Abstract
Buruli ulcer (BU) is a neglected tropical disease caused by Mycobacterium ulcerans. The tissue damage characteristic of BU lesions is known to be driven by the secretion of the potent lipidic exotoxin mycolactone. However, the molecular action of mycolactone on host cell biology mediating cytopathogenesis is not fully understood. Here we applied two-dimensional electrophoresis (2-DE) to identify the mechanisms of mycolactone's cellular action in the L929 mouse fibroblast proteome. This revealed 20 changed spots corresponding to 18 proteins which were clustered mainly into cytoskeleton-related proteins (Dync1i2, Cfl1, Crmp2, Actg1, Stmn1) and collagen biosynthesis enzymes (Plod1, Plod3, P4ha1). In line with cytoskeleton conformational disarrangements that are observed by immunofluorescence, we found several regulators and constituents of both actin- and tubulin-cytoskeleton affected upon exposure to the toxin, providing a novel molecular basis for the effect of mycolactone. Consistent with these cytoskeleton-related alterations, accumulation of autophagosomes as well as an increased protein ubiquitination were observed in mycolactone-treated cells. In vivo analyses in a BU mouse model revealed mycolactone-dependent structural changes in collagen upon infection with M. ulcerans, associated with the reduction of dermal collagen content, which is in line with our proteomic finding of mycolactone-induced down-regulation of several collagen biosynthesis enzymes. Our results unveil the mechanisms of mycolactone-induced molecular cytopathogenesis on exposed host cells, with the toxin compromising cell structure and homeostasis by inducing cytoskeleton alterations, as well as disrupting tissue structure, by impairing the extracellular matrix biosynthesis.
Author Summary
Buruli Ulcer (BU) is a neglected tropical disease caused by Mycobacterium ulcerans infection. It has been recognized for many years that BU pathogenesis is mediated by the potent exotoxin mycolactone; however, the molecular action of this toxin on the host cell biology that drives its pathogenesis is not fully understood. Here we present a proteomic-based study that explores the molecular action of mycolactone on host cells biology. Our results provide further molecular evidence for the cytoskeleton-disarrangement induced by mycolactone, and unveil its impact on cytoskeleton-dependent cellular functions. Moreover, we extend the field of action of this toxin to the biosynthesis of collagen, implicating mycolactone on the decrease of dermal collagen found on BU lesions. Given the dependence of M. ulcerans virulence on its toxin, these findings on mycolactone's molecular action on host cells and tissues are of major importance for the understanding of BU pathogenesis.
Introduction
Buruli ulcer (BU) is a neglected tropical disease caused by Mycobacterium ulcerans infection [1]. Infection usually starts in the subcutaneous tissue and initially gives rise to non-ulcerative lesions. Histologically, increasing areas of necrosis contrast with the smaller central zone, in which acid-fast bacilli concentrate [2] during both an intracellular phase as well as extracellularly [3], [4]. With disease progression, necrosis advances, radiating from the focus of infection and involving all cells and structures in its path [5]. If left untreated, necrosis extends to the corium and the lesion breaks down into a severe ulcer. In the ulcerative stage of the disease bacteria disseminate and become predominantly extracellular [3], [4], being found throughout the necrotic tissue [5]. The treatment of BU consists primarily in a lingering antibiotic protocol with a combination of rifampicin and streptomycin [6], however surgical resection of infected skin is still necessary for advanced stages [7]. Moreover, the frequent delay in treatment seeking hampers disease management and increases morbidity [8], with serious long-term sequelae [9]. Prevention is also difficult as little is known about disease transmission [10], [11], [12], [13], [14] and no vaccine is currently available [15], [16].
M. ulcerans pathogenicity and the tissue damage characteristic of BU are mediated by its toxin mycolactone, a potent cytotoxic and immunosuppressive polyketide-derived macrolide [2], [17], [18], [19], [20], [21], [22]. Mycolactone is produced as a mixture of congeners, with one major form, which is conserved within a given geographical area [23]. Mycolactone A/B is the main variant produced by African isolates; Australian isolates produce mycolactone C [23] and the Chinese isolate MU98912 used in this study produces mycolactone D [24]. Regarding mycolactone's action, in vitro studies mainly performed in the mouse fibroblast L929 cell line have shown that the toxin diffuses passively through the plasma membrane [25]. Further studies also show that cells incubated with the toxin display a distinctive cytopathicity, characterized by early actin cytoskeleton rearrangement, cell round-up and detachment from the bottom of the well, and an arrest in the G0/G1-phase [17], [26], culminating in an apoptotic cell death [19]. Recently, Guenin-Macé et al. unveiled that the toxin targets the actin-cytoskeleton regulator Wiskott-Aldrich syndrome protein (WASP), inducing its hyperactivation [27], and Hall et al. described that mycolactone inhibits co-translational translocation of proteins into the endoplasmic reticulum (ER), thus inhibiting the production of nearly all proteins that transit through the ER [28]. However, despite these advances, the molecular action of this toxin on the host cell biology that drives its pathogenesis is not fully understood.
This work had the purpose of conducting a characterization of the proteome of mycolactone-treated cells, in order to better understand the effects of this toxin on host cell biology. At first, we performed a kinetic characterization of mycolactone's cytopathic, cytostatic and cytotoxic effects on L929 cells. Based on this, specific incubation times and toxin doses were chosen for the proteomic study by two-dimensional electrophoresis (2-DE). Functional studies were performed in both in vitro and in vivo models to verify our findings in mycolactone-exposed cells and investigate their role in BU pathogenesis. The data obtained showed that cytoskeleton and collagen biosynthesis are severely affected by mycolactone, supporting the involvement of cytoskeleton on mycolactone-induced cytopathogenicity and identifying a new activity of the toxin on the decrease of the collagen content in M. ulcerans-infected tissues.
Results
Kinetic characterization of mycolactone cytopathic, cytostatic and cytotoxic effects
The time- and dose-dependent kinetics of the cytostatic and cytotoxic activities of mycolactone were investigated by an integrated analysis of cell cycle and cell death in L929 cells. Doses of mycolactone were selected through a pre-screening MTS assay based on the concentrations reported for human ulcer exudates (0–300 ng/mL) [29]. MTS assay showed a threshold around 15 ng/mL, above which mycolactone is progressively cytotoxic reaching a plateau at 50 ng/mL (data not shown). Therefore, the range of mycolactone concentrations tested in this study was narrowed to 12.5–50 ng/mL.
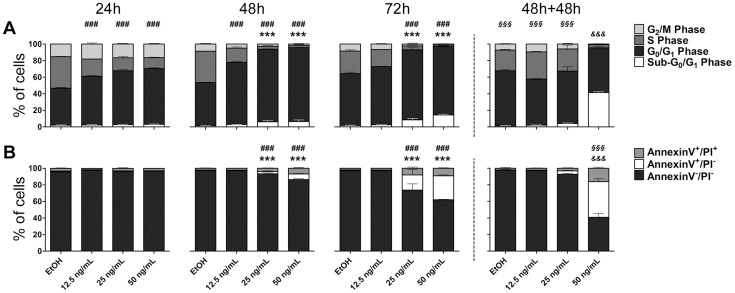
Results presented in figure 1 show that the ethanol (vehicle) equivalent (<0.002%), as well as the mycolactone concentration below the threshold (12.5 ng/mL), had no detectable cytopathic (rounding and detachment) or cytotoxic effects. Cytotoxic doses of mycolactone (>12.5 ng/mL) induced detachment, cell cycle arrest in G0/G1 phase at 48 h and 72 h of treatment and the appearance of a sub-G0/G1 population, compatible to apoptotic cells, more evident at 72 h (figure 1A). Consistent with cell cycle data, annexin-V/PI assays revealed an annexin-V+/PI− population for the highest mycolactone concentrations (25 and 50 ng/ml) at 72 h (figures 1B), indicative of apoptotic cells.
Figure 1. Kinetics of mycolactone cytostatic and cytotoxic effects.
Mouse fibroblasts L929 cells were incubated for 24, 48 or 72(referred to as 48 h+48 h). Cell cycle analysis (A) and annexin-V/PI assay (B) were performed for each-time point. Bars represent the mean + SD (n = 3) from one out of, at least, two independent experiments. Each condition was compared to EtOH-treated samples throughout each time-point (24 h, 48 h and 72 h) by Two-way ANOVA with Bonferroni posttest; statistical differences were represented by *** (P<0.001) for sub-G0/G1 and Annexin-V+/PI− , and by ### (P<0.001) for G0/G1 Phase and Annexin-V+/PI+. Each condition at 48 h+48 h time-point was compared with the same condition at the 48 h by Two-way ANOVA with Bonferroni posttest; statistical differences were represented by &&& (P<0.001) for sub-G0/G1 and Annexin-V+/PI−, and by §§§ (P<0.001) for G0/G1 Phase and Annexin-V+/PI+.
It was previously reported that cells incubated with mycolactone re-grow when mycolactone is removed from the medium, indicating that mycolactone's effect might be reversible [17]. To further investigate the reversibility of mycolactone's effect, cells were incubated with different concentrations of mycolactone for 48 h and afterwards washed and incubated in fresh media for an extra 48 h period (figure 1, 48 h+48 h). We found that cells that had previously been incubated with the lowest cytotoxic concentration of the toxin (25 ng/mL) re-adhere, recover the normal cell cycle progression (figure 1A, 48 h+48 h) and appeared to overcome the cytotoxic stimulus, since no increase in the sub-G0/G1 or annexin-V+/PI− populations was observed (figures 1A and 1B, 48 h+48 h). On the other hand, cells that had been previously incubated with the highest cytotoxic dose (50 ng/mL), while remaining in suspension, were not able to overcome the initial stress induced by mycolactone and became committed to death (figures 1A and 1B, 48 h+48 h). These data demonstrate that the reversibility of mycolactone's effect occurs within a window of concentrations around 25 ng/mL.
