Abstract
The fatty acid synthase (FAS) is a conserved primary metabolic enzyme complex capable of tolerating cross-species engineering of domains for the development of modified and overproduced fatty acids. In eukaryotes, acyl-acyl carrier protein thioesterases (TEs) off-load mature cargo from the acyl carrier protein (ACP), and plants have developed TEs for short/medium-chain fatty acids. We showed that engineering plant TEs into the green microalga Chlamydomonas reinhardtii does not result in the predicted shift in fatty acid profile. Since fatty acid biosynthesis relies on substrate recognition and protein-protein interactions between the ACP and its partner enzymes, we hypothesized that plant TEs and algal ACP do not functionally interact. Phylogenetic analysis revealed major evolutionary differences between FAS enzymes, including TEs and ketoacyl synthases (KSs), in which the former is present only in some species, whereas the latter is present in all, and has a common ancestor. In line with these results, TEs appeared to be selective towards their ACP partners whereas KSs showed promiscuous behavior across bacterial, plant and algal species. Based on phylogenetic analyses, in silico docking, in vitro mechanistic crosslinking and in vivo algal engineering, we propose that phylogeny can predict effective interactions between ACPs and partner enzymes.
Keywords: acyl carrier protein, evolution, algae, bacteria, fatty acid synthase, Chlamydomonas reinhardtii
Introduction
Fatty acids are biosynthesized by the fatty acid synthase (FAS), which consists of several catalytic domains, either present on separate proteins (type II) or on one polypeptide (type I). Type II synthases are typically found in bacteria but also in mitochondria, chloroplasts and apicoplasts of eukaryotes. Type I synthases are present in eukaryotes and thought to be a fusion of the evolutionarily older type II synthases (McCarthy and Hardie 1984). The acyl carrier protein (ACP) (Crosby and Crump 2012; Cantu et al. 2012b) is the central protein that carries the iteratively growing fatty acid on its phosphopantetheine arm, which is post-translationally added by dedicated phosphopantetheinyl transferases (PPTases) (Lambalot et al. 1996). In type II synthases, ACP buries its attached cargo within its inner hydrophobic core, and protein-protein interactions with catalytic proteins induces the acyl chain to release from the inner core and into the active site of the catalytic protein (Zornetzer et al. 2010). Productive catalysis therefore depends not only on substrate recognition, but also on favorable protein-protein interactions (Worthington et al. 2006).
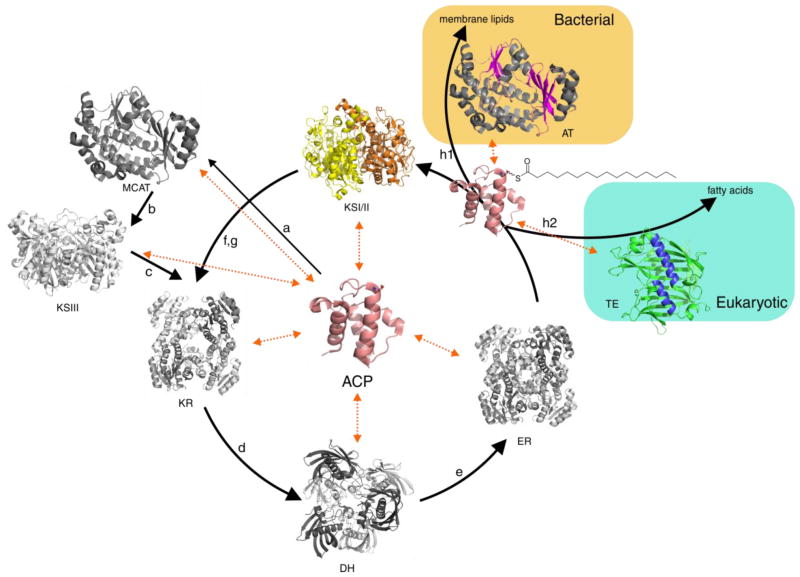
Fatty acid biosynthesis is catalyzed in iterative cycles by acyl transferases (ATs), ketoacyl synthases (KSs), a ketoreductase (KR), a dehydratase (DH), and an enoyl reductase (ER) (Figure 1). This set of enzymes is highly conserved throughout all kingdoms of life (Campbell and Cronan 2001). However, to off-load cargo from the ACP, bacteria use an acyl transferase (AT) to directly acylate glycerol-3-phosphate, whereas eukaryotes utilize an acyl-acyl carrier protein thioesterase (TE) to hydrolyze the mature fatty acid chain from the ACP (Figure 1) (Jones et al. 1995). Plants and green microalgae are noteworthy in this context, since their fatty acid biosynthesis takes place in the chloroplast, which originates from an endosymbiotic event between an early eukaryote and cyanobacteria. Although prokaryotic in nature (type II FAS), plants and algae use thioesterases, and we (Blatti et al. 2012) and others (Jing et al. 2011) have recently shown that the TE from green microalgae is distinct from plants, both phylogenetically as well as in substrate specificity. Whereas plants have developed specific TEs for different acyl chain lengths, FatA and FatB (Salas and Ohlrogge 2002), algae use one broad-specificity thioesterase, Fat1 (Blatti et al. 2012). Since TEs determine chain length, many bio-engineering efforts have been geared toward manipulating fatty acid chain length for ‘designer oils’ and biofuels by introducing foreign TEs or up-regulating endogenous ones, which has proven to be successful in bacteria (Yuan et al. 1995) and plants (Thelen and Ohlrogge 2002). Similar efforts in microalgae, however, show limited effect (Radakovits et al. 2011), suggesting that plant TEs engineered into algae do not productively interact with the endogenous FAS machinery and in particular the ACP. Why are vascular plant TEs functional in bacteria but not (or much less so) in algae? Here, we attempt to answer this question by investigating the evolutionary origin of TEs in comparison to the other FAS enzymes.
Figure 1.
Type II fatty acid biosynthesis in bacteria and eukaryotes: (a) Fatty acid biosynthesis is initiated by the formation of malonyl-ACP from malonyl-CoA and holo-ACP by malonyl-CoA acyltransferase (MCAT, FabD). (b) β-ketoacyl ACP synthase III (KSIII, FabH) is responsible for the initial condensation of malonyl-ACP with acetyl-CoA. (c) 3-oxoacyl ACP ketoreductase (KR, FabG) reduces the keto group and (d) 3-hydroxyacyl ACP dehydratase (DH, FabA/FabZ) eliminates water. (e) enoyl ACP reductase (ER, FabI) reduces the double bond using NADH. Iterative activity of KS, KR, DH and ER domains extends the chain by two carbons each cycle. β-ketoacyl ACP synthase I (KSI, FabB) extends the chain from C4 to C16 and β-ketoacyl ACP synthase II (KS2, FabF) forms C18 from C16 (f and g). In the bacterial termination pathway, fatty acids are transferred from acyl-ACP to glycerol-3-phosphate for phospholipid formation using an acyl transferase (AT) (h1), and in the eukaryotic pathway, mature fatty acids are hydrolyzed from acyl-ACP by an acyl-ACP thioesterase (TE) (h2).
