introduced two forms of miR166a duplexes into miR168a and 390b precursors in place of miR168/168* and miR390/390* duplexes. One duplex contained the authentic miR166/166* duplex-mispairing structure (miR166/166*390), and the others contained miR168a- and 390b duplex-like-mispairing structures (miR166/168*-like168 and miR166/390*-like390, Fig. 3A). Primer extension experiments showed that miR166 processed from pre-miR166a and the chimeric precursors had the correct 5′ ends (Fig. 3B). miR166 expressed from these wild type or chimeric precursors accumulated to comparable levels (Fig. 3C). However, the ability of miR166 derived from miR166/168*-like168 and miR166/390*-like390 to co -IP with AGO10 was either substantially decreased or abolished, whereas miR166 processed from miR166/166*390 maintained a strong association with AGO10 (Fig. 3C). In contrast, the differential binding abilities of miR166 from different stem-loop contexts were not obvious with AGO1 (Fig. 3D). These results indicated that the miR166/166* structure was sufficient to direct its predominant association with AGO10.
Figure 3.
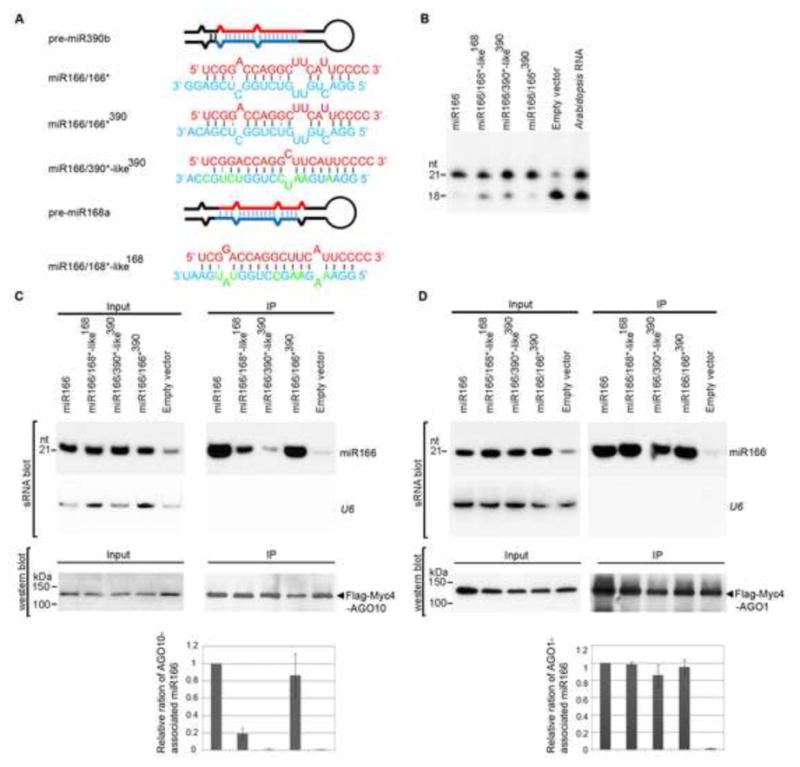
The internal structure of miR166/166* determines the specific AGO10-miR166 association. (A) Predicted foldbacks of pre-miR390b and -miR168a and chimeric precursors expressing miR166. miR166 (red) and its * strand (blue) are shown. The mutated nucleotides in the miR168*- and miR390*-like strands are shown in green. (B) Primer extension experiments were conducted with total RNAs prepared from N. bentha transfected with the indicated constructs. (C and D) Change of the miR166/166* structure dramatically decreased the loading of miR166 to AGO10 (C), but not to AGO1(D). Analyses of sRNA and western blots were conducted as in Figure 1E. The relative mean ratio of miR166/AGO10 (or AGO1) was normalized to that obtained with pre-miR166a with ±SD from five repeats (bottom panels). See also Fig. S3.
Specific mispairings and pairings in the miR166/166* duplex determine its dominant association with AGO10
The duplex regions of miR166/165 and their star strands contain more mismatches than those of the other miRNA/*s (4.5 vs. ~2.5). This distinct characteristic might contribute to the specific preference of AGO10 for miR166. To test this hypothesis, we generated mutations to remove the bulges in duplex region of miR166/166* (Fig. S3A). For assessment convenience, only nucleotides in the star strand were mutated, except for the C22U mutation, which is in an outside region of the miR166/166* duplex. Mutations in the star strand did not affect the accurate maturation of miR166 (Fig. S3B). Figure S3C shows that miR166* C15U U16C and pre-miR166 C22U did not compromise the effective loading of miR166 into AGO10. In contrast, full pairing at the 3′ end side of the miR166/166* duplex (miR166* C4A U7A U8A) dramatically decreased the loading of miR166 to AGO10. Further mutations demonstrated that miR166* C4A and U7A had limited or no obvious effect on AGO10-miR166 association, whereas miR166* U8A and miR166* U7A U8A substantially reduced their loading to AGO10, but not AGO1, indicating that miR166 12U/*8U provides a critical signature for AGO10-miR166/165 recognition (Fig. S3 D and F). To identify additional positions that might contribute to the sorting of miR166 into AGO10 complexes, we generated a quintuple mutation in the miR166* strand (miR166*C4A U7A U8A G9U U10C) (Fig. S3A). The duplex structure of this mutant mimicked the 5′ end side of miR166/390*-like390 (Fig. 3A). This quintuple mutation further compromised its ability to bind to AGO10 (Fig. S3D), whereas it showed no effect on its loading to AGO1 (Fig. S3F), indicating that the pairings of miR166 11C/*9G and 10G/*10U are also important contributors to AGO10 selectivity.