Overall, within the range of studied mycolactone concentrations, we found doses that did not induce observable cytotoxic effects, doses that induced a reversible stress, and doses that irreversibly triggered an apoptotic cell death.
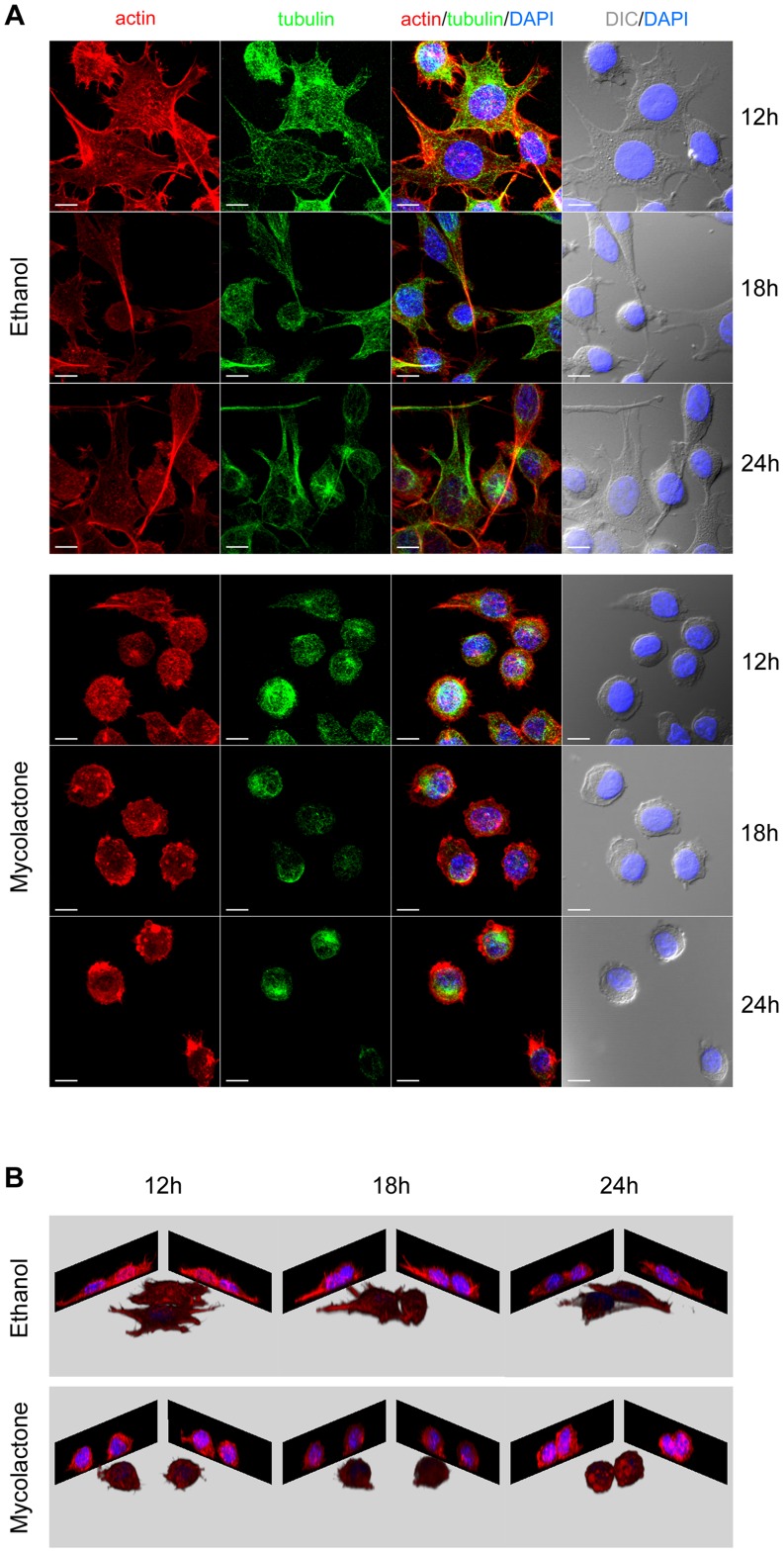
To further characterize our model, the kinetics of the cytopathic effects, namely cytoskeleton alterations and cell round-up and detachment, were also assessed. In cells incubated with mycolactone, we observed not only the previously described alterations for actin [26], but also changes in the tubulin cytoskeleton, which appeared bended into a microtubule hank (figure 2A). Within 12–18 h of exposure to the toxin, actin ultrastructures (stress fibers and lamellipodia) were lost, and, although still attached, most of the cells were completely round-up by 18–24 h (figures 2A and 2B). At 24 h, half of the cells were already in suspension, while the remaining cells eventually detached in the following 12 h [26]: detachment being probably a consequence of cell round-up and loss of adhesion structures.
Figure 2. Mycolactone induces cytoskeletal alteration, cell round up and detachment.
Mouse fibroblasts L929 cells seeded on coverslips were incubated for 12, 18 or 24/mL of mycolactone. Cytoskeletal changes were visualized by immunofluorescence microscopy using rhodamine-phalloidin conjugate (red) and a tubulin-specific antibody (green). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The cellular shape was visualized by differential interference contrast (DIC) microscopy. White horizontal bar represents a 10 µm scale (A). Blue (nucleus) and red (actin) channels were used for 3D remodeling of the respective confocal z stacks (B).
Mycolactone causes time-dependent proteomic changes related to cytoskeleton and collagen biosynthesis
To study the effects of mycolactone on host cell biology, the total cellular proteome from mouse fibroblast L929 cells incubated with 50 ng/mL of mycolactone (dose that triggers a commitment to apoptotic cell death) or the ethanol equivalent (control), was separated by 2-DE. To establish a temporal perspective of mycolactone's action, three incubation times were chosen: 24 h (when cells are detaching, but viable and mounting a response to the mycolactone-induced stress, associating with the first detectable consequences of mycolactone on the fibroblast proteome); 48 h (when cells become committed to death, coinciding with the onset of an apoptotic population); and 48 h+48 h, (when most of the cells are in the process of apoptotic cell death, with only 40% of viability) (figure 1).
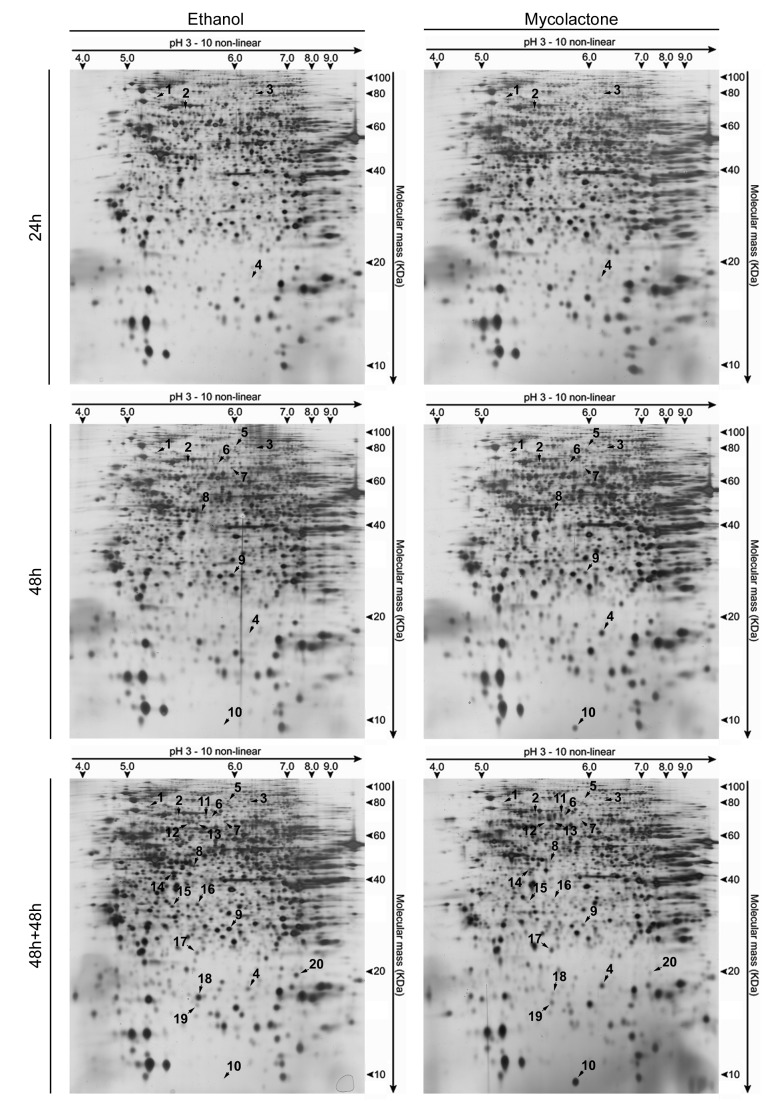
The comparison of control cells proteome at different time-points did not reveal any significantly changed spots, showing that ethanol (vehicle) represents a suitable control. In contrast, the comparison between the proteome of control and mycolactone-treated cells at each time-point revealed a time-dependent increase in the number of changed spots, with 4 spots changed at 24 h, 10 at 48 h and 20 at 48 h+48 h. All 20 spots were identified by mass spectrometry and found to correspond to 18 proteins comprising 5 up- and 13 down-regulated (figure 3 and figure 4).