In contrast to TEs, ketoacyl synthases (KSs) (Chen et al. 2011) are omnipresent, essential, and share a known common ancestor in the archaeal thiolase (Jiang et al. 2008). KSs catalyze carbon-carbon bond formation via a Claisen condensation, and typically three different KSs are found in the type II FAS synthase (Figure 1). Together with malonyl-CoA acyl transferase (MCAT), KSIII is responsible for initiation of the fatty acid catalytic cycle by condensing malonyl-ACP with acetyl-CoA. KSI elongates the fatty acid up to palmitoyl-ACP, and KSII catalyzes the biosynthesis of stearoyl-ACP from palmitoyl-ACP. By divergent evolution, these initiation and elongation enzymes have developed from the thiolase precursor. Chen et al. (2011) identified five KS families: KS1 is represented by KSIII, KS2 by eukaryotic fatty acid elongases, KS3 by KSI and KSII, KS4 by chalcone synthases and KS5 by animal very long chain fatty acid elongases. To date no structures of KS2 and KS5 have been published but KS1, KS3 and KS4 share a similar struture (Cantu et al. 2011). In this superfamily of enzymes, the nucleophilic α-anion of an acyl-thioester is generated in either a non-decarboxylative or decarboxylative fashion, the latter an evolutionarily more recent optimization of this class of enzymes, to which the KSs belong. Here, we identify algal KSs, determine their phylogenetic context, and compare engineering of KS into various species with similar efforts for TE domains. Comparing TE and KS side-by-side shines light on the apparent directionality in the ability of FAS enzymes to interact with each other. Including data on the other FAS enzymes (MCAT, KR, DH and ER) led us to the hypothesis that evolution can teach us rules on protein-protein interactions across species. Using phylogenetic analyses, in silico protein-protein modeling, in vitro mechanistic crosslinking and in vivo engineering, we showcase the compatibility between FAS enzymes and ACP from different species, focusing here on the green microalgae Chlamydomonas reinhardtii.
Materials and Methods
Sequence alignments
Structure-based alignments of bacterial, algal, and plant FAS enzymes were produced using TCoffee (Notredame et al. 2000), MUSCLE (Edgar 2004) and ESPript (Gouet et al. 1999).
Phylogenetic analyses
Phylogeny was initially analyzed using FastTree (Price et al. 2010). Some enzymes (e.g. KSIII, FabH) are not annotated in any algal genome. These putative enzymes were identified by psi-blasting (Altschul et al. 1997) known plant enzymes against microalgal genomes. Detailed phylogenetic analyses were conducted with sequences obtained from the Uniprot and NCBI databases. Sequences were aligned with MUSCLE (Edgar 2004) and Bayesian trees constructed using MrBayes (Huelsenbeck and Ronquist 2001). The ‘mixed’ amino acid analysis model (Ronquist and Huelsenbeck 2003) was used, and Markov chain Monte Carlo analysis (Larget and Simon 1999) was performed for 1 million generations with four independent chains, sampling every 500 generation. At the end of the run, average standard deviation (< 0.001) and potential scale reduction factor (PSRF, approaching 1.0000) were used as a measure for convergence of the process. Tree representations were made using FigTree (Rambaut) or Mega (Tamura et al. 2011).
Protein modeling and docking
Proteins were homology modeled using Swissmodel (Schwede et al. 2003) and docked using the Cluspro server (Comeau et al. 2004). Models were compared with the models obtained from the i-Tasser server (Zhang 2008) and no significant deviations between the two homology modeling algorithms were observed. Since KSII is dimeric, dimer models (prior to protein-protein docking) were predicted using the Cluspro server (Comeau et al. 2004). Models were manually examined using PyMol (Pymol 2012).
Protein expression and purification
CoaA, CoaD, CoaE, Sfp and Escherichia coli ACP (EcACP) (Worthington et al. 2006), C. reinhardtii chloroplastic ACP (Cr-cACP) (Blatti et al. 2012), C. reinhardtii acyl-ACP thioesterase (CrTE) (Blatti et al. 2012), Pseudomonas aeruginosa ACP hydrolase (ACPH) (Blatti et al. 2012; Kosa et al. 2012), and E. coli KSII (EcKSII) (Worthington et al. 2006; Kitagawa et al. 2006) were expressed in E. coli and purified according to standard protocols. ACPH catalyzed the formation of apo-ACPs from holo-ACPs in vitro (Kosa et al. 2012; Blatti et al. 2012).
In vitro activity-based crosslinking assay
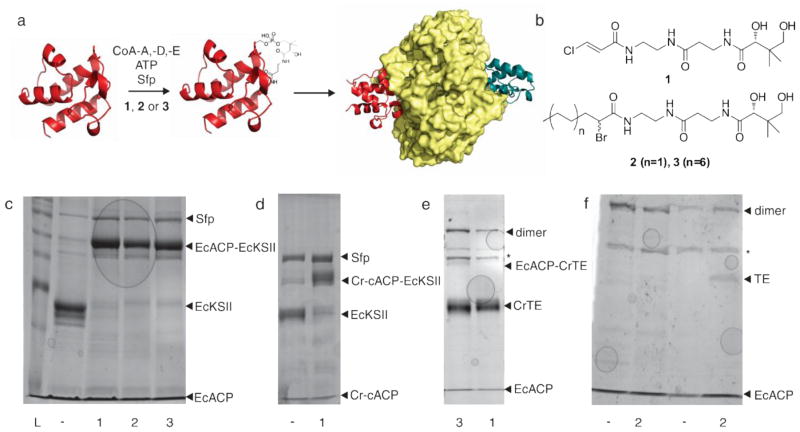
Biochemical reconstitution of a truncated CoA biosynthetic pathway formed reactive CoA-analogues in vitro (Worthington et al. 2006), and the PPTase Sfp was used to load each ACP with reactive phosphopantetheine analogues 1, 2 and 3. Reactions contained phosphate buffer, pantetheine analogue (1, 2 or 3 in DMSO, see structures in Figure 5), MgCl2, ATP, ACP, CoaA, CoaD, CoaE and Sfp (Worthington et al. 2006). Following the one-pot formation of crypto-ACPs for 1 h at 37 °C, EcKSII or CrTE/UcTE (Davies et al. 1991)/ChTE (Dörmann et al. 1993) was added and the reactions were incubated for 3 h (KS) or overnight (TE) at 37 °C (Blatti et al. 2012). In negative control reactions, pantetheine analogue was omitted. To detect crosslinked species, anti-FLAG resin or Ni-NTA resin was added directly to each reaction and incubated for 1 h at 25 °C. After washing the resin with TBS, proteins were eluted with 1 M arginine (pH 3.5) or 100 mM imidazole, respectively, and run on 8% SDS-PAGE stained with coomassie to visualize crosslinked complexes.
Figure 5.