To investigate whether any pairing in the 3′ side of the miR166* strand has an effect on miR166 loading into AGO10, we generated a series of triple mutations in the miR166* C15U U16C template (Fig. S3A). We chose miR166* C15U U16C as a template rather than miR166a because any additional mismatches introduced into the 3′ end side of the miR166* strand might distort the structure of miR166/166* due to the presence of two mismatches (4G 5A and *16U 15C) in the adjacent regions. Co-IP experiments showed that miR166* G13A C15U U16C significantly compromised AGO10-miR166 association, while miR166* G14A C15U U16C did not (Fig. S3E). Interestingly, miR166* G13 and miR166 C7 pair with each other, and mutations in either nucleotide (Figs. 2E and S3E) decreased AGO10-miR166 association, indicating that this pairing is an additional contributor to the strict selection between AGO10 and miR166. Taken together, we conclude that the predominant association of AGO10-miR166/165 is determined by the distinct structure of the miR166/166* duplex, specifically by a combination of a mismatch (12U/*8U) and several adjacent pairings in the duplex.
Deficient incorporation of miR166 into AGO10 causes a pinhead phenotype
AGO10 is involved in SAM maintenance and the establishment of leaf polarity. miR166/165 target class III HD-ZIP transcription factors, all of which participate in these developmental processes. In light of our finding that AGO10 predominantly recruits miR166/165, we hypothesized that AGO10 controls SAM maintenance through miR166/165. We reasoned that introducing into plants an excess of “dominant-negative” miR166 that could not be sorted into AGO10 might allow us to determine whether ago10 phenotypes arise from an inability to load miR166 into AGO10. To test this hypothesis, we generated transgenic plants that constitutively express miR166/390*-like390, which should be selectively loaded into AGO1, but not AGO10, in Col-0 and Ler backgrounds. Transgenic plants overexpressing miR166a, and miR166/166*390 were also generated as controls.
A majority of miR166-overexpressing transformants, regardless of the precursor context, demonstrated a diverse array of phenotypic alternations such as downward curled leaves and stunted growth; some eventually died after the appearance of a few pairs of rosette leaves (Fig. 4A). Analyses of sRNA and northern blots showed that these phenotypes correlated with miR166 over-accumulation and subsequent down-regulation of its target genes (Fig. 4B and C). Interestingly, the expression of AGO10, but not AGO1, was upregulated approximately 4-fold, suggesting that miR166 might participate in a feedback loop to enhance AGO10 expression (Fig. 4C).
Figure 4.
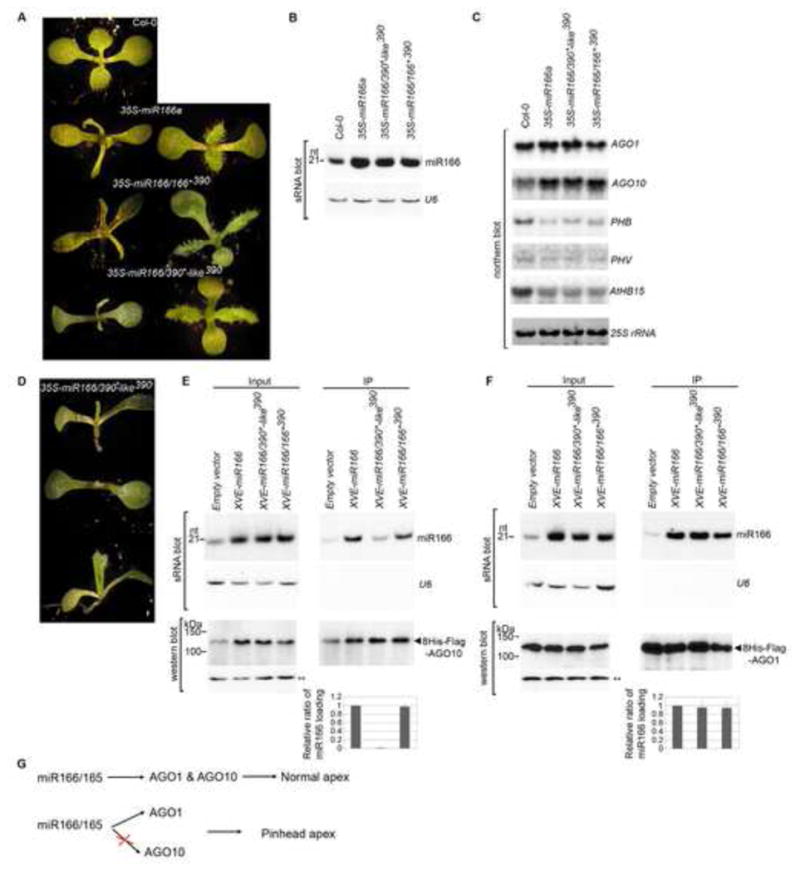
Deficient loading of miR166 into AGO10 causes pinhead phenotypes in the Col-0 background. (A) Shared morphological phenotypes of 35S-miR166a, -miR166/166*390 and -miR166/390*-like390 plants. Photographs were taken of 10-day-old seedlings. Two representative lines are shown for each construct. (B) miR166 level was measured by sRNA blot analysis. (C) Transcript levels of selected AGO and HD-ZIP family genes were measured by northern blot analysis. (D) Unique pinhead phenotypes of 35S-miR166/390*-like390 plants. (E and F) Deficient loading of miR166 from the miR166/390*-like390 precursor into AGO10 (E) but not AGO1 (F) in Arabidopsis. Analyses of sRNA blot and western blot (using an anti-Flag antibody) were conducted as in Figure 1D. The exposure times for AGO10 and AGO1 protein blots were 30 and 5 seconds, respectively. The relative mean ratio of miR166/AGO10 (or AGO1) was measured as in Figure 3 with ±SD from three experiments (bottom panels). (G ) Correlation of the imbalanced loading of miR166/165 into AGO1/AGO10 with pinhead phenotype. See also Fig. S4.