Figure 3. Mycolactone causes time-dependent proteomic changes.
Mouse fibroblasts L929 cells were incubated for 24 or 48(50 ng/mL). Additionally, an assay was performed where cells were incubated for 48 hours in the same conditions followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h). Representative silver-stained 2-D gels of total protein extracts (100 µg) are shown for the different exposure times (all analyzed gels are in Supplementary Information). Spots altered upon treatment are represented by numbers (1–20).
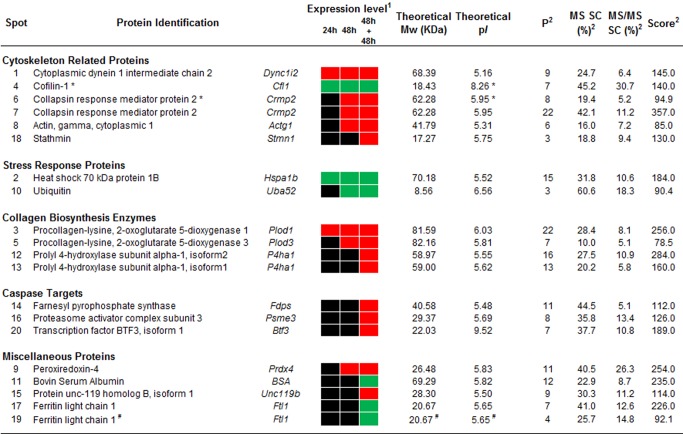
Figure 4. Altered proteins upon mycolactone treatment.
Figure shows the proteins identified for each spot (numbers according to figure 3 and figure S1) clustered into five categories. (1) Expression levels show decreased (red), increased (green) or no changes (black) in spot intensities upon mycolactone treatment. (2) MS and MS/MS parameters are presented, including the number of specific matched peptides (P), the sequence coverage (SC) in percentage, as well as the MS scores. Phosphoproteins (*) and putative protein fragments (#) are indicated. Full MS and MS/MS data in Supplementary Information.
To reveal the cellular processes altered by mycolactone, the proteins were clustered into functional groups according to UniProt database (www.uniprot.org). The major groups comprised (i) cytoskeleton-related proteins (Dync1i2, Cfl1, Crmp2, Actg1, Stmn1); (ii) stress response proteins (Hspa1b, Uba52); and (iii) collagen biosynthesis enzymes (Plod1, Plod3, P4ha1) (figure 4 and figure S1). This reveals that mycolactone results in an alteration of cytoskeleton-related proteins and down-regulation collagen biosynthesis enzymes. On the other hand, stress response proteins were up-regulated. An additional group (caspase targets) was created clustering proteins identified as caspase substrates (Fdps, Psme3, Btf3) through the web caspase substrates database CASBAH (www.casbah.ie) [30]. Consistent with being caspase substrates, these proteins were only decreased in the last time point (48 h+48 h) when most of the cells were undergoing an apoptotic death process (figure 1). Four additional proteins were classified as miscellaneous proteins (Prdx4, BSA, Unc119b, Ftl1) (figure 4 and figure S1).
Overall, the proteome of mycolactone-treated cells revealed that the intracellular structure (cytoskeleton) and the extracellular matrix (collagen) are severely affected by the M. ulcerans toxin.
Intracellular structure: Mycolactone impairs cytoskeleton dynamics and functions
The proteomic study revealed several regulators and structural components of both microfilaments and microtubules affected by mycolactone treatment after 24 h (Dync1i2, Cfl1), 48 h (Crmp2, Actg1) and 48 h+48 h (Stmn1) (figure 4 and figure S1).
Cytoplasmic dynein 1 intermediate chain 2 (Dync1i2) is a non-catalytic subunit of the microtubule-associated molecular motor dynein, which is involved in the transport of elements of the Golgi apparatus, endosomes and lysosomes [31]. The here detected early (24 h) down-regulation of Dync1i2 (spot 1) suggests that this transport may be compromised in mycolactone-treated cells.
Three other proteins altered in cells treated with the toxin are cytoskeleton regulators (Cfl1, Crmp2, Stmn1). Cofilin 1 (Cfl1, spot 4), a well-established regulator of actin dynamics, promotes microfilament assembly or disassembly depending upon the concentration of Cfl1 relative to actin and other actin-binding proteins, as well as upon its phosphorylation status [32]. Collapsin response mediator protein 2 (Crmp2), which was identified in two spots (spot 6 and 7), is a multifunctional adaptor protein which can induce microtubule assembly by binding to αβ-tubulin heterodimers [33], whereas stathmin (Stmn1, spot 18) has been described as a microtubule-destabilizing oncoprotein [34]. Interestingly, the isoelectric points (pIs) of spots 4 (Cfl1) and 6 (Crmp2) differed in the gel from their expected theoretical values (figure 3 and figure 4), suggesting posttranslational modification such as phosphorylation. Given that the regulatory activity of Cfl1 [32] and Crmp2 [33] can be modulated by phosphorylation, we studied the phospho-status of these proteins with the Pro Q Diamond phosphostaining. Indeed, the analysis of the phosphoprotein stained gel revealed that both spots were phosphorylated (figure S2). Thus, mycolactone increases the phospho-Cfl1 at 24 h, decreases both phosphorylated and non-phosphorylated forms of Crmp2 at 48 h, and decreases Stmn1 in the latest time point (48 h+48 h).
In addition to the alteration of cytoskeleton regulators, the proteomic study revealed that the cytoskeleton is also altered on its structural components. Actin gamma (Actg1), a component of microfilaments, is down-regulated at 48 h in mycolactone-treated cells.
Overall, these results show that cells exposed to mycolactone undergo a process of cytoskeleton remodeling involving regulators and structural components, providing a novel molecular basis for the effect of mycolactone on this organelle.
Mycolactone impairs autophagy, a cytoskeleton-dependent cellular function
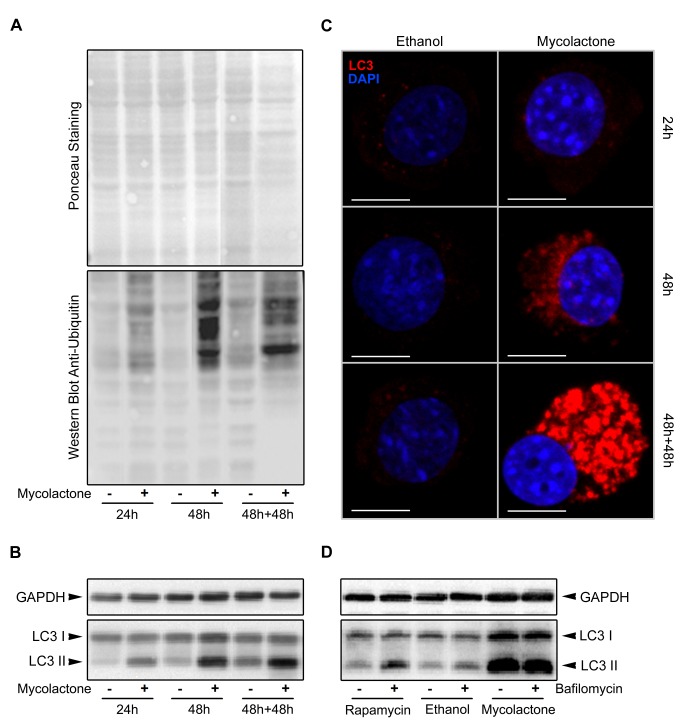
Two stress response proteins (Hspa1b, Uba52) were up-regulated upon treatment with mycolactone. Spot 10 (figure 4 and figure S1) was identified as a fusion protein (Uba52) consisting of N-terminal ubiquitin and C-terminal 60S ribosomal protein L40. The detected spot position in the gel (figure 3) in comparison with the theoretical positions for the fusion protein (pI 9.87/14.7 kDa), ubiquitin (pI 6.56/8.6 kDa) and the ribosomal protein (pI 10.32/6.2 kDa) suggest the presence of ubiquitin. Indeed, all three spot-specific peptides covered amino acids 13–55 revealing that ubiquitin is present. The here detected increase of free ubiquitin after 48 h of mycolactone treatment could result from an inhibition of ubiquitin ligases or from an up-regulation of its expression. To investigate this in more detail, protein ubiquitination was studied by western blot, which revealed that mycolactone exposure results in an increase of ubiquitinated proteins, more evident at 48 h and 48 h+48 h (figure 5A). Therefore, these data show that rather than an inhibition of ubiquitin ligases, mycolactone induces an up-regulation of the ubiquitin/proteasome system (UPS).
Figure 5. Mycolactone causes an up-regulation of the ubiquitin-proteasome pathway and an accumulation of autophagosomes.