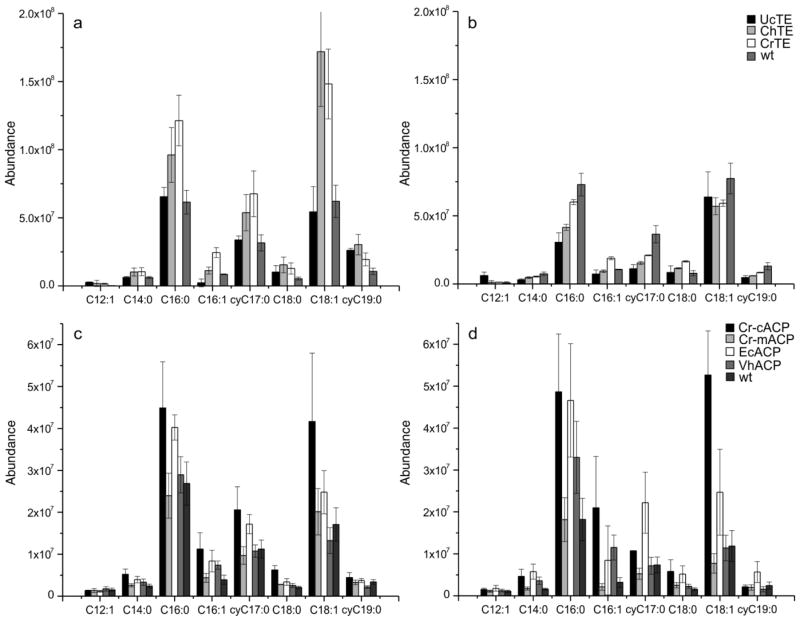
Acyl-ACP TE and ACP expression activity in E. coli strain BL21. a) Uninduced and b) induced TEs: UcTE (solid black), ChTE (light grey), CrTE (white) and control (dark grey). c) Uninduced and d) Induced ACPs: Cr-cACP (solid black), Cr-mACP (light grey), EcACP (white), VhACP (dark grey) and control (darker grey). Each data point is obtained in triplicate.
ACP complementation assay
To test whether Cr-cACP functionally interacts with E. coli FAS in vivo, we used ACP chromosomal knockout strain CY1877 (De Lay and Cronan 2007). Since ACP is essential, this strain also harbors acpp on an arabinose-inducible plasmid (De Lay and Cronan 2007). CY1877 was transformed with IPTG-inducible plasmids (proteins were cloned into pGEX-5X3 using BamH1 and Xho1) harboring Cr-cACP or FAS ACP from Vibrio harveyi (VhACP) (Volkmann et al. 2010). Transformants were selected on LB-agar plates supplemented with 50 μg/ml spectinomycin, 100 μg/ml ampicillin and 0.2% w/v arabinose after overnight incubation at 37 °C. A dense overnight culture was diluted and plated on arabinose deplete LB-glucose-agar plates supplemented with spectinomycin and ampicillin. Sterile filter discs were added to each plate, 1 pmol of IPTG spotted, and incubated at 25 °C and 37 °C overnight. In parallel, the assay was performed in liquid culture, as previously described (Volkmann et al. 2010).
Fatty acid analysis of transgenic E. coli strains
E. coli BL21 cells harboring FAS enzymes were cultured in 5 ml LB media supplemented with the appropriate antibiotics at 37 °C overnight. These starter cultures were used to inoculate 5 mL cultures, and upon reaching an OD of 0.8, 500 μM IPTG was added to the cells to induce protein expression. Cells were induced for 4 h at 37 °C and harvested by centrifugation. Both lyophilized supernatant media and bacterial cell pellet were separately resuspended in 1 M HCl in methanol, incubated for 30 min at 65 °C, and fatty acid methyl esters (FAMEs) extracted using hexanes. The fatty acid composition was determined by GC/MS analysis on an Agilent 7890A GC system connected to a 5975C VL MSD quadrupole MS (EI). Helium was used as a carrier gas. A method employing a gradient of 110 °C to 200 °C at 15 °C min−1 followed by 20 min at 200 °C on a 60 meter DB23 column was used.
Expression of RcKSII in algal chloroplast
KSII from the castor-oil plant (Ricinus communis) was engineered into the C. reinhardtii chloroplast using methods previously described (Goldschmidt-Clermont 1991). The plant RcKSII gene was codon-optimized for C. reinhardtii chloroplast and subcloned into a chloroplast expression vector bearing a FLAG epitope at the carboxy terminus. Biolistics transformed C. reinhardtii strain C137+ with the RcKSII gene, integration of the RcKSII gene into the chloroplast genome was validated by PCR, and expression of the RcKSII protein in C. reinhardtii chloroplast was detected by Western blot using M2 anti-FLAG antibody. C. reinhardtii strains were grown on TAP media (Gorman and Levine 1965) agar plates supplemented with 40 mg L−1 carbendazim, 500 mg L−1ampicillin, 100 mg L−1cefotaxime (Kan and Pan 2010) and 50 mg L−1 kanamycin under constant illumination. Liquid TAP media cultures were inoculated and grown under constant shaking in a greenhouse for 3 days. Cultures were harvested and the cell pellet and the supernatant subjected to FAME extraction as described earlier.
Results
Sequence alignment and phylogeny of ACPs
Structure-based sequence analysis of bacterial, algal, and plant ACPs (Figure S1) shows that the α-helical secondary structure, as well as the serine residue which serves as the site of post-translational modification, is conserved across species. Algal and plants genomes contain multiple copies of ACPs, including different isoforms and cellular localization (chloroplast, mitochondria and/or roots, leaves, see Table S1). C. reinhardtii has two ACPs, annotated as ACP1 and ACP2. We have at least three lines of evidence for the localization of ACP1 and ACP2 of C. reinhardtii. First, Atteia et al. (Atteia et al. 2009) identify ACP1 in the mitochondria of C. reinhardtii by a proteomics approach. Second, Predalgo (Tardif et al. 2012) (currently the only targeting prediction tool for algae) predicts mitochondrial targeting for ACP1 and chloroplastic targeting for ACP2. Third, a phylogenetic tree of ACPs shows at least 16 families (Cantu et al. 2012b) (Figure S2A) and subfamilies in FAS ACPs (Figure S2B), including the two C. reinhardtii ACPs: ACP1 clustering with mitochondrial ACPs and ACP2 with chloroplastic ACPs.