More importantly, approximately 15% of miR166/390*-like390 transformants in either the Col-0 (n>200; Fig. 4D) or Ler background (n>100; Fig. S4) showed the expected pinhead phenotype (Fig. 4D, top two panels), and 41% contained a pinhead-like structure (vertical terminate rosette leaves developed at a later stage; Fig. 4D, bottom panel). These phenotypes were much stronger than ago10 mutants in the Col-0 background which rarely have developmental defects. In contrast, less than 0.3% of transformants (n>200) overexpressing miR166a and miR166/166*390 displayed a pinhead phenotype.
To investigate whether deficient loading of miR166 accounted for the higher frequency of the ago10 phenotype in miR166/390*-like390 transformants than in other miR166 transformants, we created double transgenic plants expressing β-estradiol-inducible pER8-miR166a, -miR166/390*-like390, or -miR166/166*390 in ago10 pnh-2; PAGO10-HF-AGO10 and ago1-27; PAGO1-HF-AGO1 backgrounds (Zuo et al., 2000). Co-IP experiments showed that when expression of the precursors was induced, miR166 was efficiently processed from miR166/390*-like390, and its association with AGO10, but not with AGO1, was significantly reduced in Arabidopsis. In contrast, miR166 generated from miR166a and miR166/166*390 did not show this discrimination (Fig. 4E and F). These results indicated that inefficient AGO10-miR166 assembly led to an imbalance in the distribution of miR166/165 between AGO10 and AGO1 and a corresponding defect in the shoot apex (Fig. 4G).
Sequestering elevated miR166/165 from the expression region of AGO10 but not AGO1 rescues the ago10 phenotype
ago10 pnh-2 is a non-sense mutation (AGO10 Q885*) that abolishes AGO10 binding to miR166 due to improper folding (Figs. S1A and 5A). In ago10 mutants, miR166/165 levels are abnormally high (Fig. 5B). Moreover, these miRNAs accumulate ectopically in the developing meristem (Liu et al., 2009). To investigate whether the ectopically accumulated miR166/165 in ago-10 mutants is re-directed to AGO1, a master repressor of miRNA targets, we examined the levels of miR166 and selected miRNAs in AGO1 complexes isolated from wild type Ler, two ago10 alleles and complemented transgenic lines. sRNA blot analysis showed that the relative amount of miR166/miR159 associated with AGO1 was much higher in ago10 mutants than that in the wild type Ler and the complemented ago10pnh-2; PAGO10-HF-AGO10 plants (Fig. 5B and C). These results indicated that loss-of-function mutations of AGO10 caused a significant increase in the loading of miR166 into AGO1.
Figure 5.
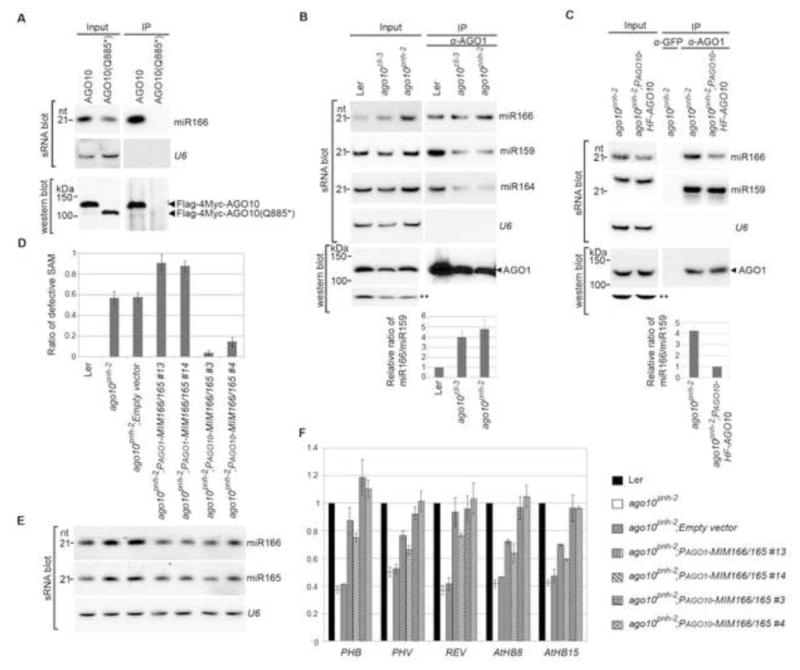
Sequestration of miR166/165 from expression domains of AGO10, but not AGO1, by target mimicry rescues the ago10pnh-2 phenotype. (A) AGO10 Q885* encoded by ago10pnh-2 did not bind to miR166 in N. bentha due to improper protein folding. Analyses of sRNA and western blots were conducted as in Figure 1E. (B and C) ago10 mutation resulted in a significant increase in miR166 binding by AGO1 in Arabidopsis. sRNA blot analyses were conducted with total RNA (input) and sRNA recovered from the AGO1 complexes (IP). Western blot assays were performed using an anti-AGO1 antibody. A cross-reacting band (**) served as a loading control. The relative signal ratio of miR166 to miR159 in AGO1 complexes was normalized to that obtained from the wild type Ler or the PAGO10-HF-AGO10 complemented lines with ±SD from three experiments (bottom panels). (D) Defective SAM was rescued in ago10pnh-2 plants by miR166/165 target mimicry expressed from the promoters of AGO10, but not AGO1. The ratios of defective SAM are shown as mean ±SD from three replicates (n > 200/each replicate). (E and F) Levels of miR166/165 and their target transcripts were measured by RNA blot assays (E) and real-time RT-PCR (F). The relative level of HD-ZIP transcripts was normalized to that in Ler plants with ±SD from four experiments. See also Fig. S5.