In A, B and C, mouse fibroblasts L929 cells were incubated for 24 or 48(−) or mycolactone (50 ng/mL, +). Additionally, an assay was performed where cells were incubated for 48 hours in the same conditions followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h). At each time-point, total protein was extracted and Western blot was performed to assess ubiquitinated proteins (A) or LC3 processing (B). Additionally, cytospins were made to assess immunofluorescence LC3 (red). White horizontal bars represent a 10 µm scale (C). In D, mouse fibroblasts L929 cells were incubated for 48 hours either with ethanol or mycolactone (50 ng/mL) and bafilomycin A1 (10 nM, +) or DMSO (vehicle control, −) was added 2 hours prior the end of the assay. Additionally, mouse fibroblast L929 cells were incubated for 48 hours without any stimuli until 2 hours prior the end of the assay, when autophagy was induced with rapamycin (1 µM), and bafilomycin A1 (10 nM, +) or DMSO (vehicle control, −) was added. At the end of the assay, total protein was extracted and Western blot was performed to assess LC3 processing (D).
The UPS and the lysosomal degradation system (autophagy) are the two main cellular degradative pathways. These systems crosstalk each other and the up-regulation of one may occur in response to a down-regulation/dysfunction of the other. Autophagy is known to be dependent on microtubule cytoskeleton [35] and dynein-driven transport [36] with dynein playing a role in the delivery of autophagosome contents to lysosomes during autophagosome-lysosome fusion [36]. Since microtubules (Figure 2) and dynein (figure 4 and figure S1) were found to be affected by mycolactone, we hypothesized that the mycolactone-induced cytoskeleton-related changes might impair the autophagic process leading to the up-regulation of the UPS. Therefore the role of mycolactone in autophagy was further investigated.
During autophagy the cytosolic form of LC3 (LC3-I) is conjugated to phosphatidylethanolamine (PE) to form LC3-PE (LC3-II), which is recruited to autophagosomal membranes. As the autophagosomes fuse with lysosomes to form autolysosomes, LC3-II is degraded together with the intra-autophagosomal components by lysosomal hydrolases. Thus, lysosomal turnover of the autophagosomal marker LC3-II reflects autophagic activity [37]. Processing of this marker was analyzed by western blot and immunofluorescence. Western blot revealed an increase of the autophagosome marker LC3-II in cells treated with mycolactone (figure 5B) compatible with an inhibition of autolysomes formation. In agreement, an increase of LC3-positive cytoplasmic vesicles upon toxin exposure was also detected with the immunofluorescence assay (figure 5C, red-stained). To further understand these results, we treated L929 cells with different stimuli and added bafilomycin A1 to inhibit the autolysosomal degradation step [38] 2 hours before protein extraction (figure 5D). The increase of LC3-II induced by mycolactone (figure 5B and 5C), together with the lack of difference in LC3-II in cells treated with mycolactone in the presence or absence of bafilomycin A1 (figure 5C), suggests a block of autophagy at the terminal stages [39]. Furthermore, the higher LC3-I levels observed in cells exposed to mycolactone, when compared with cells where autophagy was induced by rapamycin (figure 5C), suggests that most probably autophagy is being induced due to cell detachment [40] or as a feedback response to the blockage of the autophagic terminal stage. These data indicate that mycolactone inhibits autophagosome-lysosome fusion and in turn impairs autophagy.
Taken together, this reveals that mycolactone mediates up-regulation of the UPS and inhibition of autophagy. Since autophagy counteracts several stresses, including infection by intracellular pathogens [41], [42], [43], mycolactone-induced impairment of autophagy may have implications for the progress of M. ulcerans infection.
Extracellular matrix: Mycolactone impairs collagen biosynthesis
Proteomics identified several enzymes of collagen biosynthesis progressively down-regulated in mycolactone-treated cells: Plod1 (24 h), Plod3 (48 h) and two isoforms of P4ha1 (48 h+48 h). Further studies showed that these proteins were transcriptionally down-regulated after 24 h of mycolactone exposure (figure S3), thus the differential down-regulation of the different proteins probably reflects different protein stability. These enzymes catalyze the hydroxylation of lysine (Plod1 and Plod3) and proline (P4ha1) residues, which is essential for the formation and stabilization of collagen fibers [44], [45]. The here detected down-regulation of these enzymes suggests that collagen fibers stability may be compromised in mycolactone exposed cells. Interestingly, histopathological studies from the 1960's reported a collagen decrease in human BU lesions [5], [46], [47]; however this feature has been overlooked and it was never subject to studies to determine its cause or its relevance for BU.
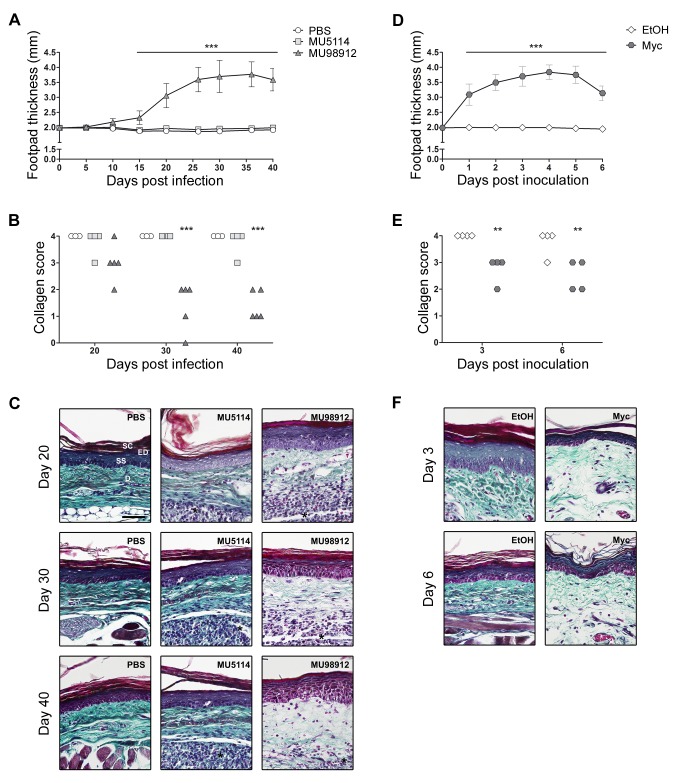
To investigate if mycolactone was responsible for a decrease in tissue collagen, an experimental model of BU disease, the mouse footpad infection with M. ulcerans, was used. Mice were challenged with virulent mycolactone-secreting (MU98912), avirulent mycolactone-negative (MU5114) strains of M. ulcerans or PBS as a control. Pathology progression was assessed by measuring footpad swelling (figure 6A) and, at different time-points, footpads were collected for histological processing and collagen scoring (figure 6B). The results showed that, in footpads infected with mycolactone-secreting M. ulcerans (MU98912), the progressive increase of pathology (figure 6A) was associated with a decrease of the collagen score (figure 6B), preceding the breakdown of the lesion into an ulcer (by day 40 post-infection). In contrast, infection with the mycolactone-negative strain (MU5114) did not induce pathology nor did it alter the collagen content in infected footpads, similar to what was observed for the PBS-injected control group (figure 6A and 6B). These results suggest that the decrease in collagen content is not a consequence of the infection or the elicited immune response, but rather caused by mycolactone. To verify this, mice were challenged with purified toxin or ethanol equivalent as control. The results showed that mycolactone induced footpad swelling associated with a decrease of the collagen score, while the vehicle did not (figure 6D and 6E). Histological samples stained with Masson's trichrome showed a decay of collagen fibers in MU98912-infected or mycolactone-treated footpads, characterized by the disorganization and thinness of collagen fibers (figure 6C and 6F).
Figure 6. Mycolactone induces a decrease in dermal collagen fibers.
Mice were infected with virulent mycolactone-secreting (MU98912: dark gray triangles) (n = 5) or avirulent mycolactone-negative (MU5114: light gray squares) (n = 4) strains of M. ulcerans, or injected with PBS (vehicle: white circles) (n = 3) as a control (A, B and C). Alternatively, mice were inoculated with 5 µg of mycolactone (dark gray hexagons) (n = 4) or ethanol (vehicle: white diamonds) (n = 4) as a control (D, E, F). Lesion progression was assessed by measurement of footpad swelling (A and D). Collagen content was assessed by a qualitative blind scoring of the amount of dermal collagen fibers (score 0 = lowest, 4 = highest) visualized in HE-stained sections by polarized light (B and E). The different groups were compared to the control groups (PBS in A and B; ethanol in C and D) by Two-way ANOVA with Bonferroni posttest; statistical differences were represented by ** (P<0.01) or *** (P<0.001). Additionally, mouse footpads exposed to the different stimuli were stained with Masson's trichrome to highlight collagen fibers (green) (C and F). On panel are depicted representative histological image each group. Black horizontal bars represent a 50 µm scale. Histological structures like dermis (D) and the stratum corneum (SC) and stratum spinosum (SS) of the epidermis (ED) are indicated on the upper-left image. Inflammatory infiltrate is indicated by an asterisk.
These results in the mouse model of infection show that the earlier described degeneration of collagen in BU lesions [5], [46], [47], [48] is a consequence of the secretion of mycolactone by the infecting strain.
Discussion
Even though, the methodology used has some limitations, since it excludes the analysis of transmembrane and secretory proteins, which were found to be severely affected by mycolactone [28], this work is the first proteomic study on the effect of mycolactone on cells, unveiling important information about the toxin action.
