Abstract
We report the synthesis of a macrocycle utilizing a novel framework of standard amino acids in combination with subunits that we have named as Linked Amino Acid Mimetics (LAAM’s). Macrocycles based on the LAAM concept provide both a peptide targeting region and two independently variable functional regions. In the prototype structure, the commonly known Arg-Gly-Asp (RGD) sequence was used for the targeting region. The functional regions contain a phenyl group, and the linkage was formed via a Ring-Closing Metathesis (RCM) reaction.
1. Introduction
Developing small molecule inhibitors of protein-protein binding has been troublesome and screening efforts have not been all that successful, with the p53-MDM2 interaction being a reasonable example of this difficulty.1 The binding surface involved in recognition of protein-protein interactions is typically large and relatively featureless in comparison to the enzyme active sites targeted by traditional drug discovery, complicating the design of small molecules.2 Nevertheless, some progress has been made, with examples being the development of inhibitors of BID-Bcl2 using a hydrocarbon-stapled helix3 and inhibitors of PSD-95-NMDA that use constrained cyclic lactams.4,5 Other advances towards inhibitors of protein-protein binding include miniature folded proteins,6 pophyrins,7 calixarenes,8 cyclodextrins,9 and proteomimetics.10
Macrocycles are common in antitumor, antibiotic and antifungal compounds, but synthetic efforts are often expensive and unpractical.11 When applied to peptides, macrocyclization can enhance the resistance to protease degradation and improve the affinity by restricting conformational flexibility.12,13 Here, we have developed the concept of Linked Amino Acids Mimetic (LAAM) subunits for the formation of a library of macrocyclic peptide structures and we envision their use in primary screening or for lead optimization, especially for protein-protein interactions.
2 Results and discussions
2.1 Description of LAAM-based macrocycles
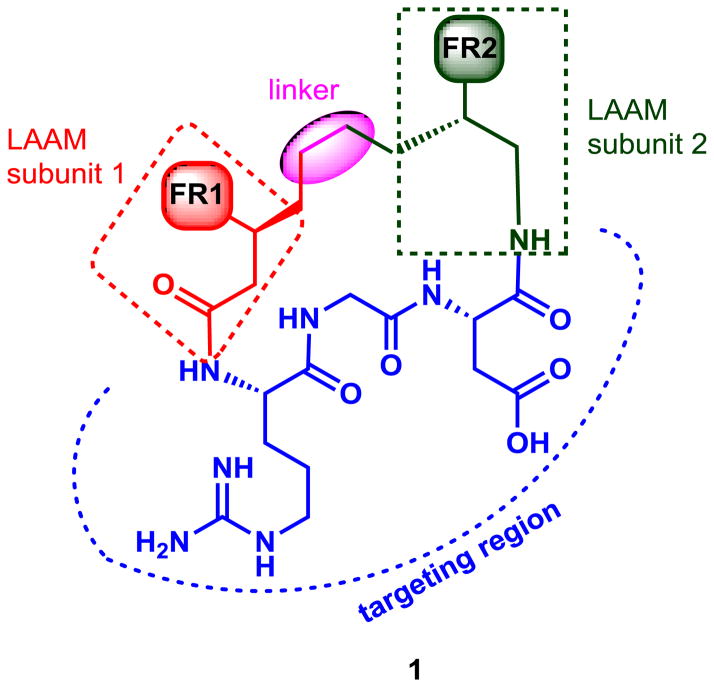
The LAAM-based macrocyclic peptide includes four components: two LAAM subunits (amino and carboxyl), the targeting peptide segment and the linker as shown in Figure 1. Two slightly different LAAM subunits are needed since there is only one attachment point, unlike standard amino acids. Nevertheless, they can still react in a manner compatible with standard solid-phase chemistry, which is important to library formation. The amino and carboxyl LAAM subunits are combined through a Ring-Closing Metathesis (RCM) to form a linker region, which would be considerably more stable in comparison to a peptide bond. This enables the macrocycle to actively participate in potential interactions with a protein target through the functional regions, an aspect which differs from alternative approaches in which the constraint is purposely placed in a non-interacting region of the molecule.4 In the first step towards LAAM based macrocycles, we selected a phenyl group for the two functional regions. This would allow the macrocycle to be an initial prototype for screening and also reasonably mimic the cyclo(RGDf-N(Me)V) peptide that actively binds to αvβ3.14 This may be easily expanded to mimics of natural acids through synthesis of the appropriate acid chloride used in the first step of synthesis.