In the ago10 mutant, the increased binding of miR166 by AGO1 might result in the down-regulation of HD-ZIP III transcripts in the AGO10 expression domain and further lead to the terminal differentiation of the SAM. According to this model, we reasoned that ago10 mutants might be rescued by hijacking the extra miR166/165 or inhibiting miR166/165 activity in the AGO10 expression domain. To test this, we generated target mimicry constructs (Franco-Zorrilla et al., 2007) expressing PAGO10-MIM166/165 to sequester miR166/165 in the expression niche of AGO10. Consistent with our expectation, when transformed into the ago10pnh-2 mutant, PAGO10-MIM166/165 largely rescued the shoot apex defects in the ago10 pnh-2 mutant (Fig. 5D). Interestingly, the steady state level of miR166/165 was also recovered in the ago10pnh-2; PAGO10-MIM166/165 transgenic plants (Fig. 5E), consistent with a recent report that the unproductive assembly of RISCs with a decoy decreases miRNA stability (Todesco et al., 2010). As a result, HD-ZIPIII transcripts were upregulated compared to those in ago10pnh-2 plants (Figs. 5F and S5).
In sharp contrast, AGO1 promoter-driven MIM166/165 could not rescue the ago10 phenotype, although it decreased the overall steady state level of miR166/165 and restored levels of HD-ZIP III transcripts to a large extent (Fig. 5D–F). These results indicated that in ago10 plants, inhibition of miR166/165 activity in the AGO1 expression domain leads to the ectopic accumulation of HD-ZIP III transcripts outside the AGO10 niche. This, however, represses SAM development, a situation reminiscent of the ago1 mutant (Kidner and Martienssen, 2004). These results further suggest that a relative higher level of HD-ZIP III transcripts in AGO10 domain than that in AGO1 domain is a prerequisite for the proper maintenance of the SAM development.
AGO10 maintains SAM development by specifically sequestering miR166/165 from AGO1
AGO10 contains the catalytic Asp-Asp-His (DDH) motif in its PIWI domain. To test its catalytic potential directly, we incubated immunoaffinity-purified AGO10 with a part of a PHV transcript containing a sequence complementary to miR166/165. The PHV mRNA was sliced by AGO10 protein, but not by AGO10 mutants with substitutions of essential catalytic residues (D709A, D793A or H935A; referred to hereafter as DDH mutants) despite comparable miR166/165-binding capacities (Fig. 6A and B).
Figure 6.
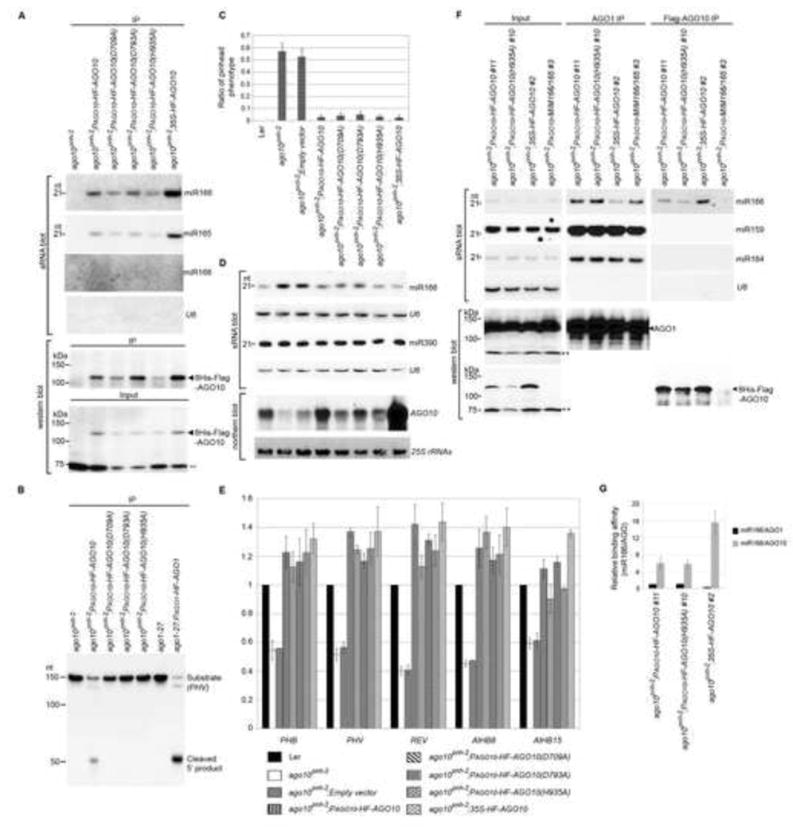
AGO10 rescues the ago10pnh-2 mutant by sequestering miR166/165 from AGO1. (A) AGO10 DDH mutants maintained miR166/165-binding capacity. Assays of sRNA blot and western blot (using anti-Flag antibody) were conducted as in Figure 1D. (B) RISC reconstitution assays of AGO10 and AGO10 DDH mutants. AGO1 was included as a positive control. (C) Non-catalytic AGO10 rescued the ago10pnh-2 mutant as efficiently as catalytic AGO10. The pinhead ratios are shown as mean ±SD from 16 lines (n > 200/line). (D and E) Levels of miR166/165, AGO10 and HD-ZIP III transcripts were measured by analyses of sRNA and northern blots (D) and real-time RT-PCR (E). The relative level of HD-ZIP transcripts was normalized as in Figure 5F.(F) AGO10 sequestered miR166 from AGO1. Analyses of sRNA blot and western blot (using the anti-AGO1 or anti-Flag antibody) were conducted as in Figure 1D. (G) The relative binding of miR166 by dual-tagged AGO10 was normalized to that of miR166/AGO1 isolated from ago10 pnh-2; PAGO10-HF-AGO10 plants with ±SD from three experiments. See also Figs. S5 and 6.
