Abstract
In several species including the buffalo cow, prostaglandin (PG) F2α is the key molecule responsible for regression of corpus luteum (CL). Experiments were carried out to characterize gene expression changes in the CL tissue at various time points after administration of luteolytic dose of PGF2α in buffalo cows. Circulating progesterone levels decreased within 1 h of PGF2α treatment and evidence of apoptosis was demonstrable at 18 h post treatment. Microarray analysis indicated expression changes in several of immediate early genes and transcription factors within 3 h of treatment. Also, changes in expression of genes associated with cell to cell signaling, cytokine signaling, steroidogenesis, PG synthesis and apoptosis were observed. Analysis of various components of LH/CGR signaling in CL tissues indicated decreased LH/CGR protein expression, pCREB levels and PKA activity post PGF2α treatment. The novel finding of this study is the down regulation of CYP19A1 gene expression accompanied by decrease in expression of E2 receptors and circulating and intra luteal E2 post PGF2α treatment. Mining of microarray data revealed several differentially expressed E2 responsive genes. Since CYP19A1 gene expression is low in the bovine CL, mining of microarray data of PGF2α-treated macaques, the species with high luteal CYP19A1 expression, showed good correlation between differentially expressed E2 responsive genes between both the species. Taken together, the results of this study suggest that PGF2α interferes with luteotrophic signaling, impairs intra-luteal E2 levels and regulates various signaling pathways before the effects on structural luteolysis are manifest.
Introduction
Corpus luteum (CL) is a transient endocrine structure formed from the remnants of the ovulating follicle. Through secretion of progesterone (P4), it plays a pivotal role in the control of reproduction in mammals. During non-pregnant cycles, regression of CL precedes initiation of new reproductive cycle to allow reovulation and another chance for conception to occur [1]. The structure and function of CL are controlled by intricate interplay between luteotrophic and luteolytic factors. In several species including bovines, the influence of lytic factors dominate over luteotropic factors in the control of luteal function. Of the many factors, prostaglandin (PG) F2α molecule is recognized as the physiological luteolysin responsible for luteal regression in mammals [2]–[4].
PGF2α or its synthetic analogues initiate a complex cascade of events within the CL leading to inhibition of steroidogenesis resulting in rapid fall in circulating P4 levels (functional luteolysis) [5]–[7] and initiation of apoptosis (structural regression) in the bovine species [6], . In several species, many of the endocrine, biochemical and morphological events observed during PGF2α-induced luteolysis have been reported to be broadly similar [11]–[20]. Disruption of transport of cholesterol due to inhibition of StAR expression has been implicated as the cause of inhibition of steroidogenesis after PGF2α treatment under both in vivo and in vitro conditions [21], [22]. However, in a more recent study, an initial increase in StAR expression post intrauterine PGF2α treatment has been observed [23], suggesting factors other than StAR are responsible for inhibition of steroidogenesis. Studies on molecular analysis of structural regression of bovine CL leading to apoptosis has previously been reported by others [24], [25] and us [6], [9], but the actual mechanisms responsible for functional luteolysis has not been systematically examined. Recent studies on transcriptome analysis of CL tissues have expanded our understanding of effects of PGF2α during luteolysis or refractory state of CL to PGF2α effects [26]–[28].
In contrast to a well-established role for pituitary LH in the control of CL function in primates and rodents [29], [30], the role of LH in the regulation of CL function in bovines has not been clearly established. In cattle, an earlier study had suggested a role for LH in CL function [31], while other studies have suggested a minor role for LH especially during pregnancy [32]–[34]. To understand mechanisms involved in the initial loss of CL function in response to PGF2α treatment, it is essential to examine molecules downstream of LH/CG receptor (LH/CGR) activation during PGF2α-induced luteolysis. The role of intraluteal molecules during luteolysis forms another crucial area for examining the PGF2α actions during the initial period of PGF2α treatment [35]. In bovines, even though expression of CYP19A1 that codes for aromatase enzyme responsible for converting androgens to estrogens, gets down regulated after ovulation, its expression is restored albeit at low levels in the developed CL and is never the less responsible for low levels of intraluteal estradiol-17β (E2) [36] in the buffalo cow. Since luteal tissue expresses both α and β receptors [37], it remains to be determined whether biosynthesis of E2 and its receptor signaling is disrupted following PGF2α treatment contributes to the loss of luteal function. Interestingly, E2 is the primary luteotrophic hormone in rabbits and experimental withdrawal of E2 has been shown to inhibit luteal function and activate apoptotic pathways [38], [39].
In view of the above, the following objectives were addressed in the present study: 1) analysis of global gene expression changes in CL of buffalo cows administered with luteolytic dose of PGF2α treatment, 2) examination of effects of PGF2α on molecules downstream of LH receptor signaling and 3) examination of the effects of PGF2α on CYP19A1 expression, E2 biosynthesis and expression of E2 responsive genes in the CL.
Materials and Methods
Ethics Statement
All procedures in animals were approved by the Institutional Animal Ethics Committee, Indian Institute of Science, Bangalore, India.
Reagents
Juramate (Cloprostenol sodium, a synthetic analogue of PGF2α) was purchased from Jurox Pty Ltd, Rutherford, Australia. P4 (GDN#337) and E2 (GDN#244) antisera was kindly provided by Prof. G. D. Niswender, Colorado State University, Fort Collins, CO. The enzyme deoxynucleotidyl transferase was purchased from Amersham Pharmacia Biotech Asia Pacific (Bangalore, India). Moloney murine leukemia virus reverse transcriptase (MMuLV RT) enzyme and 100 bp DNA ladder were obtained from MBI Fermentas GmbH (St. Leon-Rot, Germany). Oligonucleotide and Oligo dT primers were synthesized by Sigma-Genosys (Bangalore, India). DyNAzyme II DNA polymerase was purchased from Finnzymes (Espoo, Finland) and dNTPs were procured from Eppendorf, (Hamburg, Germany). Power SYBR Green PCR master mix was obtained from Applied Biosystems (Foster City, CA). Labeled α-32P dCTP (LCP 103, 3000 Ci/mmole) was procured from BRIT (Hyderabad, India). Random primer extension labeling kit (#KT04) was procured from Bangalore Genei (Bangalore, India). Nylon (Gene screen plus) and PVDF membranes were purchased from NEN life Sciences, (Boston, MA). Polyclonal antibodies specific to LH/CGR (H-50: sc-25828), SF-1 (H-60: sc-28740) and LRH-1 (H-75: sc-25389) were purchased from Santa Cruz Biotechnology Inc (Santa Cruz city, CA). Antibody for β-actin (#A3854) was purchased from Sigma-Aldrich Co. (St.Louis, MO). pCREB (#9191), CREB (#9192), pAkt (#4060), Akt (#9272), pFKHR (#9464), FKHR (#9462), pPI3k p85 (#4228), PI3k p85 (#4257) and horse radish peroxidase labeled goat anti-rabbit IgG and ECL chemiluminescence kit were purchased from Cell Signaling Technology (Beverly, MA). Antibodies for Bax (44–62: 196820) and Bcl-2 (4–21: 197207) were purchased from Calbiochem-Novabiochem Corporation (La Jolla, CA). StAR antibody was a gift from Professor D. M. Stocco (Texas Tech University Health Sciences Center, Lubbock, TX). All other reagents were purchased from Sigma-Aldrich Co. (Bangalore, India) and Life Technologies (Carlsbad, CA) or sourced from local distributors.
Experimental protocol, blood and CL collection schedule
Non-lactating buffalo cows (Bubalus bubalis, Surthi breed) aged 5–6 years, with a history of normal cyclicity were monitored daily for onset of behavioral signs of estrus such as bellowing, restlessness and mucous discharge from the vulva. The estrous cycles were synchronized with two injections of PGF2α and the day of onset of estrus was designated as day 0 of estrous cycle. To verify the presence of functional CL, blood samples were collected on days 3 to 7 of the cycle for monitoring circulating P4 concentration. In this experiment, Juramate (500 µg i.m.) was administered on day 11 of estrous cycle and blood samples (n = 5–6 animals) were collected immediately before and at various time points post PGF2α treatment. CL was collected at slaughter from untreated control animals (0 h), as well as from animals at 1, 2, 3, 6 and 18 h post PGF2α injection (n = 3–4 animals/time point). For serum E2 estimation, blood sampling has also been done at earliest time points, i.e. at 1 and 2 h post PGF2α treatment. CL tissues were also collected at varied points throughout an estrous cycle with day 5, 11, 14, 17 and 20 CL being designated as early (E), mid (M), mid-late (ML), late (L) and very late (VL) CL, respectively. Ovaries containing CL were collected and washed in sterile ice cold PBS and transferred into Dulbecco's Modified Eagles Medium supplemented with penicillin (500 U/ml) and streptomycin (50 µg/ml) and transported to the laboratory on ice within 30 min of collection. Following retrieval of CL from the ovary, CL was cut into ∼50 mg pieces and 3–4 pieces were placed in sterile cryovial and flash frozen in liquid nitrogen before storing at −70 °C for various analyses. The progesterone analysis data and a portion of CL tissue used for different analysis have been reported recently [40].
Hormone assays
For the measurement of luteal P4 and E2, wet weight of one of the CL pieces was determined before homogenization in a known volume of 1X PBS solution. Undiluted CL tissue lysate (E2) or at a dilution of 1∶500 (P4) was used in the assay. Hormone extraction from lysate was carried out using diethyl ether and reconstituted in GPBS at 37°C for 1 hour in an incubator shaker. Luteal and serum P4 and E2 concentrations were determined by radioimmunoassay method as reported previously [41]. The sensitivity of the assay was 10 pg/tube and 3.9 pg/tube for P4 and E2, respectively and the inter- and intra- assay coefficients of variation were <10%.
Isolation of genomic DNA and fragmentation analysis
Genomic DNA was extracted from individual CL using standard phenol-chloroform extraction method. The isolated genomic DNA was precipitated, dissolved and quantitated spectrophotometrically for fragmentation analysis as described elsewhere [6]. Briefly, genomic DNA from different CL tissue samples were analyzed for quantitation of low molecular weight DNA fragmentation using protocol described previously [42], [43] with few modifications. Genomic DNA (4 µg) was labeled with 50 µCi of [α32P] dCTP by incubation at 37°C for 1 h with 10 units of terminal deoxynucleotidyl transferase enzyme. The DNA samples were then resolved on a 2% agarose gel and transferred onto a nylon transfer membrane using the alkaline (0.4 M NaOH and 1 M NaCl) buffer with capillary blotting transfer method for 16 h. The membranes post transfer were cross linked using 1200 µJoules of UV for 1 min in Stratalinker. The membranes were covered with plastic wrap and were exposed to an intensifying screen at room temperature and the low molecular weight labeled DNA signals were quantitated densitometrically using a PhosphorImager (Typhoon 9210; Amersham Biosciences) with exposure of 6 h or longer.
RNA isolation
Total RNA was extracted from control and PGF2α treated CL tissues using TRI Reagent according to the procedure as reported previously [41]. RNA was quantitated spectrophotometrically using ND-1000 (NanoDrop, Thermo Scientific, Wilmington, DE). The quality and quantity of RNA were determined by electrophoresis on a 2% (w/v) formaldehyde agarose gel along with RNA samples of known concentration, and A260: A280 ratio was >1.8. Additionally, the quality of RNA samples was assessed using Agilent Bioanalyzer 2100 and RNA samples with RNA integrity number ≥9 were used for microarray analysis.
Quantitative real time PCR (qPCR)
The qPCR analysis was carried out as described previously from the laboratory [44]. The cDNA samples equivalent to 10 ng of total RNA were subjected to validation analysis on Applied Biosystems 7500 Fast Real Time PCR system with SDS v 1.4 program employing Power SYBR green 2X PCR master mix. The details of primers employed along with the annealing temperature and expected product size are provided in Table S1. Analysis of expression of each gene included a no template control (NTC) and generation of a dissociation curve. Expression level of individual gene was normalized by using L19 expression level as calibrator (internal control) for each cDNA sample. The relative expression of each gene was determined using the ΔΔCt method which calculates the fold change in gene expression using the formula: Fold change = 2−ΔΔCt, where Ct = Threshold cycle i.e. the cycle number at which the relative fluorescence of test samples increases above the background fluorescence and ΔΔCt = [Ct gene of interest (unknown sample) - Ct of L19 (unknown sample)] - [Ct gene of interest (calibrator sample) - Ct of L19 (calibrator sample)]. PCR for each sample was set up in duplicates and the average Ct value was used in the ΔΔCt equation.
Immunoblot analysis
Immunoblot analysis of the total protein lysates from CL tissues was carried out as per the procedure reported previously [6].
Protein kinase A assays
Protein kinase A assays were performed according to the manufacturer's instructions provided with the kit and as described in detail previously [45].
Microarray target preparation and hybridization
Transcriptome analysis of CL tissues was performed on GeneChip Bovine Genome Array (Affymetrix, Santa Clara, CA) that contains 24,072 probe sets representing ∼23,000 bovine transcripts and few housekeeping/control genes. Use of heterologous array hybridization screening for gene expression changes of closely related species has been validated for many species [46]. Affymetrix Bovine GeneChip platform is very efficient and can be easily extended to other species for which genetic sequences are available [47], [48]. The water buffalo and domestic cattle, both belonging to the Bovidae family, are closely related. Moreover, use of robust multiarray average (RMA) algorithm for background correction, normalization across arrays and summarization is well suited for cross species microarray analysis, since the software considers hybridization from perfect match probes and not the mismatch probes [47]. More recently, microarray analysis has been performed on buffalo granulosa cells employing GeneChip Bovine Genome Array and analysis data indicated excellent hybridization [49]. The entire procedure of microarray target preparation, hybridization and scanning were performed at Center for Integrated Biosystems, Utah State University, Utah by employing standard Affymetrix protocols (Chip Expression Analysis Technical Manual rev. 3, 2001). Further, scanning was performed using Affymetrix Genearray Scanner 3000 series.