Figure 1.
Description of LAAM based macrocycle (FR = functional region)
2.2 Synthesis of LAAM subunits
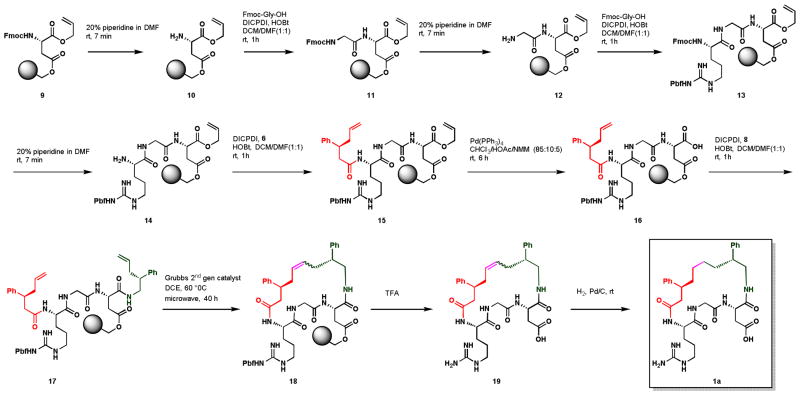
The synthesis of LAAM subunits 6, and 8, commenced with the deprotonation of the Evans oxazolidinone 2 with n-BuLi followed by the addition of trans-cinnamoylchloride 3 to give 4 in high yield (Scheme 1).15 The auxiliary directed allylation of 2 with allylmagnesium bromide in presence of Cu(I)thiophenolate to give the allylated product 5 in good yields with high diastereoselectivity.16,17 Removal of the auxiliary using H2O2-LiOH combination yielded LAAM-1 6, which can be converted in to LAAM-2 8 by Curtius rearrangement followed by acid hydrolysis of the carbomate.18 In our first prototype macrocycle, LAAM-1 and LAAM-2 were chosen to be equivalent, but this is not mandated by the design. Furthermore, the synthetic scheme provided allows for a variety of amine and acid (LAAM) groups and other examples may be synthesized from known α,β-unsaturated carbonyl compounds.
Scheme 1.

Synthesis of LAAM-1 and LAAM-2
With exception to the last two steps, generation of the macrocycle was completed through solid-phase synthesis, as shown in Scheme 2. The synthesis began with the commercially available Fmoc-Arg(OAllyl)-Wang resin 9 through a series of deprotection and coupling steps, the Arg-Gly-Asp (RGD) targeting sequence which is very well known to play a central role in cell adhesion biology as the prototype adhesion signal was added,19 resulting in intermediate compounds 10-13, and final compound 14. Compound 6 was coupled to the free amine 14 in the presence of HOBt and DICPDI (Diisopropylcarbodiimide) to afford compound 15. Deprotection of the allyl group was achieved using tetrakis(triphenylphosphine)palladium(0) to give the free acid 16, which was immediately coupled with amine 8 (LAAM-2) using the similar conditioned applied for compound 15 synthesis to give the metathesis precursor 17. In presence of catalytic about of Grubbs 2nd generation catalyst, compound 17 in dichloroethane was heated at 60 °C under microwave irradiation for 40 h to yield the macrocycle 18. Concomitant removal of Wang resin and pbf group was achieved under milder conditions using TFA, and then catalytic hydrogenation of the double bond of 19 yielded the final desired macrocycle 1a in good yield.
Scheme 2.
Solid Phase Synthesis of Macrocycle
2.3 Selection of stereochemistry for the LAAM subunits
In the synthesis on the LAAMs, we utilized the same chiral auxiliary for both LAAMs resulting in S stereochemistry for LAAM-1 and R for LAAM-2, due to the priority resulting from change of C(=O)OH to NH2. Keeping with the same chiral auxiliary was more convenient and supported by the modeling calculations. Prior to selecting the first macrocycle for synthesis, we had studied an unsaturated version of the structure with all alanine residues. Calculations on this generic macrocycle showed that the combination of R for LAAM-1 and S for LAAM-2 was lowest in energy at −423.148 kJ/mol whereas the combination actually utilized (S for LAAM-1 and R for LAAM-2) was only slightly higher in energy at −422.395 kJ/mol.