Sequence alignment and phylogeny of KSs
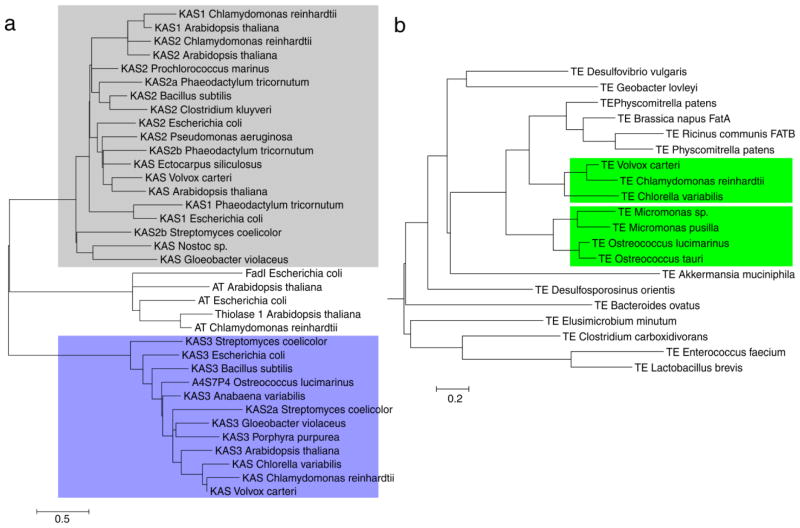
Sequence alignment of bacterial, algal, and plant KSs (KSI, II, III) illustrates a high degree of sequence conservation between KS types across species (Figure S1) (Jiang et al. 2008; Chen et al. 2011). Phylogenetic analysis reveals clustering of KSIs, KSIIs, and KSIIIs in all organisms interrogated, indicating KS domains are similar across species (Figure 2A and S3). Sequences of KSs and thiolases from algae, plants, bacteria, cyanobacteria and archaea were collected. Although KSIII is not annotated in any algal genome, BLAST analysis of known plant KSIIIs against sequenced algal genomes identifies putative KSIIIs in algae. Constructing a phylogenetic tree of KS domains clearly illustrates a division between thiolases, KSI/II, and KSIII enzymes in all species.
Figure 2.
Phylogenetic analysis of thiolase and thioesterase families. a) Thiolase phylogeny. b) Acyl-ACP thioesterase phylogeny. Full trees of all FAS enzymes can be found in Figures S2–S13. The scale bar represents branch length: the number of amino acid substitutions per 100 residues.
Sequence alignment and phylogeny of TEs
Chen et al. (2011) classified thioesterases into 23 families, from which at least six function on acyl-ACPs, including TE14 (classical hot-dog fold FatA/FatB), TE15 (polyketide enediyne specific hot-dog fold, TebC), TE16 (α/β-hydrolase part of type I FAS, PKS or NRPS), TE17 (α/β-hydrolase part of type I PKS), TE18 (α/β-hydrolase part of type II FAS, NRPS, PKS) and TE19 (α/β-hydrolase, LuxD, responsible for hydrolyzing myristoyl-ACP as precursor for the substrate of luciferase) (Cantu et al. 2010). The archetypical hot-dog fold acyl-ACP TE family TE14 has recently been described in detail, being present in anaerobic bacteria, algae and plants, but not in cyanobacteria (Jing et al. 2011). Previously, we created a homology model of CrTE, modeled after the thioesterase from an obligate anaerobic bacteria (Lactobacillus plantarum) (Blatti et al. 2012). Sequence alignment of TEs (Figure S1) from anaerobic bacteria, plants and algae shows many regions of sequence homology albeit that the putative active site cysteine residue of plant and algal TEs is missing in anaerobic bacteria. A rooted phylogenetic tree of TE sequences from anaerobic bacteria, algae, and plant FatA and FatB TEs was constructed to shine light on the evolutionary origin of the TE (Figure 2B and S4). The TE superfamily can be divided in several clades (Jing et al. 2011; Blatti et al. 2012). There are two separate clades of algal Fat1 TEs, which appear to correlate with distinct algal fatty acid profiles (Figure S20). Primitive plants, like the mosses Selaginella and Physcomitrella, form a separate clade in between green microalgae and plants (Jing et al. 2011).
FAS enzyme phylogeny
FabD is the malonyl CoA ACP acyltransferase (MCAT) that catalyzes the synthesis of malonyl-ACP from malonyl-CoA and holo-ACP. Interestingly, FabD links FAS and polyketide synthases in some bacterial species, providing the necessary starting material for both synthases (Florova et al. 2002). Surprisingly, phylogenetic analysis of MCATs between cyanobacteria, bacteria, plants and algae shows no close relationship between cyanobacterial and algal/plant enzymes(Figure S5, S6), suggesting an unusual evolutionary history (Ryall et al. 2003). Recently, Cantu et al. classified DH, ER and KRs based on primary and teriary structure (Cantu et al. 2012a), and deposited these enzymes and their families in the ThYme database (Cantu et al. 2011). Our phylogenetic analysis shows algal KRs placed between cyanobacteria and plants, having a similar close evolutionary relationship with both (Figure S7, S8). The acyl-ACP dehydratase (DH) used by most organisms is FabZ, which shows a similar evolutionary tree as KR (Figure S9, S10). FabA is another dehydratase found in E. coli that is absent in many other species (Figure S11). The phylogeny of enoyl-ACP reductase (ER, FabI) shows unusual characteristics, including its close homology between plants/algae and chlamydia, whereas the affinity between cyanobacteria and plants/algae is low (Figure S12, S13).
In silico modeling of ACP-KS and ACP-TE protein-protein interactions
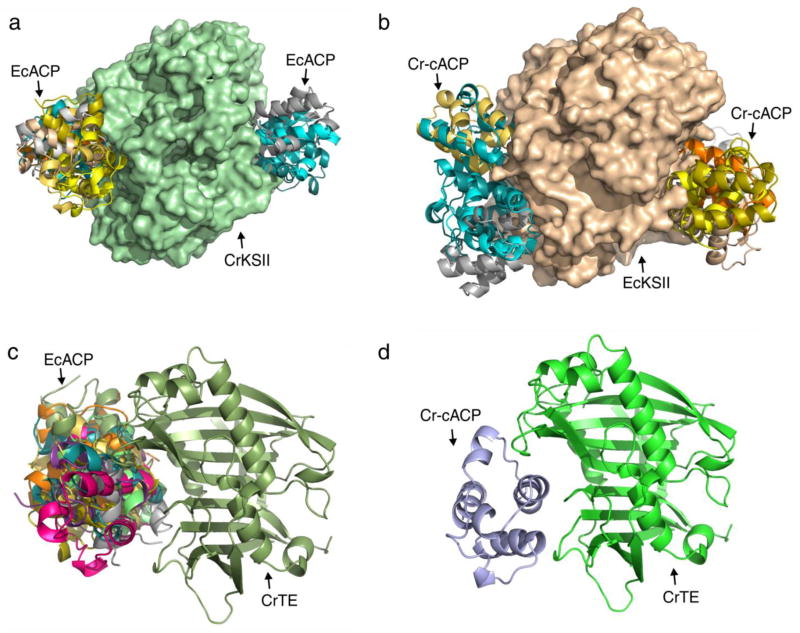
Having identified KSs and TEs in green microalgae and proposed their evolutionary origin, we next created homology models in order to screen in silico for protein-protein interactions with ACPs (Srikanta Dani et al. 2011). E. coli ACP (EcACP) was docked to KSII from E. coli, R. communis and C. reinhardtii, and all three dimeric enzymes show highly convergent and similar docking with EcACP (Figure S14 and 3a). Interestingly, an extracted EcACP structure from an EcACP-P450biol complex (Cryle 2010) shows even more energetically favorable docking (Figure S15). Docking Cr-cACP to EcKSII, RcACP to CrKSII (Figure S17) and Cr-cACP to RcKSII all show convergent docking (Figure S16 and 3b), suggesting functional protein-protein interactions between these ACPs and KSs across species. Docking EcACP to CrTE (Figure 3c) shows similar results as docking Cr-cACP to CrTE, albeit with results that are not as convergent (Figure S18 and 3d).