Analysis of microarray data
Total RNA isolated from control and PGF2α-treated CL tissue was labeled and hybridized to Affymetrix GeneChip Bovine Genome Arrays as per the manufacturer's recommendations. 3 Affymetrix GeneChip Bovine Genome Arrays were used for RNA from individual CL as biological replicates (n = 3/time point; 0, 3, 6 and 18 h post PGF2α treatment). A total of 12 arrays were used. A detailed description of procedures and subsequent generation of processed image files of microarray analysis have been reported previously [44], [49]. The microarray procedure and the data analysis were performed as per Minimum Information About Microarray Experiments (MIAME) compliance. The raw data and the completed analysis of microarray data files have been deposited at NCBI's Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27961 and are accessible through GEO series accession number GSE27961. The processed image files (.cel) were normalized across arrays using RMA software [50] and log-transformed (base 2), which allowed direct comparison of probe set values between all samples used in the experiment normalization. GeneSifter (VizX Labs; Seattle, WA, data not shown), GeneSpring GX (Agilent Technologies, Inc. Santa Clara, CA) and R/Bioconductor (FHCRC labs; Seattle, WA) microarray expression analysis softwares were used to identify differentially expressed transcripts. The differential expression of genes was calculated by averaging the normalized samples and performing a pairwise analysis. Statistical significance for differentially expressed genes among control and treatment groups was determined using Student's t-test (two tail, unpaired) employing Benjamini and Hochberg correction factor for false discovery rate [51]. As many of the original annotations for the Affymetrix Bovine Genome Chip have been reported to be erroneous [52], a computational tool, AffyProbeMiner, was employed to redefine the chip definition files taking into account the most recent genome sequence information [53]. AffyProbeMiner analysis results suggested that the original Affymetrix gene annotation were not compromised and the identified differentially expressed genes were transcript consistent and did not hybridize to multiple transcripts. Analysis of the data by Bioconductor analysis tool employing ≥2 fold change cut-off and statistical filters provided a number of differentially expressed genes. The linear regression analysis was performed on fold change expression values for selected differentially expressed genes obtained from qPCR and microarray analyses and a statistically significant (p<0.05) correlation between the two analyses was determined as reported previously [44], [49]. The list of top 15 differentially expressed (both up- and down-regulated) genes at different time points post PGF2α administration are represented in Table S2, S3, S4, S5, S6, S7. The differentially expressed genes were clustered by hierarchy analysis by GeneSpring analysis for all the probe sets at each time point post PGF2α administration and represented as dendrograms or heat map in Figure S1. Further, to examine the molecular function and genetic networks, the microarray data was analyzed using Ingenuity Pathways Analysis tool (IPA version 8.7, Ingenuity Systems Inc., Redwood City, CA, USA; http://www.ingenuity.com), a web-based software application that enables identification of biological mechanisms, pathways, and functions from the differentially expressed genes.
Statistical analysis
The data were expressed as mean ± SEM. Statistical evaluation of mean differences of E2 and P4, protein blots and qPCR fold change in expression of genes among different treatment groups were analyzed using one-way ANOVA test followed by the Newman-Keuls multiple comparison tests (PRISM GraphPad; GraphPad Software Inc., San Diego, CA). A p-value of <0.05 was considered to be significant.
Results
Effects of PGF2α on circulating P4, StAR expression and biochemical integrity of DNA (DNA laddering) in the CL tissue
Following administration of PGF2α, circulating P4 levels declined significantly within 1 h and continued to be lower at 2, 3, 6 and 18 h time points examined (p<0.05; Figure S2A). The luteal P4 level too declined significantly by 3 h and further declined by 6 and 18 h post PGF2α treatment (p<0.05; Figure S2A). Analysis of StAR protein pattern paralleled P4 decline at all-time points examined (Figure S2B). Analysis of low molecular weight DNA fragments indicated that although a slight increase in low molecular weight fragments could be visualized in CL tissues at 6 h post PGF2α treatment compared to untreated CL tissue, but a clear increase of laddering pattern of DNA was observed in CL tissues collected from animals, 18 h post PGF2α treatment (Figure S2C). These results verified that CL tissue underwent a time related rapid functional loss followed by evidence of structural loss after initiation of PGF2α treatment.
Genome wide analysis of differentially expressed genes in the CL of buffalo cow post PGF2α treatment
The mechanism of PGF2α-induced luteolysis is a complex process involving expression changes in genes associated with regulation of steroidogenesis, apoptosis, expression changes of gene products of several factors such as enzymes involved in PGF2α biosynthesis and cross talk with other signaling pathways that include luteotrophic factors. In the present study, efforts were made to identify temporal changes in the profile of transcriptome of CL tissue in response to PGF2α treatment. The microarray analysis provided a set of differentially expressed genes based on various high stringency statistical filters such as a t-test with p<0.05 and multiple hypothesis testing (Benjamini and Hochberg comparison test) to eliminate the false positives. Further, the differentially expressed genes that passed the statistical filters were classified based on whether the identified differentially expressed gene was present in all the samples examined. Another parameter employed was a sliding scale fold change cut-off filter of ≥2 (except in identification of E2 target genes, in which changes <1.5 fold was considered for analysis) with some degree of confidence in our candidate list to narrow down the list.
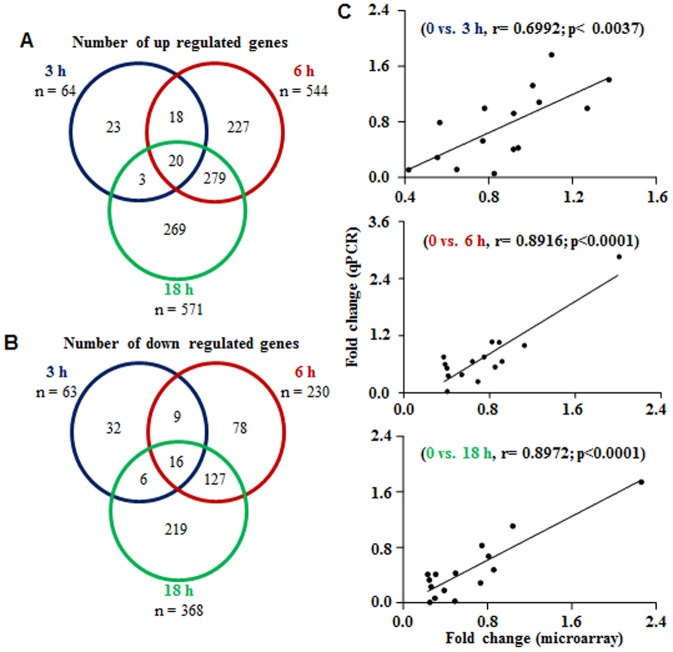
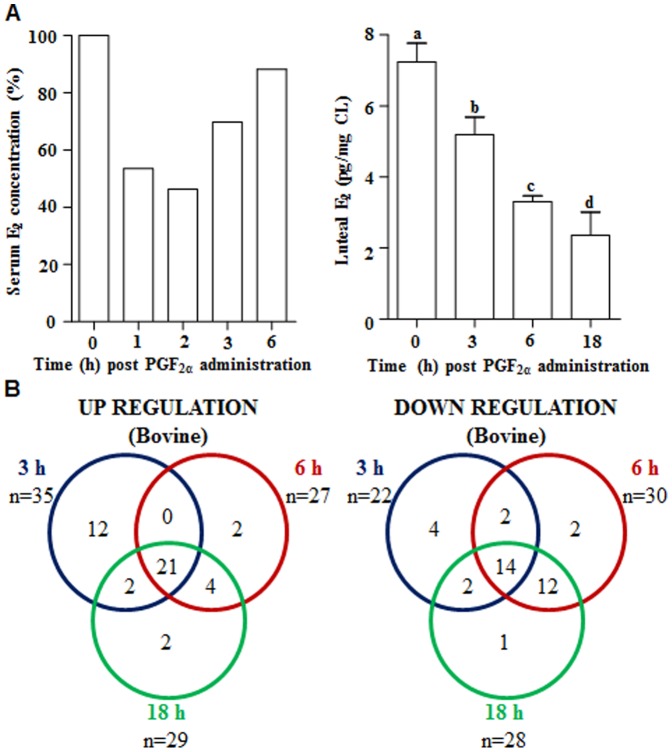
The whole transcriptome analysis data presented as Venn diagrams in Figure 1A and B revealed a total of 127, 774 and 939 genes differentially expressed in CL tissues collected at 3, 6 and 18 h post PGF2α treatment, respectively. Also, the distribution of nearly 40 differentially expressed genes which were common across different time points post PGF2α treatment were identified as up regulated (20 genes), down regulated (16 genes) or up regulated at one time point, but down regulated at another time points (4 genes) are shown in the Venn diagrams (Figure 1A&B). It was observed that most cluster of genes appeared to be modulated in a synchronous or synergistic way following PGF2α treatment, i.e. a higher percentage of the genes regulated at the 3 h time point had a similar regulation at 6 and 18 h time points.
Figure 1. Schematic representation of differentially expressed genes in the CL post PGF2α treatment.
(A and B) Venn diagrams representing the number of differentially expressed genes identified after microarray analysis of CL tissues collected before (0 h) and at different time points post PGF2α treatment. Data analyzed by Bioconductor analysis tool employing ≥2 fold change cut-off and statistical filters with Benjamini and Hochberg correction factor for false discovery rate. The comparison of total number of differentially expressed up (A) and down (B) regulated genes found common between 0 vs. 3 h (blue circle) and 0 vs. 6 h (red circle), as well as between 0 vs.18 h (green circle) post PGF2α treatment are presented. (C) Correlation analysis between expression ratios obtained from microarray and qPCR analyses of few genes post PGF2α treatment (n = 15 genes/time point). Linear regression analysis was performed for selected differentially expressed genes using qPCR fold change in expression values (2−ΔΔCT; Y-axis) with fold change expression values obtained by microarray analysis (X-axis). The r value, correlation coefficient, generated for the theoretical line of best fit (represented as solid line in each panel) and the p value indicate the significance of the correlation as determined by ‘F’ test.
qPCR validation of differentially expressed genes from the microarray analysis data
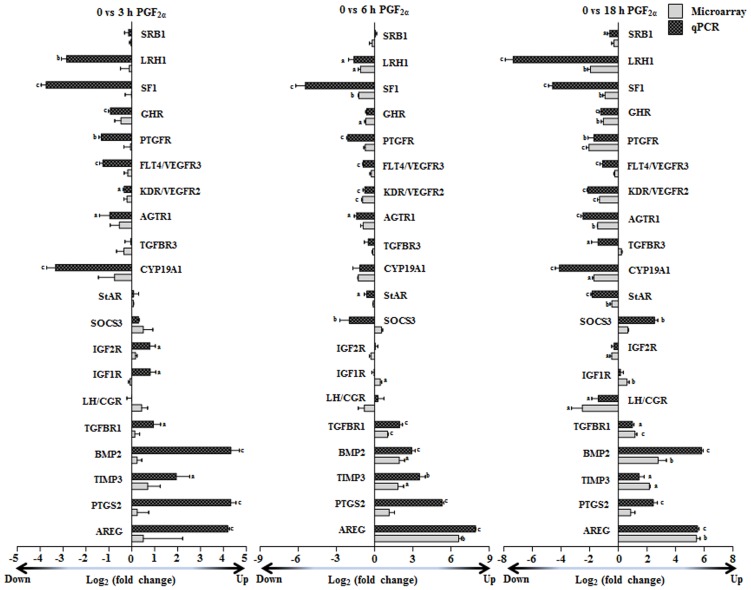
Figure 2 shows fold changes in mRNA expression of 20 differentially expressed genes of both microarray and qPCR analyses. Of the 20 differentially genes analyzed by qPCR analysis, the results of 15 genes (CYP19A1, TGFBR3, AGTR1, VEGFR2/KDR, VEGFR3/FLT4, AREG, PTGS2, TIMP3, BMP2, TGFBR1, PTGFR/FPR, GHR, IGF1R, IGF2R and StAR) were also utilized for validation of microarray data (i.e. for validation and correlation between microarray and qPCR analyses was performed and the results presented in Figure 1C). The goodness of fit analysis showed that the fold change in qPCR analysis of mRNA expression data corroborated well with the microarray analysis data (Figure 1C). At 3 h post PGF2α administration the correlation coefficient ‘r’ value was 0.69 (p<0.003, Figure 1C), while the ‘r’ value at both 6 and 18 h time points was 0.89 (p<0.0001, Figure 1C). Many of the differentially expressed genes were selected for qPCR analysis to further characterize their expression in relation to their function such as signaling pathways (for pathway activity analysis), key enzymes/proteins associated with the steroidogenesis, growth factor activation status, prostaglandin biosynthesis, activation of cytokine signaling and genes associated with extracellular matrix (Figure 2). Expression analysis data of select genes presented in Figure 2 also provide information as to the time course changes in expression post PGF2α treatment. Comparison of expression data of select genes between microarray and qPCR analyses at 3 h time point revealed that the fold expression changes by microarray analysis was lower compared to other time points, but fold change in expression of same genes by qPCR analysis was higher (Figure 2). Similar comparison analyses at 6 and 18 h time point revealed similar observation, except for the gene expression of SRB1 (at 6 h) and LHCGR, IGF1R, IGF2R, PTGFR (at 18 h) which showed higher fold change expression changes by microarray than qPCR analysis.
Figure 2. Validation of microarray data and its comparison with qPCR analysis.
Log ratio of microarray fold change expression of the selected 20 up and down regulated genes associated with specific biological process at 3, 6 and 18 h post PGF2α administration in the CL. The genes selected by microarray analysis were subjected to qPCR analysis and log ratio of fold expression changes at different time points post PGF2α administration are represented as bar graphs. Individual bar for each gene represents mean±SEM log2 (fold change) in mRNA expression value for microarray analysis and qPCR analysis at each time point (n = 3 animals/time point). For each gene, bars with different alphabets above them are significantly different (p<0.05).
The top fifteen differentially expressed genes at the three time points (3, 6 and 18 h) post PGF2α administration were determined and the data are represented as list of top most up- and down-regulated genes in a tabular format comprising of probe set ID, fold change compared to untreated control time point (0 h), gene ID and gene title [from Entrez Gene or UniGene] (Tables S2, S3, S4, S5, S6, S7).
Pathway analysis of differentially expressed genes employing Ingenuity pathway analysis (IPA) software
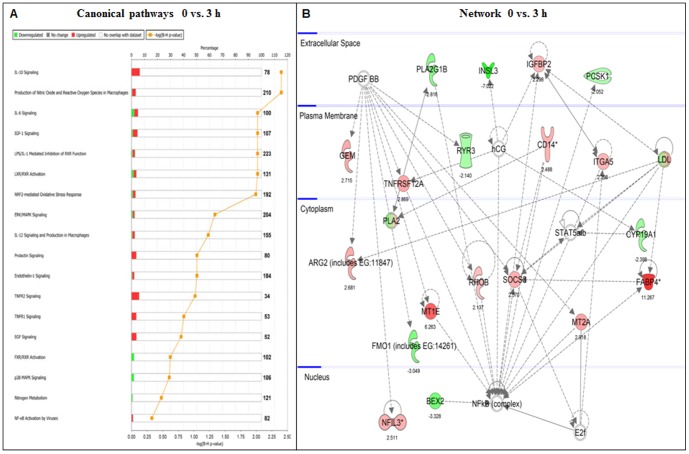
The differentially expressed genes identified in CL tissues post PGF2α treatment were classified into different functions. The IPA analysis revealed that 85–90% of the differentially expressed genes were observed to be function or pathway eligible. Gene classification according to the canonical signaling pathways indicated that many of the differentially expressed genes were associated with process and/or function of IGF-1 signaling, steroidogenesis, chemokine signaling, prolactin signaling, cellular growth and proliferation, extracellular matrix modulation and apoptosis. Further, performing IPA on the differentially expressed genes, 42 networks were identified, out of which 16 networks had 20 or more focus genes in each network. Figure 3A&B shows different canonical signaling pathways and genes identified in Network for 0 vs. 3 h time point (score 48, 21 focus molecules, Figure 3B). The network of genes are associated with growth factor signaling, cytokine signaling, steroidogenesis and extracellular matrix The classification of canonical pathways and network of genes for 6 and 18 h time points post PGF2α treatment are provided in Figure S3 and S4, respectively.