It has been known for years that the actin-cytoskeleton of mycolactone-treated cells suffers early structural rearrangements [26]. Recently, it was also shown that these changes were mediated by the mycolactone-induced hyperactivation of the actin-cytoskeleton regulator WASP [27]. In this study we show that mycolactone causes structural changes in microtubules and we identify several regulators and structural components of both microfilaments and microtubules affected by the M. ulcerans toxin. These data confirm the cytoskeleton as a major target of mycolactone and further specifies the mechanisms of the toxin's cellular action. Moreover, given the cytoskeleton's dynamic nature, with constant remodeling, it remained unclear how these changes contribute to the tissue damage characteristic of BU lesions. Since the proteomic data pointed likewise to an involvement of the UPS further experiments were performed confirming its mycolactone-dependent up-regulation. UPS and autophagy constitute the main intracellular processes of protein degradation taking part in the cellular protein quality control system. Thus, UPS and autophagy are critical in the maintenance of cellular homeostasis and their activities are strictly orchestrated. Moreover, perturbations in the flux through either pathway have been reported to affect the activity of the other system, and a number of mechanisms have been proposed to rationalize the link between the UPS and autophagy [49]. Therefore, it was investigated if the detected mycolactone-dependent changes affect autophagy. The here obtained data indicate that autophagosome-lysosome fusion is impaired in mycolactone-treated cells. Given that the delivery of autophagosome contents to lysosomes is dependent on microtubule cytoskeleton [35] and on dynein-driven transport [36], the mycolactone-induced impairment of autophagy appears to occur secondarily to mycolactone-induced cytoskeleton alterations. Further evidence of a dysfunctional vesicle-lysosome fusion is given by another altered protein in mycolactone-treated cells. Proteomics revealed that the cell culture medium constituent BSA (clustered on miscellaneous proteins) was increased in cells treated with mycolactone. As degradation of extracellular proteins occurs in lysosomes [50], the observed BSA accumulation suggests likewise that the endosomes-lysosome fusion may be compromised. Since the delivery of autophagosome to lysosomes [36] and retrograde transport of endosomes [31] is mediated by dynein the here observed down-regulation of one of its components (Dync1i2), suggest an impairment of dynein-driven transport upon mycolactone exposure. Further work is needed to explore the effect of mycolactone on cytoskeletal motors-mediated transport, however the down-regulation of a molecular motor subunit (Dync1i2) together with the microfilaments' and microtubules' architectural changes, induced by the toxin, hint at a dysfunctional cytoskeletal motors-mediated transport within mycolactone-treated cells. Other cytoskeleton dependent functions, like phagocytosis [51], [52], cell motility [27] and cell shape [17] are also described to be impaired in mycolactone treated cells. Thus, these evidences imply that mycolactone induces a nonfunctional cytoskeletal-architecture, affecting cytoskeleton-dependent functions, with consequences for cellular homeostasis. Moreover, our proteomic study revealed several regulators and structural constituents of both actin- and tubulin-cytoskeleton (Cfl1, Crmp2, Stmn1 and Actg1) affected by mycolactone. These alterations may reflect a cell feedback response to the abnormal cytoskeletal architecture as an attempt to restore the physiological cytoskeletal conformation and dynamics. In particular, the early alterations found on cofilin, a well-known regulator of actin dynamics [32], and on dynein, recently found to play a role in the production of normal bundled stress fibers [53], may represent an immediate cellular response to actin polymerization mediated by mycolactone-induced WASP hyperactivation [27]. Thus, a growing body of evidences supports a model in which the cytoskeletal disarrangement induced by mycolactone impairs multiple cytoskeleton-dependent cellular functions with cytotoxic consequences for the host cells.
These cytoskeletal changes might have also implications early on infection, during the M. ulcerans intracellular phase, when the pathogen has to survive and proliferate inside the host cell [3], [4]. Autophagy is being increasingly recognized as an important component of immunity, playing specific roles in shaping the immune system development, fuelling host innate and adaptive immune responses, and directly controlling intracellular microbes as a cell-autonomous innate defense mechanism. As an evolutionary counterpoint, intracellular pathogens have evolved to block autophagic microbicidal defenses and subvert host autophagic responses for their survival or growth [41], [42]. Importantly, studies have implicated autophagy in the control of many pathogenic bacteria [43], from which M. tuberculosis [54] should be highlighted here due to its genetic proximity to M. ulcerans. Thus, the mycolactone-induced impairment of autophagy, mediated by its action over the cytoskeleton, might represent a virulence mechanism of M. ulcerans to impair host cell immunity against intracellular pathogens.
One of the main findings of this work is the identification of a novel activity of mycolactone, with the demonstration of its role in the decrease of collagen content in M. ulcerans-infected tissues. Collagen decrease in human BU lesions was described in the 1960's, in the first histopathological studies of this disease [5], [46], [47], however, this phenomenon has been overlooked, even when, more recently, Guarner et al. described this feature as one of the most reliable criteria for the histopathological diagnosis of BU [48]. This previously unappreciated feature of the disease was never subject to studies to determine its cause or its relevance for BU progression and associated sequelae.
Here we link, for the first time, the activity of mycolactone with the collagen reduction in M. ulcerans-infected tissues. Our results from the in vivo model show that collagen decrease is not a consequence of the infection or the immune response, but of the presence of mycolactone. In fact the inoculation of the purified toxin shows an association between decrease of collagen and the presence of mycolactone. In vitro, we showed that in L929 cells mycolactone transcriptionally down-regulated several ER-resident collagen-modifying enzymes (Plod1, Plod3 and two isoforms of P4ha1). Additionally, Hall et al. described a post-transcriptional mechanism in which mycolactone blocks co-translational translocation of proteins into the ER, thus inhibiting the synthesis of the majority of ER-resident (like the collagen-modifying enzymes) and secretory proteins (like extracellular matrix proteins) [28]. Although these mechanisms have not been verified in vivo, altogether, these data suggest that mycolactone inflicts a transcriptional and post-transcriptional inhibition of the collagen biosynthesis pathway, which translate into a degeneration of collagen fibers in mycolactone-exposed tissues.
Our data also show that the mycolactone-induced collagen degeneration precedes the breakdown of the lesion into an ulcer, suggesting that collagen decrease may be involved, together with cell death, in the tissue destructuration that culminates in the emergence of an M. ulcerans-induced ulcerative lesion. Moreover, it may be a mechanism of pathogen dissemination, given that in early lesions bacteria concentrate in a smaller central zone, while in advanced lesions bacilli are dispersed throughout the necrotic area. Finally, this collagen decay in BU lesions may also be implicated in the development of the sequelae characteristic of this devastating skin disease. BU has a very high morbidity rate associated with contractures [9]. Wound contraction is a natural mechanism by which open wounds close during the healing process, but also results in significant tissue distortion with loss of joint mobility and cosmetic disfigurement. Although the mechanism of wound contraction is not fully understood, it is associated with the abnormal generation of thicker collagen fibers [55], [56], [57]. Therefore, it is conceivable that during the healing process, fibroblasts and myofibroblasts repopulating the lesion overcome the collagen-deficiency through abnormal- or over-production of collagen leading to the extreme contractures characteristic of BU [58]. In fact, a recent paper by Andreoli et al. described an increase in activated myofibroblasts and an abundant production of extracellular matrix proteins in antibiotic-treated BU lesions [59]. Further studies are needed to test this hypothesis, but if proven correct the use of collagen-based materials as a bed for the skin graft, or even as a replacement in smaller legions, may decrease the contracture and thus the morbidity in BU patients.
Overall, our results provide molecular and functional evidence of the impact of mycolactone on the cytoskeleton and cytoskeleton-dependent cellular functions, and extend our knowledge on the action of the M. ulcerans toxin to collagen biosynthesis, providing new perspectives on BU pathogenesis and paving the way for future therapeutic approaches.
Materials and Methods
Bacteria
M. ulcerans strains were selected from the Institute of Tropical Medicine collection in Antwerp, Belgium. MU5114 is a mycolactone-negative strain due to repeated subculturing, leading to the spontaneous loss of genes involved in mycolactone synthesis [23], [60]. MU98912 is highly virulent for mice [22] and produces mycolactone type D [24]. The isolates were grown on Middlebrook 7H9 medium (Becton, Dickinson and Company) with 1.5% of agar at 32°C for approximately 6–8 weeks. For the preparation of the inoculum, M. ulcerans was recovered, vortexed using glass beads and diluted in phosphate-buffered saline pH 7.4 (PBS) to a final concentration of 1 mg/ml.
Mycolactone extraction/purification
Protocol for mycolactone extraction/purification was adapted from the one previously described [17]. Briefly, MU98912 was cultured in Dubos medium supplemented with 10% oleic acid-albumin-dextrose complex, at 32°C. At late exponential growth phase, bacteria were harvested and lipids were extracted with chloroform and methanol (2∶1) for 4 hours. The organic phase was separated from bacterial debris and hydrophilic components by addition of a 0.2 volume of water, followed by centrifugation. The organic phase was dried and resuspended in ice-cold acetone. The individual lipid components of the acetone-soluble lipid fraction were separated by chromatography using the CycloGraph system (Analtech). The separated fractions were analyzed by thin layer chromatography, and the fractions corresponding to mycolactone were pooled, dried down, weighed, resuspended in absolute ethanol, and stored at −80°C under nitrogen atmosphere in the dark [61]. Purified mycolactone was analyzed by mass spectrometry (MS detector Thermo LxQ linear ion trap) and the presence of mycolactone D confirmed. Under these conditions, mycolactone was stable for at least three years.