Figure 3.
In silico docking of ACP-KS and ACP-TE across species. a) EcACP-CrKSII, b) CrACP-EcKSII, c) EcACP-CrTE and d) CrACP-CrTE. Homology models were constructed using Swissmodel (Schwede et al. 2003) and protein-protein interactions docked using the Cluspro server (Comeau et al. 2004). Details (including RMSD values) can be found in Table S1 and additional docking figures in Figures S14–S18.
Mechanistic in vitro crosslinking assay between ACPs and FAS enzymes
To screen for in vitro compatibility between ACPs and KSII or TE, we utilized a mechanistic crosslinking assay developed in our laboratory (Figure 4a) (Worthington et al. 2006). Here, pantetheine probes bearing a unique covalent warhead are post-translationally attached to ACP, and a catalytic partner protein is added. Crosslinking occurs between the ACP pantetheine warhead (Figure 4b) and the partner’s active site only upon successful protein-protein interactions. Based on sequence alignment, phylogenetic analyses of KS domains, and ACP-KS virtual docking, it seems likely that ACPs and KS domains are interchangeable across species and will show crosslinking in all performed experiments. Indeed, when Cr-cACP and EcKSII were incubated with chloroacrylamide pantetheine probe 1 (Figure 4d), the cross-linked complex was observed in high yield (as can be seen from the intense newly formed band in the gel), similar to that for EcACP with EcKSII (Figure 4c). We next sought to examine whether the algal/plant TEs interact with E. coli ACP, or whether the observed in vivo activity in bacteria and cyanobacteria is a case of substrate recognition. EcACP loaded with α-bromo-acyl-pantetheine probe(s) (2 and 3) was incubated with CrTE and the bacterial ACP crosslinks with the algal TE, although not to the same yield (as observed by a fainter band in the gel) as for EcACP/EcKSII or Cr-cACP/CrTE (Figure 4e) (Blatti et al. 2012). In contrast to ACP-TE formation with C. reinhardtii’s proteins, where a higher yield was observed with a longer (C16) probe (3), both C6 and C16 probes resulted in the same yield of crosslinked product in the EcACP-CrTE experiment. The plant TEs did not crosslink with EcACP using either 2 or 3 (Figure 4f).
Figure 4.
In vitro activity-based crosslinking between ACP and FAS enzymes. a) Schematic of activity-based crosslinking assay to screen for functional interactions between ACP and its partners. b) Pantetheine probes for mechanistic crosslinking. c) EcACP crosslinking with EcKSII. d) Cr-cACP crosslinking with EcKSII. e) EcACP crosslinking with CrTE and f) EcACP crosslinking with plant TEs (left two lanes are UcTE and right two lanes ChTE). Each lane is labeled with the probes used. (*) marks contaminations, (-) marks no probe and (L) the benchmark ladder.
In vivo E. coli ACP complementation assay
Results of the in vitro crosslinking assay between Cr-cACP and EcKSII demonstrate a strong functional interaction between the two proteins. To test whether the algal ACP is able to complement bacterial fatty acid biosynthesis and functionally interact with all seven of the bacterial FAS enzymes in vivo, we used an ACP complementation assay developed by Cronan et al. (De Lay and Cronan 2007). As a positive control we used the fatty acid synthase ACP from gram-negative bacterium V. harveyi (VhACP). Whereas VhACP complements the absence of EcACP both on plates and in liquid culture (Figure S19), Cr-cACP did not complement E. coli fatty acid synthase in vivo.
Fatty acid composition of E. coli strains engineered with ACPs, KS and TEs
In vitro, Cr-cACP and EcKSII show a strong functional interaction (Figure 5), and although not complementing the absence of endogenous ACP in E. coli, overexpression of Cr-cACP in E. coli strain BL21 increases the levels of fatty acids, similar to the overexpression of EcACP (Figure 5c–d). In contrast, overexpression of Cr-mACP does not result in an alteration of the E. coli fatty acid profile, most likely due to the fact that this protein expresses as insoluble inclusion bodies (Blatti et al. 2012). In general, when TEs were induced, fatty acid content decreased significantly (Figure 5a versus 5b). E. coli BL21 (over)expressing UcTE shows an increase in C12:1, according to its natural activity, but the same behavior is not observed for ChTE. Uninduced, E. coli harboring CrTE shows increased levels of 14:0, 16:0 (2-fold), 16:1 (3-fold), 18:1 (Δ11), and the cyclopropane-containing fatty acid cyC17:0; whereas induced CrTE leads to decreased levels of C16:0, cyC17:0 and cyC19:0 and increased levels of C16:1 and C18:0. In general, all uninduced TEs lead to increased levels of cyC17:0.
Engineering of RcKSII into C. reinhardtii
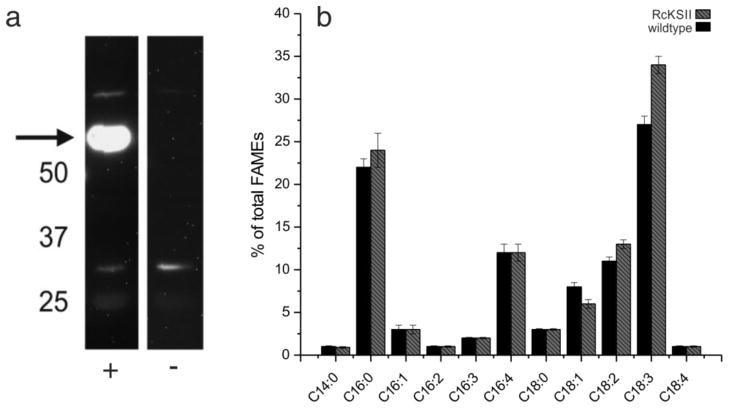
To determine whether plant KSs can interact with Cr-cACP in vivo, RcKSII was engineered into the C. reinhardtii chloroplast (Figure 6a). No major changes in fatty acid profile were observed; however a slight, but significant, increase in C18 fatty acids was observed (Figure 6b).
Figure 6.
Engineering RcKSII into the C. reinhardtii chloroplast. Engineering of RcKSII into the C. reinhardtii chloroplast was performed as previously described for the engineering of thioesterases (Blatti et al. 2012). a) Western blot analysis of transgenic C. reinhardtii strain expressing RcKSII. b) fatty acid distribution of wildtype (−) and RcKSII overexpressing (+) C. reinhardtii strain.