Figure 3. Ingenuity Pathway Analysis (IPA) of differentially expressed genes.
(A) The pathway analysis indicated that a large number of differentially expressed genes belong to canonical pathways such as IGF-1 signaling, steroidogenesis, chemokine signaling, prolactin signaling, cellular growth and proliferation, extracellular matrix modulation and apoptosis. The orange line represents score for the likelihood [-log (B-H p<0.05)] that genes belonging to a specific canonical pathway category affected at 3 h post PGF2α administration. The stacked bars indicate the percentage of genes distributed according to regulation, i.e., green (down), red (up) and open bars (no overlap with dataset) in each canonical pathway. (B) Network 0 vs. 3 h: Ingenuity Pathway Analysis of the differentially regulated genes 3 h post PGF2α administration shows a network of 21 focus molecules with a score of 48, with top biological functions of cell death, cellular growth and proliferation (immune cells), and cell-to-cell signaling. The network is displayed graphically as nodes (genes/gene products) and edges (biological relationship between nodes). The node color intensity indicates the fold change expression of genes; with red representing up-regulation, and green representing down-regulation of genes between 0 vs. 3 h post PGF2α administration. The fold change value for individual gene is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction.
Effect of PGF2α administration on expression of orphan nuclear transcription factors associated with regulation of steroidogenesis in the CL
Since the effects of PGF2α on PKC activation pathway are well recognized, its effects on steroidogenesis at the molecular and biochemical levels were examined. Both microarray and qPCR analyses of mRNA expression of orphan nuclear transcription factors NR5A1/SF-1 and NR5A2/LRH-1 involved in the regulation of expression of steroidogenic genes showed down regulation, excepting at 3 h time point for SF-1, at all-time points post PGF2α treatment (Figure 2). Immunoblot analyses for protein of both these genes also showed down regulation similar to their mRNA expression patterns (Figure S5A&B).
Effect of PGF2α treatment on LH/CGR activation
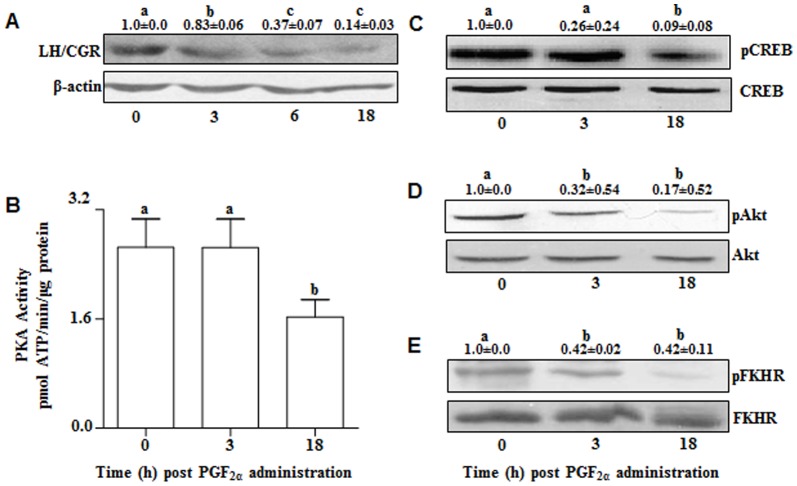
Although LH/CGR mRNA expression by microarray and qPCR analyses indicated marginal decrease at 6 and 18 h post PGF2α treatment (Figure 2), but the protein expression was significantly (p<0.05) lower post PGF2α treatment (Figure 4A). To further examine whether PGF2α treatment affected signaling molecules downstream of LH/CGR activation, PKA activity and CREB phosphorylation levels were determined and the results indicated decreased PKA activity (Figure 4B) and lowered pCREB levels (Figure 4C) at 3 and 18 h time points examined. Also, a decreased phosphorylated Akt (Figure 4D) and FKHR (Figure 4E) levels post PGF2α treatment suggesting inhibitory effects on survival signaling pathway.
Figure 4. Effects of PGF2α on expression of LH/CGR and downstream signaling molecules, Akt and FKHR.
(A) Protein levels of LH/CGR in bovine CL. Protein lysate (100 µg) prepared from CL tissue collected before (0 h) and post (3, 6 and 18 h) PGF2α treatment were resolved on 10% SDS PAGE, transferred onto PVDF membrane and immunoblot analysis was performed using anti-LHCGR and anti-β-actin antibody. A representative immunoblot for each of the antibody probed is shown. The immunoblot probed with β-actin antibody indicates loading control for each lane. Densitometric values were determined and indicated as mean±SEM (n = 3 animals/time point), relative to intensity of β-actin for each time point post PGF2α treatment. The values of immunoblot analysis has been put on top of each lane and lane with different alphabets indicate statistical significance, p<0.05. (B) PKA activity in the bovine CL before (0 h) and after (3 and 18 h) PGF2α administration. Values represent mean±SEM for each time point post PGF2α administration (n = 3 animals/time point). Individual bars with different alphabets indicate statistical significance (p<0.05). (C, D and E) Protein levels of pCREB, CREB, pAkt, Akt, pFKHR and FKHR in the CL. Protein lysate (50 µg) of each CL tissue collected at different time points was subjected to immunoblot analysis employing anti-pCREB, anti-CREB, anti-pAkt, anti-Akt, anti-pFKHR, anti-FKHR antibodies. A representative immunoblot for each of the antibody probed is shown. Densitometric values were determined and represented as mean±SEM (n = 3 animals/time point), relative to intensity of total CREB (C), Akt (D) and FKHR (E) for each time point post PGF2α treatment. The values of immunoblot analysis has been put on top of each lane and lanes with different alphabets indicate statistical significance (p<0.05).
Effect of PGF2α treatment on circulating and luteal E2 levels and expression of E2 responsive genes in the CL
Of the many steroidogenic genes which were affected by PGF2α treatment, mRNA expression of CYP19A1 gene that codes for aromatase enzyme responsible for conversion of androgens to estrogens was down regulated (see Figure 2) at all the time points examined. Figure 5A shows circulating and luteal E2 levels before and after PGF2α treatment. Due to variations in circulating E2 concentrations between animals, the pre-treatment concentration of E2 in each animal was set as 100% and values post PGF2α treatment were expressed in relation to 100%. As can be seen from Figure 5A, E 2 concentrations decreased significantly within 1 h after PGF2α treatment and continued to be lower until 6 h. Since % decrease in E2 was not seen after 6 h post treatment this was perhaps due to contribution of E2 from increased growth and development of ovarian follicles. To further validate the effects of PGF2α on E2 synthesis and secretion, luteal E2 levels were monitored and the results are presented in Figure 5A. As can be seen from the figure, luteal E2 levels continued to decline post PGF2α treatment.
Figure 5. Effects of PGF2α treatment on circulating and luteal E2 levels and E2 responsive genes.
(A) Circulating serum and luteal estradiol (E2) concentration at different time points post PGF2α treatment. Values from animals (n = 3/time point) were represented as mean±SEM and bars with different alphabets indicate statistical significance, p<0.05. (B) Venn diagrams representing the number of differentially expressed E2 responsive genes identified after microarray analysis of CL tissues collected at different time points post PGF2α treatment. Identification of potential E2 responsive genes from the differentially expressed genes. Identification of genes as target of E2 action was based on PCR array human estrogen signaling, estrogen responsive gene database and classical E2 responsive genes. Data analyzed by Bioconductor analysis tool employing ≥1 fold change cut-off and statistical filters with Benjamini and Hochberg correction factor for false discovery rate. The comparison of total number of differentially expressed up and down regulated E2 responsive genes found common between 0 vs. 3 h (blue circle) and 0 vs. 6 h (red circle), as well as between 0 vs.18 h (green circle) post PGF2α administration are presented.
Luteal expression of E2 receptors, α and β, during the luteal phase of estrous cycle and their expression patterns post PGF2α treatment are presented in Figure S6. Since estrogen has both local and systemic effects, expression of estrogen responsive genes in the CL tissue was examined. For examining the effects of PGF2α treatment on expression of E2 responsive genes in the CL tissue, in addition to the classical E2 responsive genes [54], genes identified as target of E2 action by PCR array human estrogen signaling (Qiagen, SABiosciences, PHAS-005A) as well as contents of estrogen responsive gene data base (ERGDB) were included. In all, a total of 89 genes identified as potential E2 responsive genes were considered for analysis. From the list of differentially expressed genes obtained by microarray analysis in CL tissue at different time points post PGF2α treatment, as many as 57 out of 89 E2 responsive genes (both up and down regulated) were identified to be differentially expressed at least 1 fold or more. The data is represented as Venn diagram in Figure 5B. The list of E2 responsive genes for 3 h time point is shown in Tables S8, S9. The list for other time points is not shown since expression of large number of E2 responsive genes were commonly regulated at all-time points. The list of E2 responsive genes commonly up and down regulated at all-time points post PGF2α is shown in Tables S10, S11. Twenty one of the 43 up regulated genes were observed to be commonly regulated at all-time points examined, while 37 genes identified as down regulated, 14 were found to be commonly regulated at all-time points post PGF2α treatment (Figure 5B).
The buffalo CL much like the cattle is not regarded as a site of high E2 production since much of the circulating E2 in bovines is contributed by waves of follicular growth occurring throughout the estrous cycle [55], [56]. On the other hand, in species such as primates, CL is the predominant source of E2 production throughout the luteal phase period and circulating E2 contributed by the functional CL is considerable [57]–[59]. To further validate our hypothesis that decreased estrogen production following inhibition of CYP19A1 expression might adversely affect CL function and its eventual survival, the previously published microarray data of the differentially expressed genes from the CL tissues of macaques receiving PGF2α treatment for 24 h ([44], GEO accession number GSE8371) was mined for E2 responsive genes (89 genes identified as potential E2 responsive genes, see above) for purposes of comparing the number of E2 responsive genes that were differentially expressed in CL of macaques (in which E2 is secreted in higher amounts) to that of the buffalo cow CL (in which E2 secretion is low). The mined data comprising up and down regulated genes of CL from macaques at 24 h time point (the single time point for which microarray analysis available) vs. E2 target genes identified at 3, 6 and 18 h time points of buffalo CL tissue post PGF2α treatment are shown in Figure 6 (A–C). Forty seven out of the 89 genes regarded as E2 responsive genes were identified as up regulated in the monkey CL, 24 h post PGF2α treatment and 19 of the 47 genes were also observed to be commonly regulated in CL tissues of buffalo cows post 3 h PGF2α treatment (Figure 6A). Remarkably, the number of down regulated genes identified in CL tissues at 24 h post PGF2α treatment in macaques and at 6 and 18 h post PGF2α treatment in buffalo cows were similar and 14 of the genes were found to be commonly regulated between the two species (Figure 6B&C). The list of E2 responsive genes commonly up and down regulated from macaques at 24 h time point vs. genes identified at 3, 6 and 18 h time points of buffalo CL tissue post PGF2α treatment is shown in Tables S12, S13, S14, S15, S16, S17.
Figure 6. Identification and comparison of differentially expressed E2 responsive genes in the monkey and bovine CL.
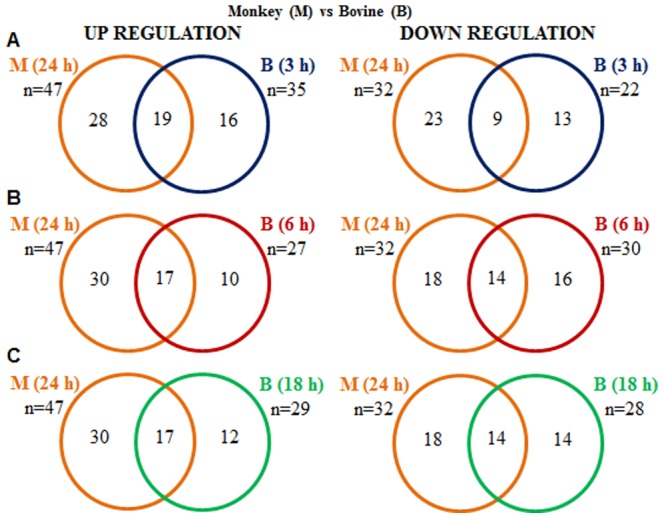
Venn diagrams representing the number of differentially expressed E2 responsive genes identified in monkey (M) and bovine (B) microarray data available from the GEO database. Data analyzed by Bioconductor analysis tool employing ≥1 fold change cut-off and statistical filters with Benjamini and Hochberg correction factor for false discovery rate. The circles designated for each time point post PGF2α is represented as orange (24 h, M), blue (3 h, B), red (6 h, B) and green (18 h, B). A comparison of total number of differentially expressed up and down regulated E2 responsive genes found common between 24 h, M vs. 3 h, B (A); 24 h, M vs. 6 h, B (B) and 24 h, M vs. 18 h, B (C) post PGF2α treatment are presented.
Effect of PGF2α administration on expression of different regulatory molecules associated with cell survival and apoptosis
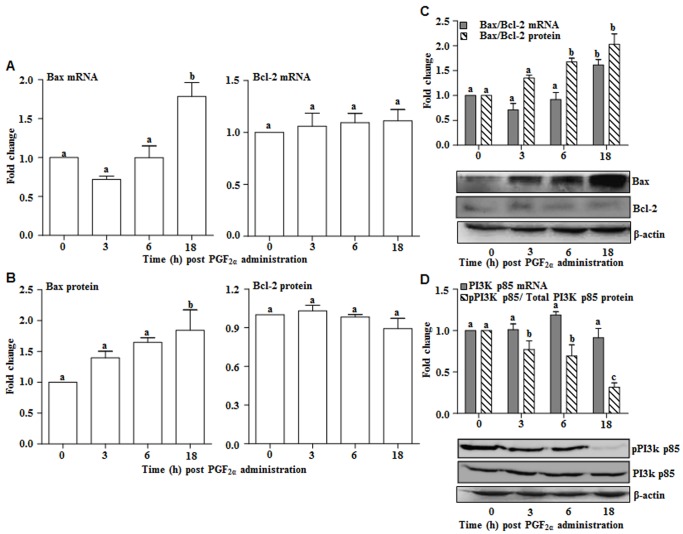
To examine whether down regulation of E2 receptor expression results in activation of apoptotic changes in the luteal tissue, expression of Bax and Bcl-2 genes were determined in the luteal tissue post PGF2α treatment. Figure 7A&B illustrates expression of (both mRNA and protein) Bcl-2 family members, Bax (pro-apoptotic) and Bcl-2 (anti-apoptotic) during PGF2α-induced luteolysis. The Bcl-2 mRNA expression was unchanged post PGF2α treatment, but a significant increase in Bax mRNA expression was seen at 18 h post PGF2α treatment (Figure 7A). Similarly, at the protein levels, Bcl-2 protein levels did not change, but a significant increase was seen for Bax protein at 18 h time point post PGF2α treatment (Figure 7B). As indices of activation or inhibition of mitochondrial permeability to apoptogenic molecules, changes in Bax and Bcl-2 mRNA expression and protein levels were expressed as Bax/Bcl-2 ratios. A significant increase in both Bax mRNA and protein levels post PGF2α treatment was observed (Figure 7A&B). As can be seen from Figure 7C, Bax/Bcl-2 ratio for both mRNA and protein levels, increased significantly by 18 h post PGF2α treatment. Also, a significant increase in Bax/Bcl-2 protein was observed by 6 h post PGF2α treatment suggesting increased mitochondrial permeability to apoptogenic molecules (Figure 7C).