Since simple sequestration or decoy of miR166/165 in the AGO10 expression domain rescued ago10 mutants (Fig. 5D), we reasoned that AGO10 DDH mutants might also complement ago10 mutants because they retain miR166/165 binding capacity and can form an unproductive RISC (Fig. 6B). To this end, we examined the T3 progeny of the transformants harboring wild type AGO10 and each DDH mutant expressed from the AGO10 promoter or the constitutive 35S promoter in the ago10pnh-2 background. Although the accumulation of the AGO10 transcript was substantially increased when transcribed from the 35S promoter compared to its native promoter (Fig. 6D), the steady state protein level was only two-fold higher (Fig. 6A and F), suggesting possible regulation of the AGO10 protein itself. Intriguingly, more than 97% of all T3 transformants (20 lines; >200 plants/line) expressing AGO10 from both the native AGO10 and the 35S promoters, or AGO10 DDH mutants from the AGO10 promoter displayed a normal shoot apex, whereas those with the empty vector did not (Fig. 6C). Moreover, miR166 accumulation was decreased in these complemented lines relative to levels in ago10pnh-2 mutants (Fig. 6D). Consistent with the re-establishment of miR166/165 levels, HD-ZIP III transcript levels were restored to those observed in wild type plants (Figs. 6E and S5). These results indicated that the slicer activity of AGO10 is unnecessary to rescue the pinhead phenotype.
Loss-of-function mutations of HD-ZIP family genes mimic the ago10 phenotype (Prigge et al., 2005). Our complemented plants expressing AGO10 or AGO10 DDH mutants showed increased or restored HD-ZIP family expression; thus, AGO10 is a positive regulator of HD-ZIP family genes and unlikely to be involved in the translational repression of HD-ZIP genes. Given the strikingly similar molecular and phenotypic characteristics of ago10pnh-2; PAGO10-MIM166/165 and ago10pnh-2; PAGO10-HF-AGO10 (DDH) plants, we propose that the main regulatory function of AGO10 in SAM maintenance is to sequester miR166/165 and to antagonize their activity. Under this model, we expect that AGO10 should have a higher affinity for miR166 than AGO1. To test this hypothesis, we first investigated the relative binding affinity of AGO1 and AGO10 to miR166 in N. bentha. When co-expressed with AGO1 or AGO10 genes, over-accumulated miR166 was readily loaded into either of the AGO complexes (Fig. S6A). However, when co-expressed with both AGO1 and AGO10, more miR166 was recruited into AGO10 than AGO1 despite a lower amount of AGO10 protein compared to that of AGO1. Imaging quantification analysis showed that the relative signal ratio of miR166/AGO10 was significantly higher than that of miR166/AGO1, indicating that miR166 was preferentially loaded into AGO10 over AGO1 when both are present (Fig. S6B). We further examined levels of miR166 in AGO1 and AGO10 complexes isolated from complemented transgenic plants expressing PAGO10-HF-AGO10 or -AGO10 (H935A) and 35S-HF-AGO10. Since the accumulation of dual-tagged AGO10 was about 40-fold less than that of endogenous AGO1 in ago10pnh-2; PAGO10-HF-AGO10 plant (Fig. S6C and D), the absolute amount of miR166 in AGO10 was slightly less than that in AGO1 complexes (Fig. 6F). However, the relative level of miR166 recovered from immunoprecipitated AGO10 was 4~6-fold higher than the amount recovered from AGO1 (Figs. 6F and G; S6E and F). More strikingly, more than 80% of miR166, but not control miRNAs, was re-directed from AGO1 to AGO10 upon expression of 35S-HF-AGO10 relative to the distribution in the plants expressing PAGO10-HF-AGO10, despite only a two-fold increase in the levels of dual-tagged AGO10 (Figs. 6F and G; S6E and F). Together, these results indicated that AGO10 binds to miR166 with a higher affinity than does AGO1, and thus possesses the capability to function as a decoy for miR166 in plants.
Finally, since AGO1 and AGO10 are the closest genetic paralogs and are believed to have functional redundancy, we directly tested whether AGO1 could replace AGO10 and vice versa by promoter swapping. We transformed PAGO10-HF-AGO1 into ago10pnh-2. Surprisingly, about half of the primary transformants harboring PAGO10-HF-AGO1 showed phenotypes suggestive of AGO1 co-suppression and the ago10 phenotype. An additional 10% of the transgenic plants exhibited a pinhead phenotype (Fig. S7A). These results indicate that AGO1 cannot substitute for AGO10 in SAM maintenance. On the other hand, PAGO1-HF-AGO10 was unable to rescue the morphological defects in ago1-27 hypomorphs (Fig. S7B) and also caused upward curled leaves. Taken together, these data suggest that AGO10 and AGO1 have functionally distinct roles and are unable to complement each other, with the additional implication that these functions may result from intrinsic differences in sRNA binding preferences (Table S1).