In vitro experimental design
Mouse fibroblasts L929 cell line was cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum (Gibco), 2 mM L-glutamine (Gibco), 10 mM HEPES (Gibco), 1 mM sodium pyruvate (Gibco) and antibiotic-antimycotic (Gibco). Cells were expanded in 175 cm2 flasks (Nunc) until 90% confluence. Then, cells were plated in 12-wells plates (Nunc) at a density of 2.5×105 cells/well, with increasing concentrations of mycolactone or with ethanol equivalent (<0.002%), as a control. Rapamycin (Calbiochen) and Bafilomycin A1 (Sigma) was used to induce autophagy and to block autolysosome degradation, respectively.
Cell cycle analysis
At each time-point, cells were collected and a pool of adherent and suspended cells was made. Cells were rinsed and resuspended in PBS. Absolute ethanol was gently added until 70% final concentration. Cells were stored in this fixing solution at 4°C. When all time-points had been collected, cells were rinsed in PBS and incubated with staining solution (0.1% triton-X-100, 20 µg/mL of propidium iodide, 250 µg/mL of RNase in PBS) for one hour in a bath at 50°C, in the dark. Samples were analyzed by flow cytometry (LSRII, BD).
Annexin-V/PI assay
The protocol was done according to the manufacturer's instructions (BD Pharmingen). At each time-point cells were collected and a pool of adherent and suspended cells was made. Cells were rinsed, stained and analyzed by flow cytometry (LSRII, BD).
Cytoskeleton imaging
L929 cells were allowed to adhere to coverslips (Nunc) overnight following the incubation in different conditions. Cells were rinsed and fixed in paraformaldehyde, for 1 hour, at room temperature. Cells were rinsed and stored in PBS at 4°C. When all time-points had been collected, cells were blocked with blocking solution (5% BSA, 0.1% triton-X-100, 0.1% tween-20 in PBS) and incubated overnight at 4°C with the mouse anti-tubulin antibody (AA4.3, developed by C. Walsh and obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biology). Cells were rinsed and incubated with secondary AF488 goat anti-mouse antibody (Invitrogen) and rhodamine-phalloidin conjugate (Invitrogen) for 1 hour, at room temperature, in the dark. Cells were visualized using a confocal microscope (FV1000, Olympus) with ×60 objective. 3D remodeling was performed using Fluoview software (Olympus).
Protein extraction
At each time-point, cells were collected and a pool of adherent and suspended cells was made. Cells were rinsed with PBS, resuspended in Lysis Buffer (50 mM Tris-HCl pH7.2, 250 mM NaCl, 2 mM EDTA, 1% NP-40, 10% Glycerol, protease inhibitor (Roche #11873580001) and phosphatase inhibitor (Roche #04906837001)) and stored at −80°C, until protein was extracted at the end of the experiment. When all time-points had been collected, samples were thawed, incubated for 30 minutes at 4°C with agitation, sonicated in a ultrasonic ice cold bath for 1 minute until no agglomerate was seen and centrifuged (30 minutes, 14000 rpm, 4°C). The supernatant was considered the total protein extract. For Western Blot analysis, protein concentration was determined (Thermo Scientific #23227) and aliquots stored at −80°C.
Two-dimensional gel electrophoresis (2-DE)
Protein was precipitated in 80% (v/v) acetone and the protein pellet resuspended in urea buffer (7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 0.15% (w/v) DTT, 0.5% [v/v] carrier ampholytes and Complete Mini protease inhibitor cocktail). The protein separation was done as previously described [62]. Briefly, 100 µg protein extract was diluted with urea buffer to a final volume of 420 µL and in-gel rehydration was performed overnight. IEF was carried out in IPG strips (pH 3–10, non-linear, 18 cm; GE Healthcare, Uppsala, Sweden) with the Multiphor II system (GE Healthcare) under paraffin oil for 55 kVh. SDS-PAGE was done overnight in polyacrylamide gels (12.5% T, 2.6% C) with the Ettan DALT II system (GE Healthcare) at 1–2 W per gel and 12°C. The gels were silver stained and analyzed with the 2-DE image analysis software Melanie 3.0 (Gene-Bio, Geneva, Switzerland). To verify the reproducibility three biological replicates for each time point and condition as well as three technical replicates were analyzed (all analyzed gels are in Supplementary Information). An expression change was considered significant if the intensity of the corresponding single spot differed reproducibly more than twofold and was reproducible for all three experiments. The expected spot position in the 2D-gel according to the known protein sequence was calculated with the Compute pI/Mw tool (http://ca.expasy.org/tools/pi_tool.html). For the detection of phosphorylated proteins 400 µg of protein were separated by 2-DE, stained with Pro-Q Diamond Phosphoprotein Gel Stain (Molecular Probes), according to the manufacturer's instructions, scanned to detect the phosphorylation signals, silver stained and rescanned. Images of both scans were matched with the 2-DE image analysis software Melanie 3.0 (Gene-Bio).
Mass spectrometry (MS)
For the protein identification, 400 µg of protein were separated by 2-DE. Selected spots were excised, digested with trypsin (recombinant; Roche), and prepared as described earlier [62]. In brief, the extracted and dried peptides were dissolved in 5 µl alpha-Cyano-3-hydroxycinnamic acid (98%, recrystallized from ethanol-water, 5 mg/ml in 50% acetonitrile and 0.1% TFA) and 0.5 µl applied onto the sample plate using the dried-droplet method. Peptide masses were measured with a UltrafleXtreme MALDI-TOF/TOF (Bruker, Billerica, MA, USA). Proteins were identified according to their spot-specific peptide mass fingerprint and/or peptide sequence with the bioinformatic tool BioTools Version 3.2 (Bruker) with the following search parameters (tolerance: MS = 10–50 ppm, MS/MS = 0.5–0.9 Da, enzyme: Trypsin, engine: Mascot, database: NCBInr, modifications: Oxidation (M)). A protein identification was accepted if at least three major peaks matched to the protein with the highest score (full MS and MS/MS data in Supplementary Information).
If the protein spot was detected at a lower molecular mass than expected, suggesting processing or fragmentation, the spot-specific peptides in the mass spectrum were also analyzed to confirm which parts of the corresponding protein sequence matched with these peptides. If the mass spectrum of the spot lacked peptides observed for the complete protein and had a different position in the 2D gel than expected it was indicated as a protein fragment. Therefore, both the spot position observed by 2-DE and the specific peptides in the corresponding mass spectrum were analyzed to indicate a putative protein fragment.
Western blot analysis
40 µg of protein were resolved in a 12% SDS-PAGE and transferred to the 0.2 µm Nitrocellulose membranes (Bio-Rad #170-4159) with the semi-dry Trans-Blot Turbo system (Bio-Rad). Membranes were blocked and subjected to immunoblotting with GAPDH antibody (CellSignaling #2118), LC3A/B antibody (CellSignaling #4108) or mono- and polyubiquitinylated conjugates antibody (Enzo Life Sciences #PW8810), followed by incubation with horseradish peroxidase linked secondary antibodies (Southern Biotech). Bands were detected with SuperSignal (Thermo Scientific #34095) in a Universal Hood II (Bio-Rad) and quantified with QuantityOne (Bio-Rad). GAPDH was used as loading control.
LC3 Immunofluorescence
At each time-point, cells were collected and a pool of adherent and suspended cells was made. Cytospins were made (Cytospin III, Shandon) and cells were fixed in paraformaldehyde for 20 minutes at room temperature and stored in ethanol 96% at 4°C. When all time-points had been collected, cells were blocked with blocking solution (5% BSA, 0.1% triton-X-100, 0.1% tween-20 in PBS) and incubated overnight at 4°C with the LC3A/B antibody (CellSignaling #4108). Normal Rabbit IgG Control (R&D Systems AB-105-C) was used as isotype control. Cells were rinsed and incubated with secondary AF568 goat anti-rabbit antibody (Invitrogen) for 1 hour, at room temperature, in the dark. Cells were visualized using a confocal microscope (FV1000, Olympus) with ×60 objective.
In vivo experimental design
Eight-weeks-old female BALB/c mice were obtained from Charles River (Barcelona, Spain) and housed under specific-pathogen-free conditions with food and water ad libitum. Mice were infected in the left hind footpad with 30 µL of M. ulcerans suspensions with 4.8 log10 AFBs, or 30 µL PBS as control. Footpad thickness was evaluated every 2–3 days. Mice were sacrificed weekly and footpads were harvested for histological studies.
Ethics statement
The in vivo studies were approved by the Portuguese national authority for animal experimentation Direção Geral de Veterinária (ID: DGV 594 from 1st June 2010). Animals were kept and handled in accordance with the guidelines for the care and handling of laboratory animals in the Directive 2010/63/EU of the European Parliament and of the Council.
Collagen assessment
Footpads were harvested, fixed in 10% phosphate-buffered formalin and embedded in paraffin. Tissue sections were stained with hematoxylin and eosin (H&E), analyzed by light microscopy with polarized light and the amount of dermal collagen fibers was blindly scored from 0 (lowest) to 4 (highest) independently by two persons in two independent experiments. Additionally, tissue sections were stained with Masson's trichrome and pictures were taken in a light microscopy.
RT-qPCR
At each time-point, cells were collected and a pool of adherent and suspended cells was made. Cells were rinsed, resuspended in TRIzol Reagent (Ambion) and stored at −80°C, until total RNA was extracted, at the end of the experiment, according to the manufacturer's protocol.