Figure 7. Effect of PGF2α on expression of Bax, Bcl-2 and Akt genes during PGF2α treatment.
(A) qPCR expression and analysis of Bax and Bcl-2 in the bovine CL. Mean (±SEM) fold expression changes before and after PGF2α treatment is presented. Bars with different alphabets above them indicate statistical `significance (p<0.05). (B) Protein levels of Bax and Bcl-2 in the bovine CL. The CL tissue lysates were resolved on 10% SDS PAGE, transferred onto PVDF membrane and subjected to immunoblot analysis employing anti-Bax, anti-Bcl-2 and anti-β-actin antibodies. The immunoblot probe with β-actin antibody was used as a loading control. Densitometric values were determined and indicated as mean±SEM (n = 3 animals/time point), relative to intensity of β-actin for each time point post PGF2α treatment. Bars with different alphabets indicate statistical significance (p<0.05). (C) Fold expression changes in mRNA and quantitative changes in protein as ratio of Bax/Bcl-2 levels in the luteal tissue during PGF2α treatment. A representative immunoblot for each of the antibody probe is shown. Individual bars with different alphabets above them indicate statistical significance (p<0.05). Individual bars in solid box and open box represent mRNA expression and protein levels, respectively. (D) mRNA and protein levels of PI3k p85 in bovine CL. Mean (±SEM) fold expression changes in PI3k p85 mRNA examined by qPCR analysis. Quantitative changes in the ratio of pPI3k p85 and PI3k p85 protein levels were estimated. For the protein loading control, blots were probed with β-actin antibody. Densitometric values were determined and represented as mean±SEM (n = 3 animals/time point), relative to intensity of total PI3k p85 for each time point post PGF2α treatment. Individual bars with different alphabets above them indicate statistical significance (p<0.05). Individual bars in solid box and open box represent mRNA expression and protein levels, respectively.
It has been reported that PI3K activity is essential for the activation of Akt for E2 actions [60], [61]. In present study, although a significant change in the expression of PI3K p85 mRNA by qPCR analysis was not observed (Figure 7D), but a significant decrease in phospho levels of PI3K p85 was observed at all-time points post PGF2α treatment (Figure 7D).
Schematic diagram illustrating a model for interaction amongst different signaling pathways during PGF2α-induced luteolysis in buffalo cows
Extensive studies carried out previously by several groups have identified different molecules of the signaling pathways for LH and PGF2α actions in the luteal tissue of various species [62]–[66]. In the present study, some of the signaling molecules of both these pathways were analysed as also genes associated with steroid biosynthesis, cell survival and apoptosis and the data for expression of various genes and their protein levels in the luteal tissue at different time points post PGF2α treatment are presented in Figure 8. A schematic diagram of LH and PGF2α signaling pathways and E2 receptor signaling pathways and their likely interactions is depicted in Figure 8. In the proposed model, an association between cell survival and E2 action could be suggested for regulating luteal function in the bovine luteal tissue. The down regulation of E2 signaling by PGF2α as suggested by the data indicates activation of apoptotic pathway (Figure 8).
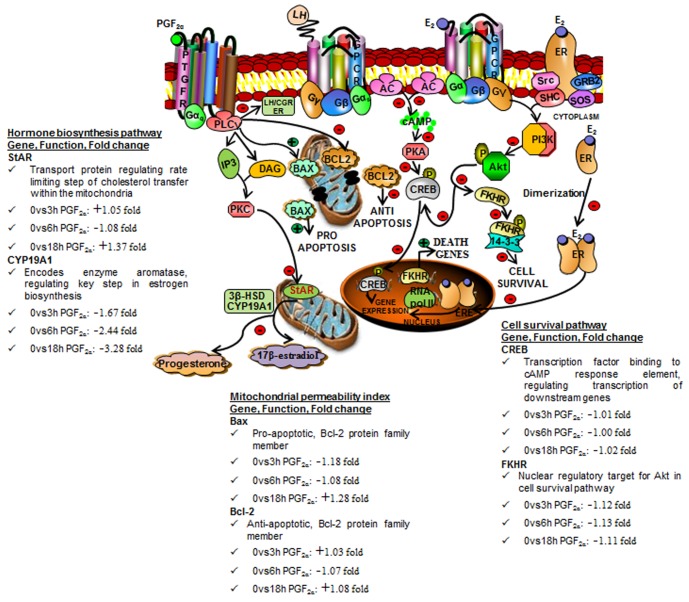
Figure 8. Schematic diagram illustrating a model for PGF2α-induced luteolysis in the bovine corpus luteum.
The model shows interaction amongst various intracellular signaling pathways activated by LH, PGF2α and E2. The luteolysis process induced by PGF2α treatment appears to be a result of distinct molecules that involve hormone biosynthesis pathways (StAR, CYP19A1) which is downstream of PTGFR signaling leading to decrease in serum and luteal P4 and E2 levels. The model also shows involvement of Bcl-2 family members, Bax (pro-apoptotic) and Bcl-2 (anti-apoptotic) that cause changes in mitochondrial permeability to apoptogenic molecules. The model also shows inhibition of cell survival pathway as shown by LH/CGR and ER (both genomic and non-genomic signaling) down regulation and in turn inhibition of downstream molecules (CREB, FKHR). Few of the selected genes out of differentially expressed genes at different time points post PGF2α treatment compared with 0 h PGF2α treatment (StAR, CYP19A1, Bax, Bcl-2, CREB, FKHR) has been provided in the Figure. The information on gene names, general function, fold change in mRNA expression and implicated pathways are represented. The stimulation and inhibition of specific molecules downstream of different signaling pathways are represented by green (+) and red (−) signs, respectively.
Discussion
In bovines, during non-conception reproductive cycles, the onset of spontaneous luteolysis is initiated by increased pulsatile secretion of PGF2α from the uterus [62], [67]. Several recent studies have compared expression changes during periods of refractoriness and responsiveness of CL to PGF2α treatment [23], [28], [68] and few studies have previously reported global gene expression changes during PGF2α induced luteolysis [23], [26]. In the present study, employing high throughput screening technique, genome-wide gene expression changes in the CL of buffalo cow during PGF2α-induced luteolysis were determined. The primary objective of this study was to examine temporal expression changes during early periods of PGF2α treatment in which functional loss in CL will be manifest. The global transcriptome changes post PGF2α treatment revealed temporal increases in the expression of a number of differentially expressed genes at 3, 6 and 18 h time points examined, and of these several of them were immediate early genes. The analysis of differentially expressed genes during induced luteolysis also revealed cluster of genes associated with steroidogenesis, angiogenesis, cell survival and apoptosis and these have major bearing on initiation of functional luteolysis and apoptotic processes. Further, comparison of differentially expressed genes data from the present study with other studies following administration of PGF2α treatment in bovine and primate species indicates gene expression changes related to angiogenesis, immune system and participation of functional changes in non-steroidogenic cells [44], [23].
Analysis of differentially expressed genes at 3 h post PGF2α treatment indicated that many of the genes were associated with canonical cell to cell signaling pathways (chemokine, IGF-I and prolactin signaling), cellular growth and proliferation. Several of the products of the up-regulated genes (TNFRSF12A, DAPL1, GEM, PPP4R4 and OSR2) were associated with activation of cellular signaling resulting in inhibition of aggregation of denatured proteins as well as processes associated with protection against metal toxicity and oxidative stress (FABP4, HSPH1, MT1A, MT2A, HP and CBR1). Many of the down-regulated genes belonged to steroidogenesis process (CYP19A1, NR5A1/SF-1, NR5A2/LRH-1, PLA2G1B and PTGR1), cell cycle regulation and tissue development (INSL3, AHSG, BEX2, CDKN1C and HSPB3). Earlier studies involving intra-uterine infusions of low [23] and pulsatile [3] administration of PGF2α also observed similar changes in many of the genes during functional luteolysis. It should be pointed out that expression of genes regulating steroidogenesis i.e. P4 biosynthesis (LH/CGR, P450scc and HMGCR) or expression of genes involved in the uptake and trafficking of cholesterol (SR-B1 and StAR) did not change significantly at 3 h post PGF2α treatment. However, changes in expression of many of these genes were observed at later time points. It has been reported that P4 decline occurs as early as 30 min after PGF2α treatment by others and us [69], [70] suggest translational or post translational changes are responsible for initiation of rapid induction of functional luteolysis.
To examine the mechanism of PGF2α-induced luteolysis, the differentially expressed genes belonging to some processes and functions of luteal cells and few top early differentially expressed genes were subjected to additional analysis. More specifically, as per the objectives of the present study stated in the introduction section, effects of PGF2α on luteotrophic signaling and role of intra luteal factors were further examined from the differentially expressed genes post PGF2α treatment. Several studies have reported inhibition of LH-stimulated P4 secretion by PGF2α [71], [72]. One of the reported actions of PGF2α was inhibition of adenylate cyclase activity [73], [74] that lie downstream of LH receptor activation, but this effect has been reported to be a transient one and moreover, this effect appears to be limited to small luteal cells [75], [76]. It is not clear to what extent PGF2α-inhibited adenylate cyclase activity will adversely affect overall P4 production in vivo, since large luteal cells, besides having large volume and being most abundant, have constitutive steroid synthesis and therefore high P4 production capacity [63]. On the other hand, the LH-responsive small luteal cells contribute very small fraction of total P4 produced from the luteal tissue. It should be pointed out that fewer pulses of LH are secreted during the luteal phase that may have little or no impact on the overall P4 production capacity of the CL [77]–[80]. It has been reported that LH/CGR expression gets inhibited post PGF2α administration in several species [71], [72]. In the present study, LH/CGR mRNA expression did not change within 3 h, but decreased at 6 h and thereafter. Surprisingly, LH/CGR protein levels decreased within 3 h post PGF2α treatment and this may be the reason for immediate inhibitory effect observed in luteal cells as reported in bovines [81] and other species [11], [72]. The decrease in PKA activity and pCREB levels at later time points post PGF2α treatment suggest that aspects of LH/CGR signaling appears to be modulated by PGF2α directly at the cellular level through inhibition of LH/CGR protein or cAMP levels due to inhibition of cAMP phosphodiesterase enzyme [82], [83]. A decrease in levels of SF1 and LRH1 associated with regulation of steroidogenesis indicates regulation of expression of early genes involved in modulation of steroidogenesis [84].
One of the novel findings of this study is the early and consistent down regulation of CYP19A1 expression post PGF2α treatment. It has been established that in response to LH surge, CYP19A1 expression gets down regulated and may even be absent post ovulation [85]–[87]. However, others [36] and we (in the present study) have observed CYP19A1 expression in the non-pregnant CL. It was of interest to examine the importance of PGF2α-induced down regulation of CYP19A1 expression. The aromatase enzyme, encoded by CYP19A1 gene, a complex composed of two proteins, an ubiquitous NADPH cytochrome P450 reductase and the cytochrome P450 aromatase, is essential for conversion of androstenedione to E2. It has been determined that the regulatory sequence, cAMP-response element (CRE) on the promoter region of CYP19A1 gene is critical for control of expression of aromatase in mammals [88]. For driving expression of CYP19 gene, many tissue specific promoters have been described for several species and in the ovary, promoter II sequence has been reported to be responsible for driving aromatase expression [89]–[91]. Although the promoter II sequence in several species including the cow presents strong homologies, differences in the regulatory sequence have been observed amongst species [92], [93]. In the cow, one nucleotide deletion and two substitutions in the CRE-like sequence (CLS) leads to inactivation of the promoter II site and this has been suggested to be responsible for the presence of very low aromatase activity in luteinized granulosa cells or in the CL tissue [92], [94]. Interestingly, CYP19 gene expression is most abundant in CL of pregnant rats [92], and this has important role since large quantities of androstenedione, the substrate for the aromatase enzyme, is synthesized in the placenta but gets converted to E2 in the luteal tissue [95], [96]. In bovines, even though CYP19 expression may be lower in non-pregnant CL, but it may be higher during pregnancy [97].
Intra luteal E2 plays an important role in the structure and function of CL ranging from hypertrophy of luteal cells to increased transport of cholesterol for steroidogenesis [98]–[100]. Estrogens have been reported to have both luteotrophic and luteolytic function across different species [15], [36], [38], [39], [64], [101], which suggest autocrine/paracrine role within the CL. The biological effects of E2 are mediated by its binding to the structurally and functionally distinct estrogen receptor (ER), α and β. It has been documented that besides classical E2 signaling, ligand activated ERα regulates a series of non-genomic events in the cytoplasm including PI3-kinase (PI3K) activation [61], [102], [103]. The present observation of decreased ER expression and inhibition of Akt phosphorylation post PGF2α treatment point to loss of PI3K activity during luteal regression as reported previously by others [25], [37]. Further, in order to gain more information on the effects of E2 on the CL tissue, the microarray data of PGF2α treatment study was mined for E2-responsive genes. The observation that a large number of E2-responsive genes was observed to be differentially expressed post PGF2α treatment was surprising since intra luteal E2 levels are considerably lower in the bovine CL tissue (and also in the circulation) and therefore, it was reasoned that influence of E2 on luteal function, if any, will be limited. However, to further explore the possible E2 effects on luteal tissue, effects of PGF2α treatment on expression of genes in CL tissues of macaques, the species considered to secrete large quantities of E2 [104] were determined. Mining of microarray data for E2-responsive genes from CL tissues of macaques receiving PGF2α treatment [44] revealed that many of the E2 responsive genes were also observed to be differentially expressed post PGF2α treatment in the macaque CL. The observation of many of the differentially expressed E2-responsive genes in CL of both macaque and bovine species post PGF2α treatment provides compelling evidence for regulation of CL function by E2. Furthermore, PGF2α appears to interfere with luteal E2 secretion and perhaps E2 actions.