DISCUSSION
By identifying sRNAs of AGO10-containing RISCs, we discovered that AGO10 predominantly associates with miR166/165 to regulate SAM development. The AGO10-miR166/165 interaction is a unique case in which an AGO protein specifically binds to a particular group of miRNAs to execute its biological function; only one other such interaction has been described (Montomery et al., 2008).
The unique secondary structure of the miR166/165 duplex determines their specific association with AGO10
How does AGO10 specifically select miR166/165 among hundreds of miRNAs and an overwhelming number of siRNAs? All miR166/165 family members have adopted distinct structures in their miRNA/* regions that are absent among the rest of the Arabidopsis miRNAs. We found that this distinct structure accounts for the specific affinity of AGO10 for miR166/165. We have further mapped the critical positions (miR166 12U/*8U and its adjacent nucleotides) that are responsible for the specific sorting of miR166/165 to AGO10. Intriguingly, miR166/165 12U/ *8U is the only mismatch conserved among the entire miR166/165 family (Fig. S3G). How can the unique mispairing and adjacent residues in miR166/165 be sensed? One possibility is that there is a factor that recognizes this particular region of miR166/165 and funnels this group of miRNAs into AGO10. However, we favor the idea that specific recognition could be conferred by AGO10 itself. It has been proposed that sRNA duplexes are unwound before loading into an AGO such that only the guide strand, which contains the less-stably paired 5′ end, is incorporated into the AGO (Tomari et al., 2004). However, in Drosophila, siRNA duplexes are indeed loaded into RISCs such that the guide strand of the siRNA duplexes directs Argonaute-catalyzed cleavage of the passenger strand (Matranga et al., 2005). Moreover, there is increasing evidence that AGOs participate directly in miRNA biogenesis and maturation (Cheloufi et al., 2010; Cifuentes et al., 2010; Diederichs and Haber, 2007). Although the RISC loading process has not yet been investigated in Arabidopsis, a previous study with suppressor 2b from Cucumber Mosaic Virus strongly suggests that miRNA/* duplexes might be loaded into AGO by a passenger-strand cleavage-assisted mechanism (Zhang et al., 2006). If so, the distinct structure of the miR166/165 duplex may be recognized and further selected by AGO10.
The mechanism by which miRNA/* structure determines its routing, which we have uncovered here in plants, is reminiscent of mechanisms that have been reported in Drosophila and C. elegans. In these systems, perfectly complementary duplexes are channeled into AGO2 as siRNAs. In contrast, the presence of mismatches in miRNA duplexes promotes their incorporation into AGO1 (Forstemann et al., 2007; Tomari et al., 2007; Steiner et al., 2007). Nevertheless, there is a fundamental difference between the AGO10-miR166/165 association in plants and the sRNA sorting mechanisms described in animals. In Arabidopsis, most miRNA duplexes harbor mismatches, whereas they were not enriched in AGO10, indicating specific mismatch recognition by AGO10. However, in animal systems, structures of sRNA duplex play a general instructive role in their sorting to various AGOs.
AGO10 mediates SAM maintenance by specifically sequestering miR166/165 to up-regulate HD-ZIP family genes
Because the primary function of AGO proteins is to repress target genes, one would imagine that AGO10 specifically recruits miR166/165 to down-regulate the HD-ZIP III transcripts. In fact, AGO10 possesses catalytic activity and can slice PHV transcripts in RISC reconstitution assays. This activity notwithstanding, AGO10 is a positive regulator of HD-ZIP genes in vivo because transcript levels of all HD-ZIP genes were decreased in the ago10 mutant relative to wild type Ler plants. Consistent with this notion is the previous observation that AGO10 and HD-ZIP III transcripts co-localize (Kidner and Martienssen, 2004). One explanation for this positive regulation might be that AGO10 increases the accumulation of HD-ZIPIII transcripts by genetically repressing miR166/165 expression (Liu et al., 2009).
We favor a notion that AGO10 positively regulates HD-ZIP family genes by acting as a specific decoy for miR166/165 (Fig. 7A). Several lines of evidence support this model: (1) In ago10 mutants, ectopically accumulated miR166/165 is redirected into AGO1, causing a reduction of HD-ZIP transcripts in the AGO10 domain during SAM development and corresponding shoot apex defects (Fig. 7B). (2) Simple sequestration of miR166/165 in the expression niche of AGO10 by target mimicry or non-catalytic AGO10 DDH can upregulate the expression of HD-ZIP family genes and rescue the ago10 phenotype (Fig. 7C and D). (3) Deficient incorporation of miR166 into AGO10 and the resulting imbalanced distribution of miR166/165 between AGO10 and AGO1 lead to a defective SAM (Fig. 4G). Interestingly, ago10 mutants display significant phenotypic difference in Ler and Col-0 backgrounds. In our study, making miR165/166 more accessible to AGO1 than to AGO10 by producing it in a different way leads to a similar phenotype in both ecotypes. This result suggests there may be a factor in Col-0 that normally prevents miR165/166 incorporation into AGO1 in the AGO10 domain in ago10 mutants. (4) AGO10 exhibits stronger binding affinity for miR166 than does AGO1, and thus possesses ability as a decoy for these miRNAs (Figs. 6G and S6). Therefore, we propose that the biological role of AGO10 is to compete with AGO1 by sequestering miR166/165, preventing them from being loaded into AGO1 and subsequently targeting HD-ZIP genes.
Figure 7.