2.4 Structure of the LAAM based macrocycle
We attempted re-crystallization of the final product under many different solvent conditions to obtain crystals necessary for X-ray analysis, but those attempts were not successful. To provide a representative three-dimensional structure that may be compared to a known RGD peptide, we predicted the solution conformation through modeling. From the modeling calculations, we identified 25 low energy configurations for structure 1a based on the mixed torsional/low-mode conformational search in MacroModel,20 The relative energy of the resulting conformations ranged from −909.222 to −894.146 kJ/mol, a difference of 15.076 kJ/mol (3.603 kcal/mol). Clustering based on the ring torsions led to nine clusters, three of which contained >2 members (clusters 1, 3, and 7; see supporting information). It is interesting to note that the lowest energy lead structures from each of those clusters also had the lowest overall energies. The lead structures from each of these clusters were then superimposed onto the prototypical RGD peptide bound to αvβ3 in structure 1L5G.21 The RMSD was 1.3780, 1.2818, and 0.8819 for clusters 1, 3, and 7 respectively. Based on this alignment, we believe that cluster 7 would be most representative of a bound configuration of macrocycle 1a, if it were to interact favorably with αvβ3 (Figure 2). The lead energy structure from this cluster is 2nd lowest in energy and only differs from lowest by 0.31 kJ/mol.
Figure 2.
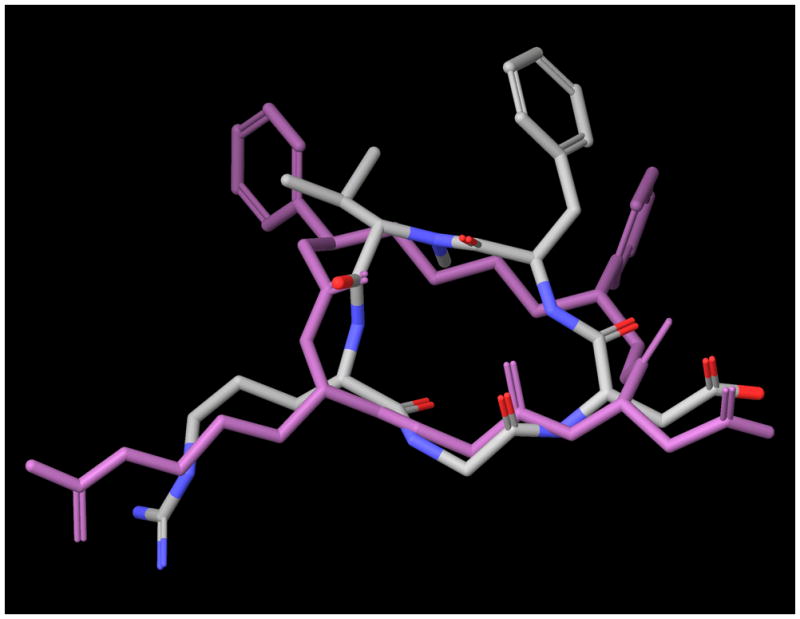
Alignment of lowest energy structure from cluster 7 (blue purple) with RGD peptide bound to αvβ3 (by-atom color)
3. Conclusion
In conclusion, we have provided a synthesis for constructing novel type of subunits that may be easily combined with natural amino acids to build unique macrocycles. We have named these subunits as Linked Amino Acid Mimetics (LAAMs) to signify their ability to link together and mimic amino acids. We have synthesized one example of each LAAM type and utilized those in the construction of a novel macrocycle containing the RGD sequence. The synthesis provided lends itself to diverse set of LAAMs that may further combined into a macrocycle library for general screening or other uses such as development of protein-protein inhibitors. We are currently exploring the synthesis of additional LAAM subunits and will report on the construction of other macrocycles in a later publication.
Supplementary Material
Acknowledgments
We acknowledge financial support from the John S. Dunn Gulf Coast Consortium for Chemical Genomics through the R. A. Welch Foundation Chemistry and Biology Collaborative Grant. The authors would also like to acknowledge the Cancer Center support grant CA016672 for the support of the translational chemistry core Facility and NMR facility at M. D. Anderson Cancer Center.