Reverse transcription was done with whole RNA using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer's instructions. qPCR was perform on the C1000TM Thermo Cycler (Bio-Rad) using TaqMan Gene Expression Assay (AB Applied Biosystems) (Plod1: Mm01255769_m1; Plod3: Mm00478798_m1; P4ha1: Mm00803137_m1; B2m: Mm00437762_m1; Gapdh: Mm99999915_g1; Hprt: Mm00446968_m1). Relative quantification was determined with CFX Manager Software (Bio-Rad) using B2m, Gapdh and Hprt as reference genes.
Statistical analysis
Differences between the means of experimental groups were analyzed using the Prism version 5.0 software (GraphPad). Percentage and fraction values were transformed to and analyzed as arcsin values. Differences were considered significant only with a P value<0.001, in the in vitro studies; or with a P value<0.01, in the in vivo study.
Supporting Information
Mycolactone treatment affects mainly cytoskeleton related proteins and collagen biosynthesis enzymes. Enlarged parts of silver-stained 2-D gel of total protein extracts (100 µg) of L929 cells treated either with ethanol or mycolactone (50 ng/mL), showing the identified altered spots clustered into five categories. Arrows and circles represent conditions where the spot intensity is increased or decreased, respectively. Phosphoproteins are indicated by an asterisk (*) and putative protein fragments are indicated by a number sign (#). For P4ha1 the number of the specific isoform is indicted in parentheses.
(TIF)
Spots 4 and 6 are phosphoproteins. Enlarged parts of silver- and phospho-stained 2-D gel showing the spot 4 (Cfl1) and the spot 6 (Crmp2) in both stainings.
(TIF)
Mycolactone treatment induces a transcriptional down-regulation of collagen biosynthesis enzymes. Mouse fibroblasts L929 cells were incubated for 24 or 48 hours either with ethanol (white) or mycolactone (50 ng/mL, gray). Additionally, an assay was performed where cells were incubated for 48 hours in the same conditions followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h). At each time-point, total RNA was extracted and Plod1, Plod3, P4ha1 mRNA levels assessed. Bars represent the mean + SD from two independent experiments with three technical replicas (n = 6). Mycolactone-treated was compared to EtOH-treated samples throughout each time-point (24 h and 48 h) by Two-way ANOVA with Bonferroni posttest; statistical differences were represented by *** (P<0.001). Each condition at 48 h+48 h time-point was compared with the same condition at the 48 h by Two-way ANOVA with Bonferroni posttest; statistical differences were represented by &&& (P<0.001).
(TIF)
2D gels from L929 cells incubated for 24 hours with ethanol.
(ZIP)
2D gels from L929 cells incubated for 48 hours with ethanol.
(ZIP)
2D gels from L929 cells incubated for 48 hours with ethanol followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h).
(ZIP)
2D gels from L929 cells incubated for 24 hours with mycolactone.
(ZIP)
2D gels from L929 cells incubated for 48 hours with mycolactone.
(ZIP)
2D gels from L929 cells incubated for 48 hours with mycolactone followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h).
(ZIP)
MS and MS/MS data.
(ZIP)
Acknowledgments
The authors would like to thank Goreti Pinto for microscopy assistance. This article is dedicated to the memory of Prof. Manuel T. Silva, deceased on 10 April 2012.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research leading to these results has received funding from the European Community's Seventh Framework Program (FP7/2007–2013) under grant agreement Nu 241500 (BuruliVac), from Fundação Calouste Gulbenkian and from Projeto Estratégico – LA 26 – 2013–2014 (PEst-C/SAU/LA0026/2013). JBG, TGM and AGF had a personal grant from the Portuguese Science and Technology Foundation (FCT) (SFRH/BD/33573/2009, SFRH/BD/41598/2007 and SFRH/BPD/68547/2010, respectively). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. MacCallum P, Tolhurst JC, Sìssons HA (1948) A new mycobacterial infection in man. J Pathol Bacteriol 60: 93–122. [PubMed] [Google Scholar]
- 2. Connor DH, Lunn HF (1965) Mycobacterium ulcerans infection (with comments on pathogenesis). Int J Lepr 33 Suppl: 698–709. [PubMed] [Google Scholar]
- 3. Silva MT, Portaels F, Pedrosa J (2009) Pathogenetic mechanisms of the intracellular parasite Mycobacterium ulcerans leading to Buruli ulcer. Lancet Infect Dis 9: 699–710. [DOI] [PubMed] [Google Scholar]
- 4. Torrado E, Fraga AG, Castro AG, Stragier P, Meyers WM, et al. (2007) Evidence for an intramacrophage growth phase of Mycobacterium ulcerans. Infect Immun 75: 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Connor DH, Lunn HF (1966) Buruli Ulceration: A clinicopathologic study of 38 Ugandans with Mycobacterium ulcerans ulceration. Arch Pathol 81: 183–199. [Google Scholar]
- 6. World Health Organization (2004) Provisional guidance on the role of specific antibiotics in the management of Mycobacterium ulcerans disease (Buruli ulcer). WHO/CDS/CPE/GBUI/2004 10: 33. [Google Scholar]
- 7. Kibadi K, Boelaert M, Fraga AG, Kayinua M, Longatto-Filho A, et al. (2010) Response to treatment in a prospective cohort of patients with large ulcerated lesions suspected to be Buruli Ulcer (Mycobacterium ulcerans disease). PLoS Negl Trop Dis 4: e736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quek TY, Henry MJ, Pasco JA, O'Brien DP, Johnson PD, et al. (2007) Mycobacterium ulcerans infection: factors influencing diagnostic delay. Med J Aust 187: 561–563. [DOI] [PubMed] [Google Scholar]
- 9. Ellen DE, Stienstra Y, Teelken MA, Dijkstra PU, van der Graaf WT, et al. (2003) Assessment of functional limitations caused by Mycobacterium ulcerans infection: towards a Buruli ulcer functional limitation score. Trop Med Int Health 8: 90–96. [DOI] [PubMed] [Google Scholar]
- 10. Marsollier L, Robert R, Aubry J, Saint Andre JP, Kouakou H, et al. (2002) Aquatic insects as a vector for Mycobacterium ulcerans. Appl Environ Microbiol 68: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marsollier L, Severin T, Aubry J, Merritt RW, Saint Andre JP, et al. (2004) Aquatic snails, passive hosts of Mycobacterium ulcerans. Appl Environ Microbiol 70: 6296–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marsollier L, Brodin P, Jackson M, Kordulakova J, Tafelmeyer P, et al. (2007) Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog 3: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Portaels F, Meyers WM, Ablordey A, Castro AG, Chemlal K, et al. (2008) First cultivation and characterization of Mycobacterium ulcerans from the environment. PLoS Negl Trop Dis 2: e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merritt RW, Walker ED, Small PL, Wallace JR, Johnson PD, et al. (2010) Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS Negl Trop Dis 4: e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nackers F, Dramaix M, Johnson RC, Zinsou C, Robert A, et al. (2006) BCG vaccine effectiveness against Buruli ulcer: a case-control study in Benin. Am J Trop Med Hyg 75: 768–774. [PubMed] [Google Scholar]
- 16. Fraga AG, Martins TG, Torrado E, Huygen K, Portaels F, et al. (2012) Cellular immunity confers transient protection in experimental Buruli ulcer following BCG or mycolactone-negative Mycobacterium ulcerans vaccination. PLoS One 7: e33406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. George KM, Chatterjee D, Gunawardana G, Welty D, Hayman J, et al. (1999) Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283: 854–857. [DOI] [PubMed] [Google Scholar]
- 18. Pahlevan AA, Wright DJ, Andrews C, George KM, Small PL, et al. (1999) The inhibitory action of Mycobacterium ulcerans soluble factor on monocyte/T cell cytokine production and NF-kappa B function. J Immunol 163: 3928–3935. [PubMed] [Google Scholar]
- 19. George KM, Pascopella L, Welty DM, Small PL (2000) A Mycobacterium ulcerans toxin, mycolactone, causes apoptosis in guinea pig ulcers and tissue culture cells. Infect Immun 68: 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Torrado E, Adusumilli S, Fraga AG, Small PL, Castro AG, et al. (2007) Mycolactone-mediated inhibition of tumor necrosis factor production by macrophages infected with Mycobacterium ulcerans has implications for the control of infection. Infect Immun 75: 3979–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torrado E, Fraga AG, Logarinho E, Martins TG, Carmona JA, et al. (2010) IFN-gamma-dependent activation of macrophages during experimental infections by Mycobacterium ulcerans is impaired by the toxin mycolactone. J Immunol 184: 947–955. [DOI] [PubMed] [Google Scholar]
- 22. Oliveira MS, Fraga AG, Torrado E, Castro AG, Pereira JP, et al. (2005) Infection with Mycobacterium ulcerans induces persistent inflammatory responses in mice. Infect Immun 73: 6299–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mve-Obiang A, Lee RE, Portaels F, Small PL (2003) Heterogeneity of mycolactones produced by clinical isolates of Mycobacterium ulcerans: implications for virulence. Infect Immun 71: 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hong H, Spencer JB, Porter JL, Leadlay PF, Stinear T (2005) A novel mycolactone from a clinical isolate of Mycobacterium ulcerans provides evidence for additional toxin heterogeneity as a result of specific changes in the modular polyketide synthase. Chembiochem 6: 643–648. [DOI] [PubMed] [Google Scholar]
- 25. Snyder DS, Small PL (2003) Uptake and cellular actions of mycolactone, a virulence determinant for Mycobacterium ulcerans. Microb Pathog 34: 91–101. [DOI] [PubMed] [Google Scholar]