Earlier studies by others and us have provided evidence supporting involvement of cohort of conserved cell death regulatory factors, required for maintenance of growth and development of CL across species [8], [9], [105], [106], [107]. Apoptosis at cellular levels is regulated by the intricate changes in the members of the Bcl-2 family proteins that are classified either as anti-apoptotic (Bcl-2 and Bcl-xL) or pro-apoptotic (Bax, Bad, Bik, Bid) molecules. Bcl-2 has been designated as estrogen responsive gene [108], [109] and up regulation of Bcl-2 expression has been identified as a critical mechanism for promoting cell survival. Akt mediated phosphorylation of cytosolic proteins is known to play a critical role in the regulation of different metabolic pathways. The promoter region of Bcl-2 contains a cAMP responsive element (CRE) site which is flanked by two estrogen response element (ERE) sites [103], [110], [111], [112]. Based on the previous studies, the transcription factor, CREB can be considered as a positive regulator of Bcl-2 expression [111], [113]. Further, Akt, a target of E2 signaling through PI3K pathway has been shown to activate CREB [103], [114] thereby regulating Bcl-2 expression via differential activation of ERs. In our present study, an increase in the Bax/Bcl-2 ratio at mRNA and protein levels further suggests an important role played by permeability of these regulatory proteins across mitochondria as reported during induced and spontaneous luteolysis in bovines [8], [9], [63]. Another regulatory molecule FKHR which is target of Akt has been proposed to participate in both metabolic and cell survival pathway [115], [116], has also been observed to decline post PGF2α treatment, activating the apoptotic pathway.
The other effects of PGF2α involve interaction with intraovarian paracrine and endocrine factors [117], [118] to mediate intracellular communication within the CL [119]. PGF2α-induced structural luteolysis depends on cell composition (immune or endothelial cell) [120] and contact [121] as the vasoconstrictor property of PGF2α may decrease ovarian blood flow and cause apoptosis of endothelial cells [65], [122]–[124] restricting access of gonadotropins and oxygen to steroidogenic cells. The possible role of apoptosis as a mechanism for structural luteolysis has received considerable attention in several species [125], [126]. The PGF2α-induced structural luteolysis mediated by apoptosis was confirmed by the presence of DNA laddering in 18 h PGF2α-treated CL, which has been demonstrated by us and others through studies involving the role of Bax, Bcl-2, Fas/FasL and caspases during spontaneous or PGF2α-induced apoptosis in ovine, bovine, rodent CL [9], [107], [127]. Thus, pathway analysis of microarray data at 6 and 18 h post PGF2α administration using IPA revealed down-regulated genes to be associated with regulation of steroidogenesis (transcription factors belonging to steroidogenesis, biosynthesis of steroids, intracellular trafficking and lipid metabolism) and angiogenesis (VEGF signaling and coagulation system). Whereas, up-regulated genes to be associated with cell-cell interaction and signaling (EGF, p38 MAP kinase, NF-κB, TGF-β and apoptotic signaling), prostaglandin metabolism and tissue remodelling. Analysis of top differentially regulated genes indicated that most genes belonged to steroidogenesis, prostaglandin metabolism, tissue remodeling or ECM modulation, angiogenesis, TGF-β signaling, cellular stress activated binding proteins, intracellular trafficking, cell survival and apoptotic signaling.
In summary, the results of the present study describe gene expression changes at early time points post PGF2α treatment. It was observed that PGF2α treatment caused down regulation of various components of LH receptor signaling, decreased CYP19A1 expression and inhibited intra luteal E2 levels. A number of E2 responsive genes were identified to be differentially expressed post PGF2α treatment. In conclusion, based on the changes of key genes encoding proteins involved in regulating CL structure and function, we propose a model depicting a cross talk between PGF2α, LH/CGR and E2 signaling during luteolysis in bovines. PGF2α-induced luteolysis involves down and up regulation of genes involved in luteal steroidognesis and susceptibility of cell system to apoptotic signals, respectively. The distinct changes that follow post PGF2α treatment is triggered by down regulation of LH/CGR and ER signaling. E2-ER/PI3K-Akt signaling regulates many transcriptional regulatory molecules such as CREB and FKHR, and members of Bcl-2 family proteins such as Bax and Bcl-2. Thus, the present study suggests key role of inhibition of E2 signaling in the regulation of various changes observed during PGF2α-induced luteolysis in bovines.
Supporting Information
Expanded tree view generated after hierarchical clustering of representative genes obtained after pairwise analysis. The expanded tree displaying the hierarchy analysis for probe sets at each time point [0 vs. 3 h (A), 0 vs. 6 h (B) and 0 vs. 18 h (C)] post PGF2α administration. The bottom of each dendrogram shows the condition color bar with the parameters in each interpretation. The legend shows the name of each condition on which clustering was performed. Header of heat map shows a normalized intensity values represented in various shades of red and green indicating relatively either up or down regulation, respectively. The row header shown on the right side represents the complete entity name or gene symbol. The upper panel shows groups of genes, whose expression was up regulated at 0 h and with the administration of PGF2α the expression is observed to be down-regulated. The lower panel shows groups of genes whose expression was down regulated at 0 h and with the administration of PGF2α the expression is observed to be up regulated.
(TIF)
Effects of PGF2α on circulating and luteal P4 levels, StAR expression and DNA fragmentation. Buffalo cows received intramuscular injection of 500 µg of PGF2α on day 11 of estrous cycle and blood and luteal tissue samples at different time intervals post PGF2α treatment. (A) Circulating mean±SEM serum and luteal progesterone (P4) concentrations immediately before (0 h) and at different time points post PGF2α treatment. Bars with different alphabets indicate statistical significance, p<0.05. (B) Protein lysate (100 µg) prepared from CL tissue collected before and post PGF2α treatment were resolved on 10% SDS PAGE, transferred onto PVDF membrane and immunoblot analysis was performed using anti-StAR and anti-β-actin antibody (β-actin was used as loading control). A representative immunoblot for each of the antibody probed is shown. Densitometric values shown on top of each lane represented were determined and represented as mean±SEM (n = 3 CL/time point), relative to intensity of β-actin for each time point post PGF2α treatment. (C) Analysis of apoptotic DNA fragmentation in luteal tissue. Genomic DNA isolated from CL tissues collected from untreated control animals (0 h) and from animals at different time points post PGF2α treatment was subjected to DNA laddering analysis. An image of a nylon membrane visualized using PhosphorImager containing the [α32P] labeled genomic DNA separated previously on 2% agarose gel and transferred on to nylon membrane is represented here. Migration (base pairs) of oligonucleosomes is indicated on the right.
(TIF)
Classification of differentially expressed genes post 6 h PGF2α administration by Ingenuity Pathway Analysis (IPA). (A) The pathway analysis indicates that a large number of differentially expressed genes belongs to canonical pathways such as p38 MAP kinase signaling, VEGF signaling, transcription factors belonging to steroidogenesis and coagulation system. The orange line represents score for the likelihood [-log (B-H P < 0.05)] that genes belonging to a specific canonical pathway category affected at 6 h post PGF2α administration. The stacked bars indicate the percentage of genes distributed according to regulation, i.e., green (down), red (up) and open bars (no overlap with dataset) in each canonical pathway. (B) Network 0 vs. 6 h: Ingenuity Pathway Analysis of the differentially regulated genes 6 h post PGF2α administration shows a network of 28 focus molecules with a score of 44, with top biological functions of cell to cell signaling, molecular transport and lipid metabolism. The network is displayed graphically as nodes (genes/gene products) and edges (biological relationship between nodes). The node color intensity indicates the fold change expression of genes; with red representing up regulation, and green down regulation of genes between 0 vs. 6 h post PGF2α administration. The fold change value for individual gene is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction.
(TIF)
Classification of differentially expressed genes post 18 h PGF2α administration by Ingenuity Pathway Analysis (IPA). (A) The pathway analysis indicates that a large number of differentially expressed genes belong to canonical pathways such as TGF-β signaling, steroid biosynthesis, NF-κB signaling and coagulation system. The orange line represents score for the likelihood [-log (B-H P<0.05)] that genes belonging to a specific canonical pathway category affected at 18 h post PGF2α administration. The stacked bars indicate the percentage of genes distributed according to regulation, i.e., green (down), red (up) and open bars (no overlap with dataset) in each canonical pathway. (B) Network 0 vs. 18 h: Ingenuity Pathway Analysis of the differentially regulated genes 18 h post PGF2α administration shows a network of 26 focus molecules with a score of 39, with top biological functions of cellular development, cell cycle and gene expression. The network is displayed graphically as nodes (genes/gene products) and edges (biological relationship between nodes). The node color intensity indicates the fold change expression of genes; with red representing up regulation, and green down regulation of genes between 0 vs. 18 h post PGF2α administration. The fold change value for individual gene is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction.
(TIF)
Effect of PGF2α administration on expression of orphan nuclear transcription factors in CL. (A and B) The orphan nuclear transcription factors associated with regulation of expression of steroidogenic genes were analyzed. Protein levels of NR5A1/SF-1(A) and NR5A2/LRH-1 (B) in bovine CL were determined. Protein lysate (100 µg) prepared from CL tissue collected before (0 h) and post (3, 6 and 18 h) PGF2α treatment were resolved on 10% SDS PAGE, transferred onto PVDF membrane and immunoblot analysis was performed using anti-SF1, anti-LRH1 and anti-β-actin antibody. A representative immunoblot for each of the antibody probed is shown. The immunoblot probed with β-actin antibody indicates loading control for each lane. Densitometric values were determined and indicated as mean±SEM (n = 3 animals/time point), relative to intensity of β-actin for each time point post PGF2α treatment. The values of immunoblot analysis has been put on top of each lane and lane with different letters indicates statistical significance, p<0.05.
(TIF)
PGF2α administration effect on expression of estrogen receptors during spontaneous and induced luteolysis in CL. Quantitative real time PCR (qPCR) fold change expression of the estrogen receptors (ERα and ERβ) during spontaneous (A) and induced (B) luteolysis. Total RNA isolated from CL was reverse transcribed and cDNA equivalent to 10 ng of total RNA was used for qPCR. The expression was normalized with L19 mRNA. The results are shown as fold changes of mRNA expression compared with that at early (E) luteal phase (A) and 0 h PGF2α (B) for bovine CL. Individual bar for each gene represents mean±SEM fold change in mRNA expression value for qPCR analysis at each time point (n = 2 animals/time point, A and n = 3 animals/time point, B). For each gene, bars with different letters above them are significantly different (p<0.05).
(TIF)
List of primer set employed for quantitative real time PCR. The list of genes and details of the primers employed in the qPCR analysis along with the annealing temperature and expected amplicon size are provided.
(TIF)
List of top 15 up regulated genes at 3 h post PGF2α administration. Microarray data analysis was carried out to obtain a set of differentially expressed genes based on statistics, a Student's t-test (two tail, unpaired) with p<0.05 and multiple hypothesis testing (Benjamini and Hochberg comparison test) to reduce the false positives. The identified differentially expressed genes were transcript consistent and did not hybridize to multiple transcripts, as suggested by the AffyProbeMiner analysis. A Bioconductor analysis was performed with ≥2 fold change as cut-off with statistical filters for identification of differentially expressed genes. Whereas, the top 15 differentially UP regulated genes at 3 h post PGF2α treatment are represented in this table. Probe Set ID: The identifier that refers to a set of probe pairs selected to represent expressed sequences on an array; Fold Change: It is a number describing changes in expression level of a gene compared between control and treatment; Gene ID: Gene symbols extracted from Entrez Gene or UniGene; Gene Title: Gene name extracted from Entrez Gene or UniGene.
(TIF)
List of top 15 down regulated genes at 3 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 3 h post PGF2α treatment are represented.
(TIF)
List of top 15 up regulated genes at 6 h post PGF2α administration. The top 15 differentially UP regulated genes at 6 h post PGF2α treatment are represented.
(TIF)
List of top 15 down regulated genes at 6 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 6 h post PGF2α treatment are represented.
(TIF)
List of top 15 up regulated genes at 18 h post PGF2α administration. The top 15 differentially UP regulated genes at 18 h post PGF2α treatment are represented.
(TIF)
List of top 15 down regulated genes at 18 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 18 h post PGF2α treatment are represented.
(TIF)
List of top 15 up regulated E2 responsive genes at 3 h post PGF2α administration. Potential E2 responsive genes in bovine CL were identified based on the available list of classical E2 responsive genes, genes employed in PCR array human estrogen signaling and the data base, ERGDB. Microarray data analysis was carried out to obtain a set of differentially expressed genes based on statistics, a Student's t-test (two tail, unpaired) with p<0.05 and multiple hypothesis testing (Benjamini and Hochberg comparison test) to reduce the false positives. The identified differentially expressed E2 responsive genes were transcript consistent and did not hybridize to multiple transcripts, as suggested by the AffyProbeMiner analysis. A Bioconductor analysis was performed with ≥1 fold change as cut-off and statistical filters for identification of differentially expressed E2 responsive genes. Whereas, the top 15 differentially UP regulated genes at 3 h post PGF2α treatment are represented in this table. Probe Set ID: The identifier that refers to a set of probe pairs selected to represent expressed sequences on an array; Fold Change: It is a number describing changes in expression level of a gene compared between control and treatment; Gene ID: Gene symbols extracted from Entrez Gene or UniGene; Gene Title: Gene name extracted from Entrez Gene or UniGene.
(TIF)
List of top 15 down regulated E2 responsive genes at 3 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 3 h post PGF2α treatment are represented. The genes are discussed in the results and discussion section.
(TIF)
List of common up regulated E2 responsive genes post 3, 6 and 18 h PGF2α administration. The common differentially UP regulated E2 responsive genes (21 genes) before (0 h) and post (3, 6 and 18 h) PGF2α treatment are represented.
(TIF)
List of common down regulated E2 responsive genes post 3, 6 and 18 h PGF2α administration. The common differentially DOWN regulated E2 responsive genes (14 genes) before (0 h) and post (3, 6 and 18 h) PGF2α treatment are represented.
(TIF)
List of common up regulated E2 responsive genes in monkey (24 h) and bovine (3 h) CL. The previously published microarray data of the differentially expressed genes from the CL tissues of macaques receiving PGF2α treatment for 24 h [GEO accession number GSE8371] was mined for E2 responsive genes for purposes of comparing the number of E2 responsive genes that were differentially expressed in CL of macaques to that of the buffalo cow CL [GEO accession number GSE27961]. The mined data comprising common UP regulated E2 responsive genes (19 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 3 h post PGF2α treatment are represented in this Table.
(TIF)
List of common down regulated E2 responsive genes in monkey (24 h) and bovine (3 h) CL. The mined data comprising common DOWN regulated E2 responsive genes (8 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 3 h post PGF2α treatment are represented in this Table.
(TIF)
List of common up regulated E2 responsive genes in monkey (24 h) and bovine (6 h) CL. The mined data comprising common UP regulated E2 responsive genes (17 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 6 h post PGF2α treatment are represented in this Table.
(TIF)
List of common down regulated E2 responsive genes in monkey (24 h) and bovine (6 h) CL. The mined data comprising common DOWN regulated E2 responsive genes (13 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 6 h post PGF2α treatment are represented in this Table.
(TIF)
List of common up regulated E2 responsive genes in monkey (24 h) and bovine (18 h) CL. The mined data comprising common UP regulated E2 responsive genes (17 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 18 h post PGF2α treatment are represented in this Table.
(TIF)
List of common down regulated E2 responsive genes in monkey (24 h) and bovine (18 h) CL. The mined data comprising common DOWN regulated E2 responsive genes (13 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 6 h post PGF2α treatment are represented in this Table.