Discussion
In contrast to bacteria and plants, engineering fatty acid biosynthesis in algae has thus far led to unpredictable results. For example, we showed recently that although expressed and active, engineering plant TEs into the C. reinhardtii chloroplast does not lead to altered levels of fatty acids (Blatti et al. 2012). In contrast, when plant TEs were engineered into E. coli (Voelker and Davies 1994), S. elongatus (Liu et al. 2011; Kay et al. 2012) and various plants (Thelen and Ohlrogge 2002), significant changes in fatty acid quantity and profile occured. Although mechanistically highly conserved, there appear to be rules governing cross-species swapping of FAS enzymes, and demonstrated that protein-protein interactions play a major role in this selection. Here, we further hypothesized that the evolutionary origin of these enzymes could be used to predict successful interchangeability.
The acyl-ACP TEs, and their phylogeny, were recently studied by us (Blatti et al. 2012), Jing et al. (2011) and Cantu et al. (2010) and although algal TEs were first annotated as plant FatA TEs, now they form a separate clade, called Fat1. Until recently, it was common belief that plant and algae TEs originated from the engulfed cyanobacterium during endosymbiosis (Jones et al. 1995), but we could not identify a typical TE, from family TE14, in any cyanobacterial genome. These TEs seem to be prevalent in obligate anaerobic bacteria like Bacteroides and Lactobacilli. Fatty acid biosynthesis occurs in the chloroplast, which is of cyanobacterial origin, and thus engulfed cyanobacterial FAS could accommodate the eukaryotic TE. This could be an explanation for the observation of plant and algal TE interchangeable function in bacteria and cyanobacteria. TE phylogeny closely matches those assembled for enzymes involved in anaerobic energy metabolism, such as FeFe-hydrogenases (also found in algal chloroplasts) (Meuser et al. 2011), and cyclopropane fatty acid synthases (Yu et al. 2011). Whereas hydrogenase and cyclopropane synthase expression in ancient anaerobic bacteria is well understood, the reason why they express acyl-ACP TEs remains unknown. Interestingly, we also identified cyclopropane fatty acid synthases in algal genomes, but so far no algae have been shown to produce cyclopropane-containing fatty acids. Based on TE phylogeny and similarity to these other two enzymes, we postulate that TEs either evolved prior to the cyanobacterial engulfment in the non-photosynthetic eukaryotic host, maybe by horizontal gene transfer (HGT) between anaerobic bacteria and the early eukaryote, or by an early HGT event between primitive green algae and anaerobic bacteria. Since we cannot identify any family TE14 TEs in diatoms, red algae or brown algae, the latter might be the case.
Although there are no acyl-ACP TEs, from family TE14, in aerobic bacteria and cyanobacteria, the E. coli genome contains the thioesterase TesA, a promiscuous enzyme with several catalytic activities ranging from TE, phospholipase to acyl transferase (Lo et al. 2003). TesA is capable of hydrolyzing C8–C18 fatty acids from EcACP in vitro, but at very low levels in vivo (Cho and Cronan 1993). Docking EcACP to TesA gives very poor in silico interactions (data not shown), suggesting that TesA mainly recognizes the fatty acid substrate. Because plant (and likely algal) TEs also recognize the fatty acid chain attached to ACP (Salas and Ohlrogge 2002), it is possible that this is the mechanism by which plant TEs alter the fatty acid profile of bacteria and cyanobacteria. Mechanistic cross-linking experiments with a C6 and C16 pantetheine probe show that both Cr-cACP (Blatti et al. 2012) and EcACP functionally bind to CrTE, but that in both cases the longer chain results in a higher yield, suggesting that the chain length of the natural substrate of CrTE lies closer to C16 (Figure 4). The plant TEs do not crosslink with either Cr-cACP (Blatti et al. 2012) or EcACP (Figure 4f), suggesting that these TEs alter the E. coli fatty acid profile by substrate recognition and not protein-protein interaction.
In a recent study, thirty-one TEs from family TE14 were expressed in E. coli strain K27, which contains a mutation in fadD, one of the enzymes responsible for degradation of fatty acids (Jing et al. 2011). The authors conclude that phylogeny alone cannot predict TE specificity. Thioesterase activity relies on protein-protein interactions and substrate binding between endogenous EcACP and engineered TEs. In contrast, we extracted fatty acid profiles from literature and compared those to a phylogenetic tree of TEs (Table S2). These profiles group well along the clades of the phylogram. This demonstrates that indeed phylogeny can be used to predict substrate specificity of TEs.
Expression of plant and algal TEs in E. coli strain BL21 shows a decrease in fatty acids upon induction of the enzymes, whereas uninduced cultures alter the E. coli fatty acid profile due to leaky expression of the T7 promoter. Similar behavior has been observed for TesA, in which moderate expression enhances fatty acid production but overexpression reduces the total amount of fatty acids. All uninduced algal and plant TEs increased cyC17:0, perhaps through increased fatty acid biosynthesis or upregulation of a cyclopropane synthase. Interestingly, plant TEs do not crosslink with either Cr-cACP or EcACP, but when plant TEs are engineered into bacteria (Voelker and Davies 1994), cyanobacteria (Liu et al. 2011; Kay et al. 2012) or other plants (Thelen and Ohlrogge 2002), fatty acid profiles change towards the specificity of the TEs. Engineering plant TEs into algae only slightly alters the fatty acid profile, but algal TEs engineered into E. coli change the fatty acid content of bacteria significantly (Radakovits et al. 2011; Gong et al. 2011). Therefore, the ACP-TE interaction seems to be selective, as over time TEs evolved to have narrower substrate specificity. For example, the further evolved plant TEs do not seem to recognize bacterial or algal ACPs, but appear to act on the fatty acid chain attached to ACP.
In contrast to TEs, KSs are derivatives of prokaryotic FAS as can be seen from our and others (Jiang et al. 2008) phylogenetic analyses. Whereas TEs are only involved in the final step of fatty acid biosynthesis, KSs are essential in every iterative cycle. We therefore hypothesized that KSs must have a more flexible acyl-ACP specificity. For example, EcKSI can take over for EcKSII in vivo. More compelling, our lab has shown that the ACP-KS interaction is permissive across prokaryotic species, but unrelated carrier proteins from non-ribosomal peptide synthases do not interact with KSs from E. coli FAS (Worthington et al. 2008). In silico protein-protein docking between bacterial, algal and plant ACPs and KSs shows similar convergent docking behavior in all cases. In vitro, Cr-cACP functionally interacts with EcKSII and forms a crosslinked complex, similarly as EcACP with its cognate KS partner, suggesting highly tolerant protein-protein interactions between ACP and KS, already predicted by its phylogeny.
Although ACPs across species are conserved in structure, they show highly divergent sequences. Sequence, structure and function are intimately linked and influence productive catalysis. Cr-cACP is expressed as a soluble protein in E. coli, alters the E. coli fatty acid profile, and is post-translationally modified by the E. coli PPTase AcpS. However, Cr-cACP is unable to complement an EcACP knockout strain, suggesting that it cannot interact with all endogenous FAS enzymes, similar to spinach ACP (De Lay and Cronan 2007). The phylogeny of ACPs (Figure S2),(Cantu et al. 2012b) shows –surprisingly- distinct families, even within the previously identified ACP1 family (Cantu et al. 2012b), suggesting that the match between ACP and partner protein also depends on the evolutionary origin of the ACP itself.