Footnotes
Supplementary data associated with this article can be found for free of charge via the internet at http://dx.doi.org/
References and notes
- 1.(a) Chene P. Mol Cancer Res. 2004;2:20–28. [PubMed] [Google Scholar]; (b) Vousden KH, Lu X. Nat Rev Cancer. 2001;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]; (c) Momand J, Jung D, Wilczynski S, Niland J. Nucleic Acid Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher S, Hamilton AD. J R Soc Interface. 2006;3:215–233. doi: 10.1098/rsif.2006.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li T, Saro D, Spaller MR. Bioorg Med Chem Lett. 2004;14:1385–1388. doi: 10.1016/j.bmcl.2003.09.103. [DOI] [PubMed] [Google Scholar]
- 5.Piserchio A, Salinas GD, Li T, Marshall J, Spaller MR, Mierke DF. Chem Biol. 2004;11:469–473. doi: 10.1016/j.chembiol.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 6.(a) Chin JW, Schepartz A. J Am Chem Soc. 2001;123:2929–2930. doi: 10.1021/ja0056668. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rutledge SE, Volkman HM, Schepartz A. J Am Chem Soc. 2003;125:14 336–14 347. doi: 10.1021/ja034508o. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gemperli AC, Rutledge SE, Maranda A, Schepartz A. J Am Chem Soc. 2005;127:1596–1597. doi: 10.1021/ja0441211. [DOI] [PubMed] [Google Scholar]; (d) Schneider TL, Mathew RS, Rice KP, Tamaki K, Wood JL, Schepartz A. Org Lett. 2005;7:1695–1698. doi: 10.1021/ol050179o. [DOI] [PubMed] [Google Scholar]
- 7.(a) Jain RK, Hamilton AD. Org Lett. 2000;2:1721–1723. doi: 10.1021/ol005871s. [DOI] [PubMed] [Google Scholar]; (b) Jain RK, Hamilton AD. Angew Chem Int Ed Engl. 2002;41:641–643. [Google Scholar]; (c) Groves K, Wilson AJ, Hamilton AD. J Am Chem Soc. 2004;126:12833–12842. doi: 10.1021/ja0317731. [DOI] [PubMed] [Google Scholar]
- 8.Blaskovich MA, Lin Q, Delarue FL, Sun J, Park HS, Coppola D, Hamilton AD, Sebti SM. Nat Biotechnol. 2000;18:1065–1070. doi: 10.1038/80257. [DOI] [PubMed] [Google Scholar]
- 9.Leung DK, Yang Z, Breslow R. Proc Natl Acad Sci USA. 2000;97:5050–5053. doi: 10.1073/pnas.97.10.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H, Lee GI, Park HS, Payne GA, Rodriguez JM, Sebti SM, Hamilton AD. Angew Chem Int Ed Engl. 2005;44:2704–2707. doi: 10.1002/anie.200462316. and references cited in. [DOI] [PubMed] [Google Scholar]
- 11.Gradillas A, Perez-Castells J. Angew Chem Int Ed Engl. 2006;45:6086–6101. doi: 10.1002/anie.200600641. [DOI] [PubMed] [Google Scholar]
- 12.Levengood MR, van der Donk WA. Bioorg Med Chem Lett. 2008;18:3025–3028. doi: 10.1016/j.bmcl.2008.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Udugamasooriya DG, Spaller MR. Biopolymers. 2008;89:653–667. doi: 10.1002/bip.20983. [DOI] [PubMed] [Google Scholar]
- 14.Dechantsreiter MA, Planker E, Matha B, Lohof E, Holzemann G, Jonczyk A, Goodman SL, Kessler H. J Med Chem. 1999;42:3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 15.Chan CKL, Zhang M-Q, Moinet C, Courchesne M, Reddy TJ. WO2005023809 (A1) International Patent. 2005
- 16.(a) Choi WJ, Kang S-U, Burke TR., Jr Lett Org Chem. 2007;4:433–439. [Google Scholar]; (b) Han Y, Hruby VJ. Tetrahedron Lett. 1997;38:7317–7320. [Google Scholar]
- 17.Gao M, Wang DX, Zheng QY, Wang MX. J Org Chem. 2006;71:9532–9535. doi: 10.1021/jo061664f. [DOI] [PubMed] [Google Scholar]
- 18.(a) Nagumo S, Nishida A, Yamazaki C, Matoba A, Murashige K, Kawahara N. Tetrahedron. 2002;58:4917–4924. [Google Scholar]; (b) Ninomiya K, Shioiri T, Yamada S. Tetrahedron. 1974;30:2151. [Google Scholar]
- 19.(a) Pierschbacher MD, Ruoslahti E. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]; (b) Ruoslahti E. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 20.MacroModel, Version 9.7. 9.7. Schrödinger, LLC; New York, NY: 2009. [Google Scholar]
- 21.Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.