(TIF)
Funding Statement
The work was supported by DBT(350), Government of India. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Weinbauer GF, Niehoff M, Niehaus M, Srivastav S, Fuchs A, et al. (2008) Physiology and Endocrinology of the Ovarian Cycle in Macaques. Toxicol Pathol 36: 7S–23S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pharriss BB, Tillson SA, Erickson RR (1972) Prostaglandins in luteal function. Recent Prog Horm Res 28: 51–89. [PubMed] [Google Scholar]
- 3. Ginther OJ, Araujo RR, Palhão MP, Rodrigues BL, Beg MA (2009) Necessity of sequential pulses of prostaglandin F2alpha for complete physiologic luteolysis in cattle. Biol Reprod 80: 641–648. [DOI] [PubMed] [Google Scholar]
- 4. Stevenson JS, Phatak AP (2010) Rates of luteolysis and pregnancy in dairy cows after treatment with cloprostenol or dinoprost. Theriogenology 73: 1127–1138. [DOI] [PubMed] [Google Scholar]
- 5. Schallenberger E, Schams D, Bullermann B, Waletrs DL (1984) Pulsatile secretion of gonadotrophins, ovarian steroids and ovarian oxytocin during prostaglandin-induced regression of the corpus luteum in the cow. J Reprod Fertil 71: 493–501. [DOI] [PubMed] [Google Scholar]
- 6. Yadav VK, Sudhagar RR, Medhamurthy R (2002) Apoptosis during spontaneous and prostaglandin F (2alpha)-induced luteal regression in the buffalo cow (Bubalus bubalis): involvement of mitogen-activated protein kinases. Biol Reprod 67: 752–759. [DOI] [PubMed] [Google Scholar]
- 7. Berisha B, Meyer HHD, Schams D (2010) Effect of prostaglandin F2 alpha on local luteotropic and angiogenic factors during induced functional luteolysis in the bovine corpus luteum. Biol Reprod 82: 940–947. [DOI] [PubMed] [Google Scholar]
- 8. Rueda BR, Tilly KI, Botros IW, Jolly PD, Hansen TR, et al. (1997) Increased bax and interleukin-1beta-converting enzyme messenger ribonucleic acid levels coincide with apoptosis in the bovine corpus luteum during structural regression. Biol Reprod 56: 186–193. [DOI] [PubMed] [Google Scholar]
- 9. Yadav VK, Lakshmi G, Medhamurthy R (2005) Prostaglandin F2alpha-mediated activation of apoptotic signaling cascades in the corpus luteum during apoptosis: involvement of caspase-activated DNase. J Biol Chem 280: 10357–10367. [DOI] [PubMed] [Google Scholar]
- 10. Berisha B, Schams D (2005) Ovarian function in ruminants. Domest Anim Endocrinol 29: 305–317. [DOI] [PubMed] [Google Scholar]
- 11. Sotrel G, Helvacioglu A, Dowers S, Scommegna A, Auletta FJ (1981) Mechanism of luteolysis: effect of estradiol and prostaglandin F2 alpha on corpus luteum luteinizing hormone/human chorionic gonadotropin receptors and cyclic nucleotides in the rhesus monkey. Am J Obstet Gynecol 139: 134–140. [DOI] [PubMed] [Google Scholar]
- 12. Auletta FJ, Flint APF (1988) Mechanisms controlling corpus luteum function in sheep, cows, nonhuman primates, and women especially in relation to the time of luteolysis. Endocr Rev 9: 88–105. [DOI] [PubMed] [Google Scholar]
- 13. Battye KM, Fairclough RJ, Cameron AWN, Trounson AO (1988) Evidence for prostaglandin involvement in early luteal regression of the superovulated nanny goat (Capra hircus). J Reprod Fertil 84: 425–430. [DOI] [PubMed] [Google Scholar]
- 14.Lahav M, Davis JS, Rennert H (1989) Mechanism of the luteolytic action of prostaglandin F-2 alpha in the rat. J Reprod Fertil Suppl 37: 233–240. [PubMed]
- 15. Schams D, Berisha B (2002) Steroids as local regulators of ovarian activity in domestic animals. Domest Anim Endocrinol 23: 53–65. [DOI] [PubMed] [Google Scholar]
- 16. Minegishi K, Tanaka M, Nishimura O, Tanigaki S, Miyakoshi K, et al. (2002) Reactive oxygen species mediate leukocyte-endothelium interactions in prostaglandin F2alpha -induced luteolysis in rats. Am J Physiol Endocrinol Metab 283: E1308–E1315. [DOI] [PubMed] [Google Scholar]
- 17. Sugino N, Okuda K (2007) Species-related differences in the mechanism of apoptosis during structural luteolysis. J Reprod Dev 53: 977–986. [DOI] [PubMed] [Google Scholar]
- 18. Diaz FJ, Luo W, Wiltbank MC (2011) Effect of decreasing intraluteal progesterone on sensitivity of the early porcine corpus luteum to the luteolytic actions of prostaglandin F2alpha. Biol Reprod 84: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Waclawik A (2011) Novel insights into the mechanisms of pregnancy establishment: regulation of prostaglandin synthesis and signaling in the pig. Reproduction 142: 389–399. [DOI] [PubMed] [Google Scholar]
- 20. Noda Y, Ota K, Shirasawa T, Shimizu T (2012) Copper/zinc superoxide dismutase insufficiency impairs progesterone secretion and fertility in female mice. Biol Reprod 86: 1–8. [DOI] [PubMed] [Google Scholar]
- 21. Juengel JL, Meberg BM, Turzillo AM, Nett TM, Niswender GD (1995) Hormonal regulation of messenger ribonucleic acid encoding steroidogenic acute regulatory protein in ovine corpora lutea. Endocrinology 136: 5423–5429. [DOI] [PubMed] [Google Scholar]
- 22. Pescador N, Soumano K, Stocco DM, Price CA, Murphy BD (1996) Steroidogenic acute regulatory protein in bovine corpora lutea. Biol Reprod 55: 485–491. [DOI] [PubMed] [Google Scholar]
- 23. Atli MO, Bender RW, Mehta V, Bastos MR, Luo W, et al. (2012) Patterns of gene expression in the bovine corpus luteum following repeated intrauterine infusions of low doses of prostaglandin F2alpha. Biol Reprod 86: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiltbank MC, Diskin MG, Flores JA, Niswender GD (1990) Regulation of the corpus luteum by protein kinase C. II. Inhibition of lipoprotein-stimulated steroidogenesis by prostaglandin F2 alpha. Biol Reprod 42: 239–245. [DOI] [PubMed] [Google Scholar]
- 25. Arvisais E, Hou X, Wyatt TA, Shirasuna K, Bollwein H, et al. (2010) Prostaglandin F2alpha represses IGF-I-stimulated IRS1/phosphatidylinositol-3-kinase/AKT signaling in the corpus luteum: role of ERK and P70 ribosomal S6 kinase. Mol Endocrinol 24: 632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Casey OM, Morris DG, Powell R, Sreenan JM, Fitzpatrick R (2005) Analysis of gene expression in non-regressed and regressed bovine corpus luteum tissue using a customized ovarian cDNA array. Theriogenology 64: 1963–1976. [DOI] [PubMed] [Google Scholar]
- 27. Goravanahally MP, Salem M, Yao J, Inskeep EK, Flores JA (2009) Differential gene expression in the bovine corpus luteum during transition from early phase to midphase and its potential role in acquisition of luteolytic sensitivity to prostaglandin F2 alpha. Biol Reprod 80: 980–988. [DOI] [PubMed] [Google Scholar]
- 28. Mondal M, Schilling B, Folger J, Steibel JP, Buchnick H, et al. (2011) Deciphering the luteal transcriptome: potential mechanisms mediating stage-specific luteolytic response of the corpus luteum to prostaglandin F2α . Physiol Genomics 43: 447–456. [DOI] [PubMed] [Google Scholar]
- 29. Benyo DF, Zeleznik AJ (1997) Cyclic adenosine monophosphate signaling in the primate corpus luteum: maintenance of protein kinase A activity throughout the luteal phase of the menstrual cycle. Endocrinology 138: 3452–3458. [DOI] [PubMed] [Google Scholar]
- 30. Bjurulf E, Selstam G (1996) Rat luteinizing hormone receptor messenger ribonucleic acid expression and luteolysis: inhibition by prostaglandin F2 alpha. Biol Reprod 54: 1350–1355. [DOI] [PubMed] [Google Scholar]
- 31. Snook RB, Saatman RR, Hansel W (1971) Serum progesterone and luteinizing hormone levels during the bovine estrous cycle. Endocrinology 88: 678–686. [DOI] [PubMed] [Google Scholar]
- 32. Okuda K, Uenoyama Y, Naito C, Sakabe Y, Kawate N (1999) Luteinizing hormone receptors in the bovine corpus luteum during the oestrous cycle and pregnancy. Reprod Fertil Dev 11: 147–151. [DOI] [PubMed] [Google Scholar]
- 33. Ginther OJ, Fuenzalida MJ, Pugliesi G, Hannan MA, Beg MA (2011) Effect of luteinizing hormone oscillations on progesterone concentrations based on treatment with a gonadotropin-releasing hormone antagonist in heifers. Domest Anim Endocrinol 40: 119–127. [DOI] [PubMed] [Google Scholar]
- 34. Luo W, Gumen A, Haughian JM, Wiltbank MC (2011) The role of luteinizing hormone in regulating gene expression during selection of a dominant follicle in cattle. Biol Reprod 84: 369–378. [DOI] [PubMed] [Google Scholar]
- 35. Wiltbank MC, Salih SM, Atli MO, Luo W, Bormann CL, et al. (2012) Comparison of endocrine and cellular mechanisms regulating the corpus luteum of primates and ruminants. Anim Reprod 9: 242–259. [PMC free article] [PubMed] [Google Scholar]
- 36. Okuda K, Uenoyama Y, Berisha B, Lange IG, Taniguchi H, et al. (2001) Estradiol-17beta is produced in bovine corpus luteum. Biol Reprod 65: 1634–1639. [DOI] [PubMed] [Google Scholar]
- 37. Shibaya M, Matsuda A, Hojo T, Acosta TJ, Okuda K (2007) Expressions of estrogen receptors in the bovine corpus luteum: cyclic changes and effects of prostaglandin F2alpha and cytokines. J Reprod Dev 53: 1059–1068. [DOI] [PubMed] [Google Scholar]
- 38. Goodman SB, Kugu K, Chen SH, Preutthipan S, Tilly KI, et al. (1998) Estradiol-mediated suppression of apoptosis in the rabbit corpus luteum is associated with a shift in expression of bcl-2 family members favoring cellular survival. Biol Reprod 59: 820–827. [DOI] [PubMed] [Google Scholar]
- 39. Maranesi M, Zerani M, Lilli L, Dall'Aglio C, Brecchia G, et al. (2010) Expression of luteal estrogen receptor, interleukin-1, and apoptosis-associated genes after PGF2alpha administration in rabbits at different stages of pseudopregnancy. Domest Anim Endocrinol 39: 116–130. [DOI] [PubMed] [Google Scholar]
- 40. Tripathy S, Kumarasamy A, Medhamurthy R (2013) Analysis of 20alpha-hydroxysteroid dehydrogenase expression in the corpus luteum of the buffalo cow: effect of prostaglandin F2-alpha treatment on circulating 20alpha-hydroxyprogesterone levels. Reprod Biol Endocrinol 11: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jyotsna UR, Medhamurthy R (2009) Standardization and validation of an induced ovulation model system in buffalo cows: Characterization of gene expression changes in the periovulatory follicle. Anim Reprod Sci 113: 71–81. [DOI] [PubMed] [Google Scholar]
- 42. Tilly JL, Hsueh A (1993) Microscale autoradiographic method for the qualitative and quantitative analysis of apoptotic DNA fragmentation. J Cell Physiol 154: 519–526. [DOI] [PubMed] [Google Scholar]
- 43. Uma J, Muraly P, Verma-Kumar S, Medhamurthy R (2003) Determination of onset of apoptosis in granulosa cells of the preovulatory follicles in the bonnet monkey (Macaca radiata): correlation with mitogen-activated protein kinase activities. Biol Reprod 69: 1379–1387. [DOI] [PubMed] [Google Scholar]
- 44. Priyanka S, Jayaram P, Sridaran R, Medhamurthy R (2009) Genome-wide gene expression analysis reveals a dynamic interplay between luteotropic and luteolytic factors in the regulation of corpus luteum function in the bonnet monkey (Macaca radiata). Endocrinology 150: 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Priyanka S, Medhamurthy R (2007) Characterization of cAMP/PKA/CREB signaling cascade in the bonnet monkey corpus luteum: expressions of inhibin-alpha and StAR during different functional status. Mol Hum Reprod 13: 381–390. [DOI] [PubMed] [Google Scholar]
- 46. Oshlack A, Chabot AE, Smyth GK, Gilad Y (2007) Using DNA microarrays to study gene expression in closely related species. Bioinformatics 23: 1235–1242. [DOI] [PubMed] [Google Scholar]
- 47. Ji W, Zhou W, Gregg K, Yu N, Davis S, et al. (2004) A method for cross-species gene expression analysis with high-density oligonucleotide arrays. Nucleic Acids Res 32: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nazar RN, Chen P, Dean D, Robb J (2010) DNA chip analysis in diverse organisms with unsequenced genomes. Mol Biotechnol 44: 8–13. [DOI] [PubMed] [Google Scholar]
- 49. Jyotsna UR, Shah KB, Jayaram P, Medhamurthy R (2011) Gene expression profiling of preovulatory follicle in the buffalo cow: effects of increased IGF-I concentration on periovulatory events. PLoS One 6: e20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 51. Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375. [DOI] [PubMed] [Google Scholar]
- 52. Dai M, Wang P, Boyd AD, Kostov G, Athey B, et al. (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu H, Zeeberg BR, Qu G, Koru AG, Ferrucci A, et al. (2007) AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics 23: 2385–2390. [DOI] [PubMed] [Google Scholar]
- 54. Lin CY, Ström A, Vega VB, Kong SL, Yeo AL, et al. (2004) Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol 5: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bryner RW, Gracia-Winder M, Lewis PE, Inskeep EK, Butcher RL (1990) Changes in hormonal profiles during the estrous cycle in old lactating beef cows. Domest Anim Endocrinol 7: 181–189. [DOI] [PubMed] [Google Scholar]
- 56. Noseir WM (2003) Ovarian follicular activity and hormonal profile during estrous cycle in cows: the development of 2 versus 3 waves. Reprod Biol Endocrinol 1: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shutt DA, Shearman RP, Lyneham RC, Clarke AH, McMahon GR, et al. (1975) Radioimmunoassay of progesterone, 17-hydroxyprogesterone, estradiol-17beta and prostaglandin F in human corpus luteum. Steroids 26: 299–310. [DOI] [PubMed] [Google Scholar]
- 58. Maas S, Jarry H, Teichmann A, Rath W, Kuhn W, et al. (1992) Paracrine actions of oxytocin, prostaglandin F2 alpha, and estradiol within the human corpus luteum. J Clin Endocrinol Metab 74: 306–312. [DOI] [PubMed] [Google Scholar]
- 59. Suresh PS, Medhamurthy R (2013) Studies on reactivation of regressing bonnet monkey corpus luteum on day 1 of menses: A pilot study. Syst Biol Reprod Med 59: 1–4. [DOI] [PubMed] [Google Scholar]
- 60. Coleman KM, Smith CL (2001) Intracellular signaling pathways: nongenomic actions of estrogens and ligand-independent activation of estrogen receptors. Front Biosci 6: D1379–D1391. [DOI] [PubMed] [Google Scholar]
- 61. Barton M (2012) Position paper: The membrane estrogen receptor GPER-Clues and questions. Steroids 77: 935–942. [DOI] [PubMed] [Google Scholar]
- 62. McCracken JA, Custer EE, Lamsa JC (1999) Luteolysis: a neuroendocrine-mediated event. Physiol Rev 79: 263–323. [DOI] [PubMed] [Google Scholar]
- 63. Niswender GD, Juengel JL, Silva PJ, Rollyson MK, McIntush EW (2000) Mechanisms controlling the function and life span of the corpus luteum. Physiol Rev 80: 1–29. [DOI] [PubMed] [Google Scholar]
- 64. Stocco C, Telleria C, Gibori G (2007) The molecular control of corpus luteum formation, function, and regression. Endocr Rev 28: 117–149. [DOI] [PubMed] [Google Scholar]
- 65. Shirasuna K, Sasahara K, Matsui M, Shimizu T, Miyamoto A (2010) Prostaglandin F2alpha differentially affects mRNA expression relating to angiogenesis, vasoactivation and prostaglandins in the early and mid corpus luteum in the cow. J Reprod Dev 56: 428–436. [DOI] [PubMed] [Google Scholar]
- 66. Stouffer RL, Bishop CV, Bogan RL, Xu F, Hennebold JD (2013) Endocrine and local control of the primate corpus luteum. Reprod Biol Endocrinol 13: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee J, McCracken JA, Banu SK, Rodriguez R, Nithy TK, et al. (2010) Transport of prostaglandin F (2alpha) pulses from the uterus to the ovary at the time of luteolysis in ruminants is regulated by prostaglandin transporter-mediated mechanisms. Endocrinology 151: 3326–3335. [DOI] [PubMed] [Google Scholar]