An additional complexity is feedback regulation by acyl-ACPs (Andre et al. 2012). For example, spinach ACP loaded with palmitic acid does not inhibit E. coli acetyl CoA carboxylase (EcACCase) whereas any acyl-EcACP does (Davis and Cronan 2001), either due to the presence of a different regulatory mechanism or due to a missing allosteric interaction between spinach ACP and EcACCase.
Since the ACP-KS interaction seems to be permissive across species, we also tested this in the reverse evolutionary direction using a plant KSII engineered into C. reinhardtii. As with the bacterial ACP-KS interaction, Cr-cACP-CrKSII and Cr-cACP-RcKSII show almost identical in silico docking. Engineering RcKSII into C. reinhardtii’s chloroplast has minimal effects on the fatty acid profile, but a slight, but significant, increase in C18 fatty acids corresponds to KSII natural activity. Engineering another thiolase, Δ5-elongase from alga Pavlova sp. into the moss Physcomitrella patens gave high levels of adrenic acid corresponding to the enzyme’s activity in the alga (Kaewsuwan et al. 2010). Taken together, it can be concluded that the ACP-KS interaction is permissive across species, directly following from the phylogenetic relatedness and common ancestry of thiolases.
Plant and algal ACPs are not able to complement the absence of endogenous EcACP in E. coli, suggesting that the ACPs are not able to interact productively with one or more partner proteins in the catalytic cycle. Since KSs show promiscuity towards their ACPs, most likely one of the other enzymes is responsible for the inability to complement. Indeed, the other FAS enzymes, MCAT, DH, KR and ER, showcase a variety in evolutionary origin and relatedness, suggesting that not all of these enzymes will productively interact with any ACP.
For example, the E. coli genome contains another acyl-ACP dehydratase (DH) besides FabZ, FabA, which is responsible for the direct biosynthesis of unsaturated fatty acids in some bacteria. FabA is bifunctional and responsible for the synthesis of 10:1Δ2 from HO-10:0-β-hydroxydecanoyl-ACP. This trans-10:1-Δ2-ACP is either isomerized into cis-10:1-Δ3-ACP by FabA, or it is further processed to 16:0-ACP by KSI/KSII. The cis-10:1-Δ3-ACP is ultimately transformed by KSI to C16:1Δ9 (Feng and Cronan 2009), which is elongated by KSII to C18:1Δ11. Interestingly, this dehydratase is absent from cyanobacteria, obligate anaerobic bacteria, plants or algae. When EcFabA was engineered into tobacco, no significant changes in fatty acid profile were observed (Saito et al. 1995). Previously, it was shown that spinach ACP interacts productively with EcFabA in vitro, even to the same extent as EcACP, suggesting that FabA mainly recognizes the acyl-chain and not the ACP (Guerra and Browse 1990). Since Western blot analysis shows EcFabA in tobacco’s chloroplast, the purified dehydratase is active and the protein levels are relatively high, most likely in tobacco’s chloroplast no β-hydroxydecanoyl-ACP is available for EcFabA to act upon, adding an additional complexity to engineering FAS.
In this work, we began with the hypothesis that evolution can predetermine successful FAS engineering. Based on phylogenetic analyses, in silico structure based predictions, in vitro mechanistic crosslinking assays and in vivo engineering efforts, we are optimistic that phylogeny indeed can set rules for compatibility between FAS enzymes across species, to a certain extend. This finding paves the way for engineering of FAS enzymes for the production of designer oils, biofuel and other fatty acid and lipid commodities in various organisms.
Supplementary Material
Acknowledgments
Strain CY1877 was a generous gift of John E. Cronan (University of Illinois). The plasmid harboring VhACP was a generous gift of Peter Murphy and David Byers (Dalhousie University). This work was supported by California Energy Commission CILMSF 500-10-039; Department of Energy DE-EE0003373; National Institute of Health R01GM094924 and R01GM095970.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Haslam RP, Shanklin J. Feedback regulation of plastidic acetyl-CoA carboxylase by 18: 1-acyl carrier protein in Brassica napus. Proc Nat Acad Sci. 2012;109:10107–10112. doi: 10.1073/pnas.1204604109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A, Adrait A, Brugière S, Tardif M, Van Lis R, Deusch O, Dagan T, Kuhn L, Gontero B, Martin W. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol Biol Evol. 2009;26:1533–1548. doi: 10.1093/molbev/msp068. [DOI] [PubMed] [Google Scholar]
- Blatti JL, Beld J, Behnke CA, Mendez M, Mayfield SP, Burkart MD. Manipulating fatty acid biosynthesis in microalgae for biofuel through protein-protein interactions. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0042949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JW, Cronan JE. Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery. Annu Rev Microbiol. 2001;55:305–332. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- Cantu DC, Chen Y, Lemons ML, Reilly PJ. ThYme: a database for thioester-active enzymes. Nucleic Acids Res. 2011;39 (suppl 1):D342–D346. doi: 10.1093/nar/gkq1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu DC, Chen Y, Reilly PJ. Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu DC, Dai T, Beversdorf ZS, Reilly PJ. Structural classification and properties of ketoacyl reductases, hydroxyacyl dehydratases and enoyl reductases. Protein Eng Des Sel. 2012a;25:803–811. doi: 10.1093/protein/gzs050. [DOI] [PubMed] [Google Scholar]
- Cantu DC, Forrester MJ, Charov K, Reilly PJ. Acyl carrier protein structural classification and normal mode analysis. Protein Sci. 2012b;21:655–666. doi: 10.1002/pro.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kelly EE, Masluk RP, Nelson CL, Cantu DC, Reilly PJ. Structural classification and properties of ketoacyl synthases. Protein Sci. 2011;20:1659–1667. doi: 10.1002/pro.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Cronan JE. Escherichia coli thioesterase I, molecular cloning and sequencing of the structural gene and identification as a periplasmic enzyme. J Biol Chem. 1993;268:9238–9245. [PubMed] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- Crosby J, Crump MP. The structural role of the carrier protein--active controller or passive carrier. Nat Prod Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- Cryle MJ. Selectivity in a barren landscape: the P450BioIACP complex. Biochem Soc Trans. 2010;38:934. doi: 10.1042/BST0380934. [DOI] [PubMed] [Google Scholar]
- Davies HM, Anderson L, Fan C, Hawkins DJ. Developmental induction, purification, and further characterization of 12:0-ACP thioesterase from immature cotyledons of Umbellularia californica. Arch Biochem Biophys. 1991;290:37–45. doi: 10.1016/0003-9861(91)90588-a. [DOI] [PubMed] [Google Scholar]
- Davis MS, Cronan JE. Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J Bacteriol. 2001;183:1499–1503. doi: 10.1128/JB.183.4.1499-1503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay NR, Cronan JE. In vivo functional analyses of the type II acyl carrier proteins of fatty acid biosynthesis. J Biol Chem. 2007;282:20319–20328. doi: 10.1074/jbc.M703789200. [DOI] [PubMed] [Google Scholar]