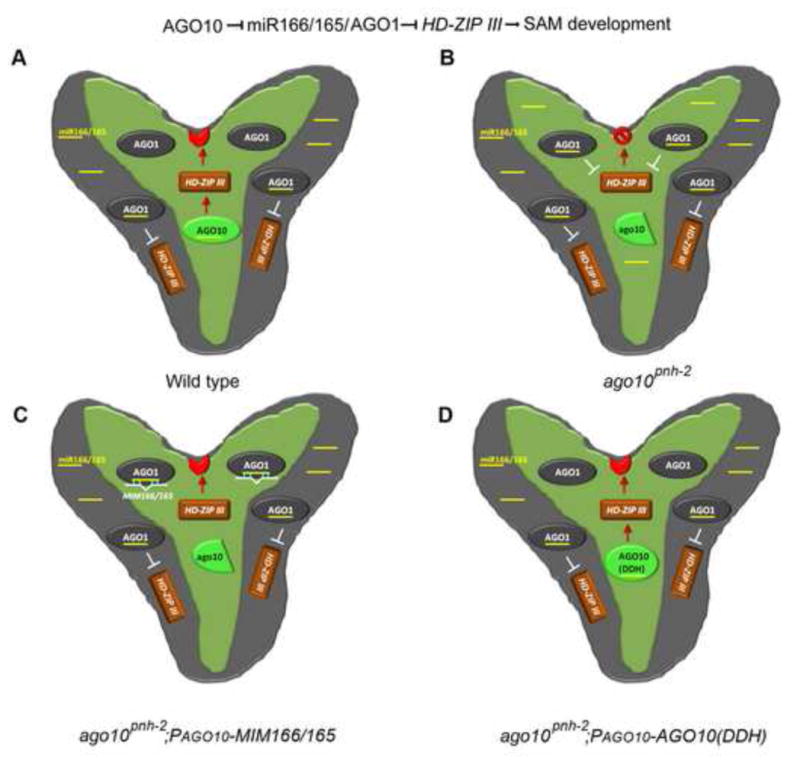
AGO10 maintains the SAM by specifically decoying miR166/165 to upregulate HD-ZIP family genes. SAM (red crescent) is specified by the HD-ZIP transcription factors (brown rectangle) located within the AGO10 expression niche. AGO10 expression (light green) is limited to the provasculature and the adaxial side of the cotyledons. AGO1 is expressed ubiquitously in the whole embryo (grey), partially overlapping the AGO10 expression domain. AGO10 is a positive regulator of HD-ZIP family genes (red arrow), whereas AGO1 is a negative regulator (white T). The terminated SAM is indicated by a red stop sign. miR166/165 and MIM166/165 are shown as yellow and white bars.
Given that AGO10 retains catalytic activity, how is its slicing activity avoided while acting as a decoy for miR166/165 in plants? Several non-exclusive models may apply. First, we envision that there may be some unidentified AGO10-interacting partners that inhibit the slicing function of AGO10. A recent report has shown that leucine-rich repeat kinase 2 (LRRK2) associates with Drosophila and human AGOs and represses their activities (Gehrke et al., 2010). The Arabidopsis genome encodes numerous LRRK proteins, some of which might interfere with AGO10 function. Alternatively, AGO10 may sequester miR166/165 away from the developing meristem into a particular niche in planta to prevent the ectopic accumulation of these sRNAs in the SAM. Consistent with this view is the finding that AGO10 is specifically required in the vasculature below the SAM (Tucker et al., 2008). Considering the cell-autonomous accumulation of AGO proteins and non-cell-autonomous functions of miRNAs, AGO10 may sequester miR166/165 in the vasculature and prevent their movement into the meristem above (Chitwood and Timmermans, 2010). Third, AGO10 slicing activity may be less efficient than that of AGO1 in vivo, resulting in a net reduction of miR166/165 potency when sequestered into AGO10. Finally, AGO10 may trigger miR166/165 turnover in addition to its sequestration, lowering the effective miR166/165 population.
Functional diversification and redundancy of AGO10 and other AGOs in Arabidopsis
In addition to SAM maintenance, AGO10 also plays a critical role in organ polarity and vascular development. Although we have not examined these developmental processes in our study, it is likely that AGO10 regulates these biological events through its association with miR166/165. Of course, we have no reason to exclude the possibility that AGO10 might bind to other sRNAs to regulate their targets. Our sequencing results revealed that AGO10 does recruit a spectrum of miRNAs and numerous ta-siRNAs, although their relative ratios are very low. A previous study with Arabidopsis AGO4 suggest that a single AGO protein may function as a catalytic engine of RNA cleavage whilst it can also execute slicing-independent regulation of smRNA targets (Qi, et al., 2006). Given that AGO10 does possess slicer activity, it will be intriguing to investigate whether or not this activity is required for regulation of sRNA targets other than HD-ZIP family genes.
Can the role of AGO10 in SAM maintenance be performed by other AGOs? Here, we showed that AGO1 could not replace AGO10 with regards to SAM maintenance and vice versa, in terms of leaf morphology. This functional diversification between AGO1 and AGO10 is apparently determined by differential binding capacities for different spectrums of miRNAs. Intriguingly, AGO1 and AGO10 have 86% similarity and 78% identity in their PAZ/PIWI domains, but less than 20% similarity in their N-terminal regions; yet their miRNA-binding preferences are distinct. Recently, an elegant study showed that only the PAZ domain, which is thought to bind to the 3′ end of sRNA, is exchangeable between both proteins, whereas the MID-PIWI and N-terminal domains appear to contribute to their functional specificity (Mallory et al., 2009). Further dissection of the AGO10 and AGO1 protein structures will advance our insight into the mechanisms underlying the differential miRNA-binding preferences of these AGOs.