- 68. Zalman Y, Klipper E, Farberov S, Mondal M, Wee G, et al. (2012) Regulation of angiogenesis-related prostaglandin f2alpha-induced genes in the bovine corpus luteum. Biol Reprod 86: 1–10. [DOI] [PubMed] [Google Scholar]
- 69. Acosta TJ, Yoshijawa N, Ohtani M, Miyamoto A (2002) Local changes in blood flow within the early and midcycle corpus luteum after prostaglandin F(2 alpha) injection in the cow. Biol Reprod 66: 651–658. [DOI] [PubMed] [Google Scholar]
- 70. Utt MD, Acosta TJ, Wiltbank MC, Ginther OJ (2007) Acute effects of prostaglandin F(2alpha) on systemic oxytocin and progesterone concentrations during the mid- or late-luteal phase in mares. Anim Reprod Sci 97: 63–73. [DOI] [PubMed] [Google Scholar]
- 71. Guy MK, Juengel JL, Tandeski TR, Niswender GD (1995) Steady-state concentrations of mRNA encoding the receptor for luteinizing hormone during the estrous cycle and following prostaglandin F(2α) treatment of ewes. Endocrine 3: 585–589. [DOI] [PubMed] [Google Scholar]
- 72. Smith GW, Gentry PC, Roberts RM, Smith MF (1996) Ontogeny and regulation of luteinizing hormone receptor messenger ribonucleic acid within the ovine corpus luteum. Biol Reprod 54: 76–83. [DOI] [PubMed] [Google Scholar]
- 73. Agudo LS, Zahler WL, Smith MF (1984) Effect of prostaglandin F2 alpha on the adenylate cyclase and phosphodiesterase activity of ovine corpora lutea. J Anim Sci 58: 955–962. [DOI] [PubMed] [Google Scholar]
- 74. Garverick HA, Smith MF, Elmore RG, Morehouse GL, Agudo LS, et al. (1985) Changes and interrelationships among luteal LH receptors, adenylate cyclase activity and phosphodiesterase activity during the bovine estrous cycle. J Anim Sci 61: 216–223. [DOI] [PubMed] [Google Scholar]
- 75. Fitz TA, Mayan MH, Sawyer HR, Niswender GD (1982) Characterization of two steroidogenic cell types in the ovine corpus luteum. Biol Reprod 27: 703–711. [DOI] [PubMed] [Google Scholar]
- 76. Balapure AK, Caicedo IC, Kawada K, Watt DS, Rexroad CE Jr, et al. (1989) Multiple classes of prostaglandin F2 alpha binding sites in subpopulations of ovine luteal cells. Biol Reprod 41: 385–392. [DOI] [PubMed] [Google Scholar]
- 77. Hall JE, Bhatta N, Adams JM, Rivier JE, Vale WW, et al. (1991) Variable tolerance of the developing follicle and corpus luteum to gonadotropin-releasing hormone antagonist-induced gonadotropin withdrawal in the human. J Clin Endocrinol Metab 72: 993–1000. [DOI] [PubMed] [Google Scholar]
- 78. Webley GE, Hodges JK, Given A, Hearn JP (1991) Comparison of the luteolytic action of gonadotrophin-releasing hormone antagonist and cloprostenol, and the ability of human chorionic gonadotrophin and melatonin to override their luteolytic effects in the marmoset monkey. J Endocrinol 128: 121–129. [DOI] [PubMed] [Google Scholar]
- 79. Ravindranath N, Little-Ihrig LL, Zeleznik AJ (1992) Characterization of the levels of messenger ribonucleic acid that encode for luteinizing hormone receptor during the luteal phase of the primate menstrual cycle. J Clin Endocrinol Metab 74: 779–785. [DOI] [PubMed] [Google Scholar]
- 80. Peters KE, Bergfeld EG, Cupp AS, Kojima FN, Mariscal V, et al. (1994) Luteinizing hormone has a role in development of fully functional corpora lutea (CL) but is not required to maintain CL function in heifers. Biol Reprod 51: 1248–1254. [DOI] [PubMed] [Google Scholar]
- 81. Behrman HR, Grinwich DL, Hichens M, MacDonald GJ (1978) Effect of hypophysectomy, prolactin, and prostaglandin F2 alpha on gonadotropin binding in vivo and in vitro in the corpus luteum. Endocrinology 103: 349–357. [DOI] [PubMed] [Google Scholar]
- 82. Michael AE, Webley GE (1991) Prostaglandin F2 alpha stimulates cAMP phosphodiesterase via protein kinase C in cultured human granulosa cells. Mol Cell Endocrinol 82: 207–214. [DOI] [PubMed] [Google Scholar]
- 83. Michael AE, Webley GE (1993) Roles of cyclic AMP and inositol phosphates in the luteolytic action of cloprostenol, a prostaglandin F2 alpha analogue, in marmoset monkeys (Callithrix jacchus). J Reprod Fertil 97: 425–431. [DOI] [PubMed] [Google Scholar]
- 84. Taniguchi H, Komiyama J, Viger RS, Okuda K (2009) The expression of the nuclear receptors NR5A1 and NR5A2 and transcription factor GATA6 correlates with steroidogenic gene expression in the bovine corpus luteum. Mol Reprod Dev 76: 873–880. [DOI] [PubMed] [Google Scholar]
- 85. Tsumagari S, Kamata J, Takagi K, Tanemura K, Yosai A, et al. (1993) Aromatase activity and oestrogen concentrations in bovine cotyledons and caruncles during gestation and parturition. J Reprod Fertil 98: 631–636. [DOI] [PubMed] [Google Scholar]
- 86. Nimz M, Spitschak M, Schneider F, Fürbass R, Vanselow J (2009) Down-regulation of genes encoding steroidogenic enzymes and hormone receptors in late preovulatory follicles of the cow coincides with an accumulation of intrafollicular steroids. Domest Anim Endocrinol 37: 45–54. [DOI] [PubMed] [Google Scholar]
- 87. Vanselow J, Spitschak M, Nimz M, Fürbass R (2010) DNA methylation is not involved in preovulatory down-regulation of CYP11A1, HSD3B1, and CYP19A1 in bovine follicles but may have a role in permanent silencing of CYP19A1 in large granulosa lutein cells. Biol Reprod 82: 289–298. [DOI] [PubMed] [Google Scholar]
- 88. Ghosh S, Wu Y, Li R, Hu Y (2005) Jun proteins modulate the ovary-specific promoter of aromatase gene in ovarian granulosa cells via a cAMP-responsive element. Oncogene 24: 2236–2246. [DOI] [PubMed] [Google Scholar]
- 89. Lenz S, Pöhland R, Becker F, Vanselow J (2004) Expression of the bovine aromatase cytochrome P450 gene (Cyp19) is primarily regulated by promoter 2 in bovine follicles and by promoter 1.1 in corpora lutea. Mol Reprod Dev 67: 406–413. [DOI] [PubMed] [Google Scholar]
- 90. Vanselow J, Pöhland R, Fürbass R (2005) Promoter-2-derived Cyp19 expression in bovine granulosa cells coincides with gene-specific DNA hypo-methylation. Mol Cell Endocrinol 233: 57–64. [DOI] [PubMed] [Google Scholar]
- 91. Bulun SE, Lin Z, Imir G, Amin S, Demura M, et al. (2005) Regulation of aromatase expression in estrogen-responsive breast and uterine disease: from bench to treatment. Pharmacol Rev 57: 359–383. [DOI] [PubMed] [Google Scholar]
- 92. Stocco C (2008) Aromatase expression in the ovary: hormonal and molecular regulation. Steroids 73: 473–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hinshelwood MM, Liu Z, Conley AJ, Simpson ER (1995) Demonstration of tissue-specific promoters in nonprimate species that express aromatase P450 in placentae. Biol Reprod 53: 1151–1159. [DOI] [PubMed] [Google Scholar]
- 94. Hinshelwood MM, Michael MD, Simpson ER (1997) The 5′-flanking region of the ovarian promoter of the bovine CYP19 gene contains a deletion in a cyclic adenosine 3′,5′-monophosphate-like responsive sequence. Endocrinology 138: 3704–3710. [DOI] [PubMed] [Google Scholar]
- 95. Jackson JA, Albrecht ED (1985) The development of placental androstenedione and testosterone production and their utilization by the ovary for aromatization to estrogen during rat pregnancy. Biol Reprod 33: 451–457. [DOI] [PubMed] [Google Scholar]
- 96. Warshaw ML, Johnson DC, Khan I, Eckstein B, Gibori G (1986) Placental secretion of androgens in the rat. Endocrinology 119: 2642–2648. [DOI] [PubMed] [Google Scholar]
- 97. Vanselow J, Fürbass R, Rehbock F, Klautschek G, Schwerin M (2004) Cattle and sheep use different promoters to direct the expression of the aromatase cytochrome P450 encoding gene, Cyp19, during pregnancy. Domest Anim Endocrinol 27: 99–114. [DOI] [PubMed] [Google Scholar]
- 98. Gibori G, Chen YD, Khan I, Azhar S, Reaven GM (1984) Regulation of luteal cell lipoprotein receptors, sterol contents, and steroidogenesis by estradiol in the pregnant rat. Endocrinology 114: 609–617. [DOI] [PubMed] [Google Scholar]
- 99. Azhar S, Khan I, Gibori G (1989) The influence of estradiol on cholesterol processing by the corpus luteum. Biol Reprod 40: 961–971. [DOI] [PubMed] [Google Scholar]
- 100. Berisha B, Pfaffl MW, Schams D (2002) Expression of estrogen and progesterone receptors in the bovine ovary during estrous cycle and pregnancy. Endocrine 17: 207–214. [DOI] [PubMed] [Google Scholar]
- 101. Diaz FJ, Wiltbank MC (2004) Acquisition of luteolytic capacity: changes in prostaglandin F2alpha regulation of steroid hormone receptors and estradiol biosynthesis in pig corpora lutea. Biol Reprod 70: 1333–1339. [DOI] [PubMed] [Google Scholar]
- 102. Simoncini T, Hafezi-Moghadam A, Brazil DP, Ley k, Chin WW, et al. (200) Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature 407: 538–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bratton MR, Duong BN, Elliott S, Weldon CB, Beckman BS, et al. (2010) Regulation of ERalpha-mediated transcription of Bcl-2 by PI3K-AKT crosstalk: implications for breast cancer cell survival. Int J Oncol 37: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sanders SL, Stouffer RL (1997) Localization of steroidogenic enzymes in macaque luteal tissue during the menstrual cycle and simulated early pregnancy: immunohistochemical evidence supporting the two-cell model for estrogen production in the primate corpus luteum. Biol Reprod 56: 1077–1087. [DOI] [PubMed] [Google Scholar]
- 105. Rodger FE, Fraser HM, Duncan WC, Illingworth PJ (1995) Immunolocalization of bcl-2 in the human corpus luteum. Hum Reprod 10: 1566–1570. [DOI] [PubMed] [Google Scholar]
- 106. Kugu K, Ratts VS, Piquette GN, Tilly KI, Tao XJ, et al. (1998) Analysis of apoptosis and expression of bcl-2 gene family members in the human and baboon ovary. Cell Death Differ 5: 67–76. [DOI] [PubMed] [Google Scholar]
- 107. Peluffo MC, Bussmann L, Stouffer RL, Tesone M (2006) Expression of caspase-2, -3, -8 and -9 proteins and enzyme activity in the corpus luteum of the rat at different stages during the natural estrous cycle. Reproduction 132: 465–475. [DOI] [PubMed] [Google Scholar]
- 108. Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, et al. (2010) BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer 103: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Vaillant F, Merino D, Lee L, Breslin K, Pal B, et al. (2013) Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell 24: 120–129. [DOI] [PubMed] [Google Scholar]
- 110. Dong L, Wang W, Wang F, Stoner M, Reed JC, et al. (1999) Mechanisms of transcriptional activation of bcl-2 gene expression by 17beta-estradiol in breast cancer cells. J Biol Chem 274: 32099–32107. [DOI] [PubMed] [Google Scholar]
- 111. Pugazhenthi S, Nesterova A, Sable C, Heidenreich KA, Boxer LM, et al. (2000) Akt/protein kinase B up-regulates Bcl-2 expression through cAMP-response element-binding protein. J Biol Chem 275: 10761–10766. [DOI] [PubMed] [Google Scholar]
- 112. Perillo B, Sasso A, Abbondanza C, Palumbo G (2000) 17beta-estradiol inhibits apoptosis in MCF-7 cells, inducing bcl-2 expression via two estrogen-responsive elements present in the coding sequence. Mol Cell Biol 20: 2890–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wilson BE, Mochon E, Boxer LM (1996) Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol 16: 5546–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Du K, Montminy M (1998) CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem 273: 32377–32379. [DOI] [PubMed] [Google Scholar]
- 115. Tang ED, Nuñez G, Barr FG, Guan KL (1999) Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem 274: 16741–16746. [DOI] [PubMed] [Google Scholar]
- 116. Rena G, Guo S, Cichy SC, Unterman TG, Cohen P (1999) Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem 274: 17179–17183. [DOI] [PubMed] [Google Scholar]
- 117. Devoto L, Vega M, Kohen P, Castro A, Castro O, et al. (2000) Endocrine and paracrine-autocrine regulation of the human corpus luteum during the mid-luteal phase. J Reprod Fertil Suppl 55: 13–20. [PubMed] [Google Scholar]
- 118. Webb R, Woad KJ, Armstrong DG (2002) Corpus luteum (CL) function: local control mechanisms. Domest Anim Endocrinol 23: 277–285. [DOI] [PubMed] [Google Scholar]
- 119. Pate JL (1996) Intercellular communication in the bovine corpus luteum. Theriogenology 45: 1381–1397. [DOI] [PubMed] [Google Scholar]
- 120. Townson DH (2006) Immune cell-endothelial cell interactions in the bovine corpus luteum. Integr Comp Biol 46: 1055–1059. [DOI] [PubMed] [Google Scholar]
- 121. Korzekwa AJ, Jaroszewski JJ, Woclawek-Potocka I, Bah MM, Skarzynski DJ (2008) Luteolytic effect of prostaglandin F 2 alpha on bovine corpus luteum depends on cell composition and contact. Reprod Domest Anim 43: 464–472. [DOI] [PubMed] [Google Scholar]
- 122. Miyamoto A, Shirasuna K, Wijayagunawardane MP, Watanabe S, Hayashi M, et al. (2005) Blood flow: a key regulatory component of corpus luteum function in the cow. Domest Anim Endocrinol 29: 329–339. [DOI] [PubMed] [Google Scholar]
- 123. Shirasuna K, Watanabe S, Asahi T, Wijayagunawardane MP, Sasahara K, et al. (2008) Prostaglandin F2alpha increases endothelial nitric oxide synthase in the periphery of the bovine corpus luteum: the possible regulation of blood flow at an early stage of luteolysis. Reproduction 135: 527–539. [DOI] [PubMed] [Google Scholar]
- 124. Shirasuna K, Asahi T, Sasaki M, Shimizu T, Miyamoto A (2010) Distribution of arteriolovenous vessels, capillaries and eNOS expression in the bovine corpus luteum during the estrous cycle: a possible implication of different sensitivity by luteal phase to PGF(2alpha) in the increase of luteal blood flow. J Reprod Dev 56: 124–130. [DOI] [PubMed] [Google Scholar]
- 125. Knickerbocker JJ, Wiltbank MC, Niswender GD (1988) Mechanisms of luteolysis in domestic livestock. Domest Anim Endocrinol 5: 91–107. [DOI] [PubMed] [Google Scholar]
- 126. Juengel JL, Garverick HA, Johnson AL, Youngquist RS, Smith MF (1993) Apoptosis during luteal regression in cattle. Endocrinology 132: 249–254. [DOI] [PubMed] [Google Scholar]
- 127. Rueda BR, Wegner JA, Marion SL, Wahlen DD, Hoyer PB (1995) Internucleosomal DNA fragmentation in ovine luteal tissue associated with luteolysis: in vivo and in vitro analyses. Biol Reprod 52: 305–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded tree view generated after hierarchical clustering of representative genes obtained after pairwise analysis. The expanded tree displaying the hierarchy analysis for probe sets at each time point [0 vs. 3 h (A), 0 vs. 6 h (B) and 0 vs. 18 h (C)] post PGF2α administration. The bottom of each dendrogram shows the condition color bar with the parameters in each interpretation. The legend shows the name of each condition on which clustering was performed. Header of heat map shows a normalized intensity values represented in various shades of red and green indicating relatively either up or down regulation, respectively. The row header shown on the right side represents the complete entity name or gene symbol. The upper panel shows groups of genes, whose expression was up regulated at 0 h and with the administration of PGF2α the expression is observed to be down-regulated. The lower panel shows groups of genes whose expression was down regulated at 0 h and with the administration of PGF2α the expression is observed to be up regulated.