- Dörmann P, Spener F, Ohlrogge JB. Characterization of two acyl-acyl carrier protein thioesterases from developing Cuphea seeds specific for medium-chain- and oleoyl-acyl carrier protein. Planta. 1993;189:425–432. doi: 10.1007/BF00194441. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J Biol Chem. 2009;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florova G, Kazanina G, Reynolds KA. Enzymes involved in fatty acid and polyketide biosynthesis in Streptomyces glaucescens: Role of FabH and FabD and their acyl carrier protein specificity. Biochemistry. 2002;41:10462–10471. doi: 10.1021/bi0258804. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Guo X, Wan X, Liang Z, Jiang M. Characterization of a novel thioesterase (PtTE) from Phaeodactylum tricornutum. J Basic Microbiol. 2011;51:666–672. doi: 10.1002/jobm.201000520. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Métoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Guerra DJ, Browse JA. Escherichia coli β-hydroxydecanoyl thioester dehydrase reacts with native C10 acyl-acyl-carrier proteins of plant and bacterial origin. Arch Biochem Biophys. 1990;280:336–345. doi: 10.1016/0003-9861(90)90339-z. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jiang C, Kim SY, Suh DY. Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol Phylogen Evol. 2008;49:691–701. doi: 10.1016/j.ympev.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Jing F, Cantu DC, Tvaruzkova J, Chipman JP, Nikolau BJ, Yandeau-Nelson MD, Reilly PJ. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 2011;12:44. doi: 10.1186/1471-2091-12-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A, Davies HM, Voelker TA. Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewsuwan S, Bunyapraphatsara N, Cove DJ, Quatrano RS, Chodok P. High level production of adrenic acid in Physcomitrella patens using the algae Pavlova sp. Δ5-elongase gene. Bioresour Technol. 2010;101:4081–4088. doi: 10.1016/j.biortech.2009.12.138. [DOI] [PubMed] [Google Scholar]
- Kan Y, Pan J. A one-shot solution to bacterial and fungal contamination in the green alga Chlamydomonas reinhardtii culture by using an antibiotic cocktail. J Phycol. 2010;46:1356–1358. [Google Scholar]
- Kay SA, Lis E, Golden S, Melnick M, Adin DM, Golden JW. Methods and compositions for the production of fatty acids in photosynthetic prokaryotic microorganisms. 2012/0184004. US Patent Application. 2012:A1.
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2006;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Kosa NM, Haushalter RW, Smith AR, Burkart MD. Reversible labeling of native and fusion-protein motifs. Nat Methods. 2012;9:981–984. doi: 10.1038/nmeth.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - The phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- Larget B, Simon DL. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol Biol Evol. 1999;16:750–759. [Google Scholar]
- Liu X, Sheng J, Curtiss R., III Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci USA. 2011;108:6899–6904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Lin SC, Shaw JF, Liaw YC. Crystal structure of Escherichia coli thioesterase I/protease I/lysophospholipase L1: Consensus sequence blocks constitute the catalytic center of SGNH-hydrolases through a conserved hydrogen bond network. J Mol Biol. 2003;330:539–551. doi: 10.1016/s0022-2836(03)00637-5. [DOI] [PubMed] [Google Scholar]
- McCarthy AD, Hardie DG. Fatty acid synthase - an example of protein evolution by gene fusion. Trends Biochem Sci. 1984;9:60–63. [Google Scholar]
- Meuser JE, Boyd ES, Ananyev G, Karns D, Radakovits R, Murthy UMN, Ghirardi ML, Dismukes GC, Peters JW, Posewitz MC. Evolutionary significance of an algal gene encoding an [FeFe]-hydrogenase with F-domain homology and hydrogenase activity in Chlorella variabilis NC64A. Planta. 2011;234:829–843. doi: 10.1007/s00425-011-1431-y. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3) doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pymol. The PyMOL Molecular Graphics System, Version 1504. Schrödinger, LLC; 2012. [Google Scholar]
- Radakovits R, Eduafo PM, Posewitz MC. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab Eng. 2011;13:89–95. doi: 10.1016/j.ymben.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Rambaut A. http://treebioedacuk/software/figtree/
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ryall K, Harper JT, Keeling PJ. Plastid-derived Type II fatty acid biosynthetic enzymes in chromists. Gene. 2003;313:139–148. doi: 10.1016/s0378-1119(03)00671-1. [DOI] [PubMed] [Google Scholar]
- Saito K, Hamajima A, Ohkuma M, Murakoshi I, Ohmori S, Kawaguchi A, Teeri TH, Cronan JE., Jnr Expression of the Escherichia coli fabA gene encoding beta-hydroxydecanoyl thioester dehydrase and transport to chloroplasts in transgenic tobacco. Transgenic Res. 1995;4:60–69. doi: 10.1007/BF01976503. [DOI] [PubMed] [Google Scholar]
- Salas JJ, Ohlrogge JB. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch Biochem Biophys. 2002;403:25–34. doi: 10.1016/S0003-9861(02)00017-6. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanta Dani KG, Hatti KS, Ravikumar P, Kush A. Structural and functional analyses of a saturated acyl ACP thioesterase, type B from immature seed tissue of Jatropha curcas. Plant Biol. 2011;13:453–461. doi: 10.1111/j.1438-8677.2010.00410.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G. PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol. 2012;29:3625–3639. doi: 10.1093/molbev/mss178. [DOI] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB. Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng. 2002;4:12–21. doi: 10.1006/mben.2001.0204. [DOI] [PubMed] [Google Scholar]
- Voelker TA, Davies HM. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J Bacteriol. 1994;176:7320–7327. doi: 10.1128/jb.176.23.7320-7327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann G, Murphy PW, Rowland EE, Cronan JE, Jr, Liu XQ, Blouin C, Byers DM. Intein-mediated cyclization of bacterial acyl carrier protein stabilizes its folded conformation but does not abolish function. J Biol Chem. 2010;285:8605–8614. doi: 10.1074/jbc.M109.060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington AS, Hur GH, Meier JL, Cheng Q, Moore BS, Burkart MD. Probing the compatibility of type II ketosynthase-carrier protein partners. ChemBioChem. 2008;9:2096–2103. doi: 10.1002/cbic.200800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. Mechanism-based protein cross-linking probes to investigate carrier protein-mediated biosynthesis. ACS Chem Biol. 2006;1:687–691. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- Yu XH, Rawat R, Shanklin J. Characterization and analysis of the cotton cyclopropane fatty acid synthase family and their contribution to cyclopropane fatty acid synthesis. BMC Plant Biol. 2011;11:97. doi: 10.1186/1471-2229-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Voelker TA, Hawkins DJ. Modification of the substrate specificity of an acyl-acyl carrier protein thioesterase by protein engineering. Proc Natl Acad Sci USA. 1995;92:10639–10643. doi: 10.1073/pnas.92.23.10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zornetzer GA, Tanem J, Fox BG, Markley JL. The length of the bound fatty acid influences the dynamics of the acyl carrier protein and the stability of the thioester bond. Biochemistry. 2010;49 (3):470–477. doi: 10.1021/bi9014659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.