Experimental Procedures
Tandem-affinity purification of AGO10 complexes, sRNA cloning and Illumina Sequencing
Homozygous T2 progeny of complemented plants expressing ago10-3; PAGO10-HF-AGO10 and ago1-27; PAGO1-HF-AGO1 were used for preparation of AGO complexes. Flower samples including floral buds, open flowers, and newly set siliques (1–2 day old) were collected for protein extraction and isolation of dual-tagged AGO complexes using a two-step affinity purification (Supplemental Information). The isolated AGO complexes were divided into two parts, one aliquot was used for sRNA extraction with Trizol reagent, whereas the other part was used for monitoring protein purity by Gelcode blue staining and western blot using a monoclonal anti-Flag antibody (Sigma) as previous described (Zhang et al., 2006).
sRNA libraries were prepared as described (Hafner et al., 2008; also in Supplemental Information). The cDNA libraries, which were generated within the exponential phase of amplification, were used for high-throughput sequencing using Genome Analyzer II (Illumina).
Co-immunoprecipitation experiments
Total protein was extracted in the IP buffer containing 50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 4 mM MgCl2, 5 mM DTT, 0.1% Triton-100 and the complete protease inhibitor cocktail (Roche). Cleared protein extracts were immunoprecipitated with agarose-conjugated monoclonal antibodies against Flag or Myc or HA (Sigma). For the immunoprecipitation of YFP-AGO10 or endogenous AGO1, protein extracts were mixed with a monoclonal anti-YFP (Invitrogen) or a polyclonal anti-AGO1 antibody together with Protein A beads and incubated for 2 hr. Beads were washed four times with the same buffer before recovery of sRNAs and analyses of sRNA and western blots.
RNA blot and western blot analyses
Assays of sRNA, northern and western blots were performed as described (Zhang et al., 2006; also in Supplemental Information and Table S2).
Supplementary Material
Acknowledgments
We are grateful to K. Barton for the ago10 pnh-2 mutant, T. Laux for the YFP-AGO10 lines, H. Vaucheret for the ago1-27 mutant, and D. Baulcombe for providing an anti-AGO1 antibody. We thank Drs. D. Shippen, G. Kapler, F. Qiao, T. Devarenne and C. Kaplan for stimulating discussions and critical review of the manuscript. The work was supported by a grant from NSF (MCB-0951120) and a startup from Texas A&M University to X.Z. L. L and A.B were supported by NSF-REU (MCB-1037884).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barton MK. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol. 2010;341:95–113. doi: 10.1016/j.ydbio.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos C, Chong M, Hannon GJ. A Dicer-independent miRNA biogenesis pathway that requires AGO catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood H, Timmermans C. Small RNAs are on the move. Nature. 2010;467:415–419. doi: 10.1038/nature09351. [DOI] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor D, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon G, Lawson N, et al. A Novel miRNA Processing Pathway Independent of Dicer Requires Argonaute 2 Catalytic Activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in MicroRNA processing and Posttranscriptional regulation of MicroRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich M, Wee L, Tomari Y, Zamore P. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J, Valli A, Todesco M, Mateos I, Puga M, Rubio-Somoza I, Leyva A, Weigel D, Garcia J, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker E, Wallbridge L, Hardcastle T, Bush M, Kelly K, Dunn R, Schwach F, Doonan J, Baulcombe D. The Arabidopsis RNA-Directed DNA Methylation Argonautes Functionally Diverge Based on Their Expression and Interaction with Target Loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Park CM. MiR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta. 2007;225:1327–1338. doi: 10.1007/s00425-006-0439-1. [DOI] [PubMed] [Google Scholar]
- Kidner C, Martienssen R. Spatially restricted microRNA directs leaf polarity through ARGONAUTE 1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H. The ARGONAUTE 10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 2009;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton K. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE 1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Mallory A, Hinze A, Tucker M, Bouche N, Gasciolli V, Elmayan T, Lauressergues D, Jauvion V, Vaucheret H, Laux T. Redundant and Specific Roles of the ARGONAUTE Proteins AGO1 and ZLL in Development and Small RNA-Directed Gene Silencing. PLoS Genet. 2009;5:e1000646. doi: 10.1371/journal.pgen.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A, Reinhart B, Jones-Rhoades M, Tang G, Zamore P, Barton M, Bartel D. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5 ′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel D, Zamore P. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5 ′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T, Howell M, Cuperus J, Li D, Hansen J, Alexander A, Chapman E, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE 7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D, Slotkin R, Martienssen R, Vielle-Calzada J. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M, Otsuga D, Alonso J, Ecker J, Drews G, Clark S. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang X, Kohany O, Jurka J, Hannon G. Distinct catalytic and non-catalytic roles of ARGONAUTE 4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Song J, Smith S, Hannon G, Joshua-Tor L. Crystal structure of argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Steiner F, Hoogstrate S, Okihara K, Thijssen K, Ketting R, Plasterk R, Sijen T. Structural features of small RNA precursors determine Argonaute loading in C. elegans. Nat Struc & Mol Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in Plant Science. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz D, Bennett R, Cook H, Koppetsch B, Theurkauf W, Zamore P. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tucker M, Hinze A, Tucker E, Takada S, Jurgens G, Laux T. Vascular signaling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. Plant ARGONAUTES. Trends in Plant Science. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan Y, Pei Y, Lin S, Tuschl T, Patel D, Chua NH. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute 1 cleavage activity to counter plant defense. Gene Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Kubo M, Zhong R, Demura T, Ye Z. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007;48:391–404. doi: 10.1093/pcp/pcm008. [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu Q, Chua N. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