- 26. George KM, Barker LP, Welty DM, Small PL (1998) Partial purification and characterization of biological effects of a lipid toxin produced by Mycobacterium ulcerans. Infect Immun 66: 587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guenin-Mace L, Veyron-Churlet R, Thoulouze MI, Romet-Lemonne G, Hong H, et al. (2013) Mycolactone activation of Wiskott-Aldrich syndrome proteins underpins Buruli ulcer formation. J Clin Invest 123: 1501–12 doi: 10.1172/JCI66576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hall BS, Hill K, McKenna M, Ogbechi J, High S, et al. (2014) The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, mycolactone, depends on blockade of protein translocation into the ER. PLoS Pathog 10: e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sarfo FS, Le Chevalier F, Aka N, Phillips RO, Amoako Y, et al. (2011) Mycolactone diffuses into the peripheral blood of Buruli ulcer patients–implications for diagnosis and disease monitoring. PLoS Negl Trop Dis 5: e1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luthi AU, Martin SJ (2007) The CASBAH: a searchable database of caspase substrates. Cell Death Differ 14: 641–650. [DOI] [PubMed] [Google Scholar]
- 31. Allan VJ (2011) Cytoplasmic dynein. Biochem Soc Trans 39: 1169–1178. [DOI] [PubMed] [Google Scholar]
- 32. Bernstein BW, Bamburg JR (2010) ADF/cofilin: a functional node in cell biology. Trends Cell Biol 20: 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidt EF, Strittmatter SM (2007) The CRMP family of proteins and their role in Sema3A signaling. Adv Exp Med Biol 600: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belletti B, Baldassarre G (2011) Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets 15: 1249–1266. [DOI] [PubMed] [Google Scholar]
- 35. Webb JL, Ravikumar B, Rubinsztein DC (2004) Microtubule disruption inhibits autophagosome-lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. Int J Biochem Cell Biol 36: 2541–2550. [DOI] [PubMed] [Google Scholar]
- 36. Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, et al. (2005) Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet 37: 771–776. [DOI] [PubMed] [Google Scholar]
- 37. Tanida I, Ueno T, Kominami E (2008) LC3 and Autophagy. Methods Mol Biol 445: 77–88. [DOI] [PubMed] [Google Scholar]
- 38. Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC (2008) Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4: 849–850. [DOI] [PubMed] [Google Scholar]
- 39. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, et al. (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fung C, Lock R, Gao S, Salas E, Debnath J (2008) Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 19: 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deretic V, Levine B (2009) Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5: 527–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sumpter R Jr, Levine B (2010) Autophagy and innate immunity: triggering, targeting and tuning. Semin Cell Dev Biol 21: 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gong L, Devenish RJ, Prescott M (2012) Autophagy as a macrophage response to bacterial infection. IUBMB Life 64: 740–747. [DOI] [PubMed] [Google Scholar]
- 44. Krane SM (2008) The importance of proline residues in the structure, stability and susceptibility to proteolytic degradation of collagens. Amino Acids 35: 703–710. [DOI] [PubMed] [Google Scholar]
- 45. Yamauchi M, Sricholpech M (2012) Lysine post-translational modifications of collagen. Essays Biochem 52: 113–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clancey JK, Dodge OG, Lunn HF, Oduori ML (1961) Mycobacterial skin ulcers in Uganda. Lancet 2: 951–954. [DOI] [PubMed] [Google Scholar]
- 47. Dodge OG (1964) Mycobacterial Skin Ulcers in Uganda: Histopathological and Experimental Aspects. J Pathol Bacteriol 88: 169–174. [DOI] [PubMed] [Google Scholar]
- 48. Guarner J, Bartlett J, Whitney EA, Raghunathan PL, Stienstra Y, et al. (2003) Histopathologic features of Mycobacterium ulcerans infection. Emerg Infect Dis 9: 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Korolchuk VI, Menzies FM, Rubinsztein DC (2009) A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 5: 862–863. [DOI] [PubMed] [Google Scholar]
- 50. Buktenica S, Olenick SJ, Salgia R, Frankfater A (1987) Degradation and regurgitation of extracellular proteins by cultured mouse peritoneal macrophages and baby hamster kidney fibroblasts. Kinetic evidence that the transfer of proteins to lysosomes is not irreversible. J Biol Chem 262: 9469–9476. [PubMed] [Google Scholar]
- 51. Adusumilli S, Mve-Obiang A, Sparer T, Meyers W, Hayman J, et al. (2005) Mycobacterium ulcerans toxic macrolide, mycolactone modulates the host immune response and cellular location of M. ulcerans in vitro and in vivo. Cell Microbiol 7: 1295–1304. [DOI] [PubMed] [Google Scholar]
- 52. Coutanceau E, Marsollier L, Brosch R, Perret E, Goossens P, et al. (2005) Modulation of the host immune response by a transient intracellular stage of Mycobacterium ulcerans: the contribution of endogenous mycolactone toxin. Cell Microbiol 7: 1187–1196. [DOI] [PubMed] [Google Scholar]
- 53. Meiri D, Marshall CB, Greeve MA, Kim B, Balan M, et al. (2012) Mechanistic insight into the microtubule and actin cytoskeleton coupling through dynein-dependent RhoGEF inhibition. Mol Cell 45: 642–655. [DOI] [PubMed] [Google Scholar]
- 54. Songane M, Kleinnijenhuis J, Netea MG, van Crevel R (2012) The role of autophagy in host defence against Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 92: 388–396. [DOI] [PubMed] [Google Scholar]
- 55. Ehrlich HP (1988) Wound closure: evidence of cooperation between fibroblasts and collagen matrix. Eye (Lond) 2 Pt 2: 149–157. [DOI] [PubMed] [Google Scholar]
- 56. Berry DP, Harding KG, Stanton MR, Jasani B, Ehrlich HP (1998) Human wound contraction: collagen organization, fibroblasts, and myofibroblasts. Plast Reconstr Surg 102: 124–131 discussion 132–124. [DOI] [PubMed] [Google Scholar]
- 57. Ehrlich HP, Hunt TK (2012) Collagen Organization Critical Role in Wound Contraction Advances in Wound Care. 1: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.World Health Organization (2006) Buruli ulcer. Prevention of disability (POD). WHO/CDS/NTD/GBUI/200612.
- 59. Andreoli A, Ruf MT, Sopoh GE, Schmid P, Pluschke G (2014) Immunohistochemical monitoring of wound healing in antibiotic treated Buruli ulcer patients. PLoS Negl Trop Dis 8: e2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stinear TP, Hong H, Frigui W, Pryor MJ, Brosch R, et al. (2005) Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J Bacteriol 187: 1668–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marion E, Prado S, Cano C, Babonneau J, Ghamrawi S, et al. (2013) Photodegradation of the Mycobacterium ulcerans toxin, mycolactones: considerations for handling and storage. PLoS One 7: e33600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ohlmeier S, Mazur W, Salmenkivi K, Myllarniemi M, Bergmann U, et al. (2010) Proteomic studies on receptor for advanced glycation end product variants in idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease. Proteomics Clin Appl 4: 97–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mycolactone treatment affects mainly cytoskeleton related proteins and collagen biosynthesis enzymes. Enlarged parts of silver-stained 2-D gel of total protein extracts (100 µg) of L929 cells treated either with ethanol or mycolactone (50 ng/mL), showing the identified altered spots clustered into five categories. Arrows and circles represent conditions where the spot intensity is increased or decreased, respectively. Phosphoproteins are indicated by an asterisk (*) and putative protein fragments are indicated by a number sign (#). For P4ha1 the number of the specific isoform is indicted in parentheses.
(TIF)
Spots 4 and 6 are phosphoproteins. Enlarged parts of silver- and phospho-stained 2-D gel showing the spot 4 (Cfl1) and the spot 6 (Crmp2) in both stainings.
(TIF)
Mycolactone treatment induces a transcriptional down-regulation of collagen biosynthesis enzymes. Mouse fibroblasts L929 cells were incubated for 24 or 48 hours either with ethanol (white) or mycolactone (50 ng/mL, gray). Additionally, an assay was performed where cells were incubated for 48 hours in the same conditions followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h). At each time-point, total RNA was extracted and Plod1, Plod3, P4ha1 mRNA levels assessed. Bars represent the mean + SD from two independent experiments with three technical replicas (n = 6). Mycolactone-treated was compared to EtOH-treated samples throughout each time-point (24 h and 48 h) by Two-way ANOVA with Bonferroni posttest; statistical differences were represented by *** (P<0.001). Each condition at 48 h+48 h time-point was compared with the same condition at the 48 h by Two-way ANOVA with Bonferroni posttest; statistical differences were represented by &&& (P<0.001).
(TIF)
2D gels from L929 cells incubated for 24 hours with ethanol.
(ZIP)
2D gels from L929 cells incubated for 48 hours with ethanol.
(ZIP)
2D gels from L929 cells incubated for 48 hours with ethanol followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h).
(ZIP)
2D gels from L929 cells incubated for 24 hours with mycolactone.
(ZIP)
2D gels from L929 cells incubated for 48 hours with mycolactone.
(ZIP)
2D gels from L929 cells incubated for 48 hours with mycolactone followed by a 48 hour incubation period in fresh medium (referred to as 48 h+48 h).
(ZIP)
MS and MS/MS data.
(ZIP)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.