(TIF)
Effects of PGF2α on circulating and luteal P4 levels, StAR expression and DNA fragmentation. Buffalo cows received intramuscular injection of 500 µg of PGF2α on day 11 of estrous cycle and blood and luteal tissue samples at different time intervals post PGF2α treatment. (A) Circulating mean±SEM serum and luteal progesterone (P4) concentrations immediately before (0 h) and at different time points post PGF2α treatment. Bars with different alphabets indicate statistical significance, p<0.05. (B) Protein lysate (100 µg) prepared from CL tissue collected before and post PGF2α treatment were resolved on 10% SDS PAGE, transferred onto PVDF membrane and immunoblot analysis was performed using anti-StAR and anti-β-actin antibody (β-actin was used as loading control). A representative immunoblot for each of the antibody probed is shown. Densitometric values shown on top of each lane represented were determined and represented as mean±SEM (n = 3 CL/time point), relative to intensity of β-actin for each time point post PGF2α treatment. (C) Analysis of apoptotic DNA fragmentation in luteal tissue. Genomic DNA isolated from CL tissues collected from untreated control animals (0 h) and from animals at different time points post PGF2α treatment was subjected to DNA laddering analysis. An image of a nylon membrane visualized using PhosphorImager containing the [α32P] labeled genomic DNA separated previously on 2% agarose gel and transferred on to nylon membrane is represented here. Migration (base pairs) of oligonucleosomes is indicated on the right.
(TIF)
Classification of differentially expressed genes post 6 h PGF2α administration by Ingenuity Pathway Analysis (IPA). (A) The pathway analysis indicates that a large number of differentially expressed genes belongs to canonical pathways such as p38 MAP kinase signaling, VEGF signaling, transcription factors belonging to steroidogenesis and coagulation system. The orange line represents score for the likelihood [-log (B-H P < 0.05)] that genes belonging to a specific canonical pathway category affected at 6 h post PGF2α administration. The stacked bars indicate the percentage of genes distributed according to regulation, i.e., green (down), red (up) and open bars (no overlap with dataset) in each canonical pathway. (B) Network 0 vs. 6 h: Ingenuity Pathway Analysis of the differentially regulated genes 6 h post PGF2α administration shows a network of 28 focus molecules with a score of 44, with top biological functions of cell to cell signaling, molecular transport and lipid metabolism. The network is displayed graphically as nodes (genes/gene products) and edges (biological relationship between nodes). The node color intensity indicates the fold change expression of genes; with red representing up regulation, and green down regulation of genes between 0 vs. 6 h post PGF2α administration. The fold change value for individual gene is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction.
(TIF)
Classification of differentially expressed genes post 18 h PGF2α administration by Ingenuity Pathway Analysis (IPA). (A) The pathway analysis indicates that a large number of differentially expressed genes belong to canonical pathways such as TGF-β signaling, steroid biosynthesis, NF-κB signaling and coagulation system. The orange line represents score for the likelihood [-log (B-H P<0.05)] that genes belonging to a specific canonical pathway category affected at 18 h post PGF2α administration. The stacked bars indicate the percentage of genes distributed according to regulation, i.e., green (down), red (up) and open bars (no overlap with dataset) in each canonical pathway. (B) Network 0 vs. 18 h: Ingenuity Pathway Analysis of the differentially regulated genes 18 h post PGF2α administration shows a network of 26 focus molecules with a score of 39, with top biological functions of cellular development, cell cycle and gene expression. The network is displayed graphically as nodes (genes/gene products) and edges (biological relationship between nodes). The node color intensity indicates the fold change expression of genes; with red representing up regulation, and green down regulation of genes between 0 vs. 18 h post PGF2α administration. The fold change value for individual gene is indicated under each node. The shapes of nodes indicate the functional class of the gene product and the lines indicate the type of interaction.
(TIF)
Effect of PGF2α administration on expression of orphan nuclear transcription factors in CL. (A and B) The orphan nuclear transcription factors associated with regulation of expression of steroidogenic genes were analyzed. Protein levels of NR5A1/SF-1(A) and NR5A2/LRH-1 (B) in bovine CL were determined. Protein lysate (100 µg) prepared from CL tissue collected before (0 h) and post (3, 6 and 18 h) PGF2α treatment were resolved on 10% SDS PAGE, transferred onto PVDF membrane and immunoblot analysis was performed using anti-SF1, anti-LRH1 and anti-β-actin antibody. A representative immunoblot for each of the antibody probed is shown. The immunoblot probed with β-actin antibody indicates loading control for each lane. Densitometric values were determined and indicated as mean±SEM (n = 3 animals/time point), relative to intensity of β-actin for each time point post PGF2α treatment. The values of immunoblot analysis has been put on top of each lane and lane with different letters indicates statistical significance, p<0.05.
(TIF)
PGF2α administration effect on expression of estrogen receptors during spontaneous and induced luteolysis in CL. Quantitative real time PCR (qPCR) fold change expression of the estrogen receptors (ERα and ERβ) during spontaneous (A) and induced (B) luteolysis. Total RNA isolated from CL was reverse transcribed and cDNA equivalent to 10 ng of total RNA was used for qPCR. The expression was normalized with L19 mRNA. The results are shown as fold changes of mRNA expression compared with that at early (E) luteal phase (A) and 0 h PGF2α (B) for bovine CL. Individual bar for each gene represents mean±SEM fold change in mRNA expression value for qPCR analysis at each time point (n = 2 animals/time point, A and n = 3 animals/time point, B). For each gene, bars with different letters above them are significantly different (p<0.05).
(TIF)
List of primer set employed for quantitative real time PCR. The list of genes and details of the primers employed in the qPCR analysis along with the annealing temperature and expected amplicon size are provided.
(TIF)
List of top 15 up regulated genes at 3 h post PGF2α administration. Microarray data analysis was carried out to obtain a set of differentially expressed genes based on statistics, a Student's t-test (two tail, unpaired) with p<0.05 and multiple hypothesis testing (Benjamini and Hochberg comparison test) to reduce the false positives. The identified differentially expressed genes were transcript consistent and did not hybridize to multiple transcripts, as suggested by the AffyProbeMiner analysis. A Bioconductor analysis was performed with ≥2 fold change as cut-off with statistical filters for identification of differentially expressed genes. Whereas, the top 15 differentially UP regulated genes at 3 h post PGF2α treatment are represented in this table. Probe Set ID: The identifier that refers to a set of probe pairs selected to represent expressed sequences on an array; Fold Change: It is a number describing changes in expression level of a gene compared between control and treatment; Gene ID: Gene symbols extracted from Entrez Gene or UniGene; Gene Title: Gene name extracted from Entrez Gene or UniGene.
(TIF)
List of top 15 down regulated genes at 3 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 3 h post PGF2α treatment are represented.
(TIF)
List of top 15 up regulated genes at 6 h post PGF2α administration. The top 15 differentially UP regulated genes at 6 h post PGF2α treatment are represented.
(TIF)
List of top 15 down regulated genes at 6 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 6 h post PGF2α treatment are represented.
(TIF)
List of top 15 up regulated genes at 18 h post PGF2α administration. The top 15 differentially UP regulated genes at 18 h post PGF2α treatment are represented.
(TIF)
List of top 15 down regulated genes at 18 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 18 h post PGF2α treatment are represented.
(TIF)
List of top 15 up regulated E2 responsive genes at 3 h post PGF2α administration. Potential E2 responsive genes in bovine CL were identified based on the available list of classical E2 responsive genes, genes employed in PCR array human estrogen signaling and the data base, ERGDB. Microarray data analysis was carried out to obtain a set of differentially expressed genes based on statistics, a Student's t-test (two tail, unpaired) with p<0.05 and multiple hypothesis testing (Benjamini and Hochberg comparison test) to reduce the false positives. The identified differentially expressed E2 responsive genes were transcript consistent and did not hybridize to multiple transcripts, as suggested by the AffyProbeMiner analysis. A Bioconductor analysis was performed with ≥1 fold change as cut-off and statistical filters for identification of differentially expressed E2 responsive genes. Whereas, the top 15 differentially UP regulated genes at 3 h post PGF2α treatment are represented in this table. Probe Set ID: The identifier that refers to a set of probe pairs selected to represent expressed sequences on an array; Fold Change: It is a number describing changes in expression level of a gene compared between control and treatment; Gene ID: Gene symbols extracted from Entrez Gene or UniGene; Gene Title: Gene name extracted from Entrez Gene or UniGene.
(TIF)
List of top 15 down regulated E2 responsive genes at 3 h post PGF2α administration. The top 15 differentially DOWN regulated genes at 3 h post PGF2α treatment are represented. The genes are discussed in the results and discussion section.
(TIF)
List of common up regulated E2 responsive genes post 3, 6 and 18 h PGF2α administration. The common differentially UP regulated E2 responsive genes (21 genes) before (0 h) and post (3, 6 and 18 h) PGF2α treatment are represented.
(TIF)
List of common down regulated E2 responsive genes post 3, 6 and 18 h PGF2α administration. The common differentially DOWN regulated E2 responsive genes (14 genes) before (0 h) and post (3, 6 and 18 h) PGF2α treatment are represented.
(TIF)
List of common up regulated E2 responsive genes in monkey (24 h) and bovine (3 h) CL. The previously published microarray data of the differentially expressed genes from the CL tissues of macaques receiving PGF2α treatment for 24 h [GEO accession number GSE8371] was mined for E2 responsive genes for purposes of comparing the number of E2 responsive genes that were differentially expressed in CL of macaques to that of the buffalo cow CL [GEO accession number GSE27961]. The mined data comprising common UP regulated E2 responsive genes (19 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 3 h post PGF2α treatment are represented in this Table.
(TIF)
List of common down regulated E2 responsive genes in monkey (24 h) and bovine (3 h) CL. The mined data comprising common DOWN regulated E2 responsive genes (8 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 3 h post PGF2α treatment are represented in this Table.
(TIF)
List of common up regulated E2 responsive genes in monkey (24 h) and bovine (6 h) CL. The mined data comprising common UP regulated E2 responsive genes (17 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 6 h post PGF2α treatment are represented in this Table.
(TIF)
List of common down regulated E2 responsive genes in monkey (24 h) and bovine (6 h) CL. The mined data comprising common DOWN regulated E2 responsive genes (13 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 6 h post PGF2α treatment are represented in this Table.
(TIF)
List of common up regulated E2 responsive genes in monkey (24 h) and bovine (18 h) CL. The mined data comprising common UP regulated E2 responsive genes (17 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 18 h post PGF2α treatment are represented in this Table.
(TIF)
List of common down regulated E2 responsive genes in monkey (24 h) and bovine (18 h) CL. The mined data comprising common DOWN regulated E2 responsive genes (13 genes) of macaques CL at 24 h vs. E2 responsive genes of bovine CL at 6 h post PGF2α treatment are represented in this Table.
(TIF)