Key Points
Juvenile zebrafish tolerate widespread coagulopathy due to complete ablation of antithrombin III, but develop lethal thrombosis as adults.
In vivo structure/function analysis of antithrombin III in zebrafish reveals limited roles for heparin-binding and anti-IXa/Xa activity.
Abstract
Pathologic blood clotting is a leading cause of morbidity and mortality in the developed world, underlying deep vein thrombosis, myocardial infarction, and stroke. Genetic predisposition to thrombosis is still poorly understood, and we hypothesize that there are many additional risk alleles and modifying factors remaining to be discovered. Mammalian models have contributed to our understanding of thrombosis, but are low throughput and costly. We have turned to the zebrafish, a tool for high-throughput genetic analysis. Using zinc finger nucleases, we show that disruption of the zebrafish antithrombin III (at3) locus results in spontaneous venous thrombosis in larvae. Although homozygous mutants survive into early adulthood, they eventually succumb to massive intracardiac thrombosis. Characterization of null fish revealed disseminated intravascular coagulation in larvae secondary to unopposed thrombin activity and fibrinogen consumption, which could be rescued by both human and zebrafish at3 complementary DNAs. Mutation of the human AT3-reactive center loop abolished the ability to rescue, but the heparin-binding site was dispensable. These results demonstrate overall conservation of AT3 function in zebrafish, but reveal developmental variances in the ability to tolerate excessive clot formation. The accessibility of early zebrafish development will provide unique methods for dissection of the underlying mechanisms of thrombosis.
Introduction
Venous thrombosis affects approximately 1 in 1000 individuals, resulting in over 300 000 hospitalizations per year. There are many well-described coagulation factor defects which are associated with thrombosis, including deficiencies of the natural anticoagulant proteins, protein C (PROC), protein S (PROS1), and antithrombin III (AT3).1 Homozygous deficiency of PROC or PROS1 is exceedingly rare and results in neonatal purpura fulminans in humans.1 However, mouse knockout models display more severe phenotypes, with neonatal and embryonic loss for PROC and PROS1 deficiency, respectively.2-4 Complete loss of AT3 has not been described in humans, and is presumed to be in utero lethal.1 This is supported by the observation that homozygous deletion of AT3 in mice results in prenatal lethality, with widespread fibrin deposition in the heart and liver.5 AT3-deficient embryos also exhibit paradoxical hemorrhage, suggesting a consumptive coagulopathy,5 but this has been difficult to study given the relative inaccessibility of mammalian embryogenesis.
AT3 (also known as SERPINC1) is a serpin (serine protease inhibitor) that regulates many coagulation proteases including thrombin and factors IXa and Xa,6,7 and consists of 2 functional domains. The reactive center loop (RCL) presents the Arg393-Ser394 bond for cleavage, resulting in the formation of a stable inactive AT3-protease complex. The heparin-binding site binds endogenous glycosaminoglycans or infused heparin, acting as a bridge to the protease and inducing allosteric changes in AT3, which accelerates inhibition.6,7 Deficiency of AT3 is classified into 2 categories: type I with reduced plasma antigen levels and type II with functional defects, but normal AT3 levels.1 The latter is further subdivided into reactive site, heparin binding, and pleiotropic defects. Over 200 different mutations from patients with AT3 deficiency have been identified,8 and rapid advances in sequencing technologies guarantee that there are many more to come. Although in vitro assays are available for assessment of AT3 function, in some patients the underlying mechanism of AT3 deficiency remains unknown.
Teleost fish possess highly conserved orthologs of nearly all blood coagulation factors.9-12 Zebrafish (Danio rerio) have been widely used to study hemostasis, demonstrating conservation and function of the structural components of coagulation, including fibrinogen13,14 and von Willebrand factor,15,16 and the ability to develop thrombosis in response to a laser-induced injury.14,17 Embryonic development is external, rapid, and transparent, allowing unfettered access to studies of the circulatory system. Individual breeding pairs can produce hundreds of offspring with each pairing, facilitating high-throughput genetic and small-molecule screens.18
Recent advances in genome editing technologies have enabled robust platforms for genetic modification.19-21 We now report ablation of at3 using zinc finger (ZF) nucleases (ZFNs). Although homozygous mutants recapitulate the consumptive coagulopathy and thrombotic phenotypes of the mouse knockout, the timing demonstrates significant differences. at3−/− zebrafish are able to tolerate spontaneous thrombosis and rampant disseminated intravascular coagulation (DIC) early in development, but die in adulthood with associated intracardiac thrombosis. Furthermore, using in vivo genetic complementation, we reveal insight into the hemostatic consequences of human AT3 variants.
Methods
Animals
Zebrafish were raised in accordance with animal care guidelines as approved by the University of Michigan Animal Care and Use Committee. Embryos are defined as 0 to 2 days postfertilization (dpf), and larvae as 3 to 29 dpf.
Targeted mutagenesis using ZFNs
ZFN pairs were designed to target exon 5 of the at3 genomic locus. ZF arrays were constructed using OPEN (Oligomerized Pool ENgineering) as described.22,23 Targeted mutagenesis with the constructed ZFNs was completed essentially as described.19,24 ZF arrays were cloned into expression vectors pMLM290 and 292, and transcribed in vitro using T7 mMESSAGE mMACHINE (Ambion). One nanogram of each left and right ZFN messenger RNA (mRNA) was injected into 1-cell AB × TL F1 embryos, as described.14 At 2 to 3 dpf, 8 morphologically normal embryos were pooled and lysed in buffer (10 mM Tris, pH 8.0, 200 mM NaCl, 10 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 100 μg/mL Proteinase K) at 50°C for 2 to 3 hours, and genomic DNA isolated by phenol:chloroform extraction and ethanol precipitation. Polymerase chain reaction (PCR) of the target site was performed, and products cloned into pCR-Blunt (Invitrogen). PCR of 96 colonies was performed using M13 reverse and T7 primers, and products sequenced. The remaining injected F0 embryos were raised to adulthood and crossed to wild-type fish. Sixteen to 24 F1 embryos from each F0 founder were screened for ZFN-induced mutations by PCR of the target site with a 6-FAM–labeled gene-specific primer, after lysis in PCR extraction buffer24 (10 mM Tris pH 8.0, 2 mM EDTA, 0.2% Triton X-100 and 100 μg/ mL Proteinase K). PCR products were resolved by capillary electrophoresis on a 3730XL DNA Analyzer (Applied Biosystems). Data were processed using GeneMarker (Softgenetics LLC) for mutant identification. The remaining F1 offspring were raised to adulthood and heterozygotes identified by fin clip genotyping.
Genotyping of mutant offspring
Fin clip biopsies were obtained from adult fish after anesthesia in tricaine (0.16 mg/mL; Western Chemical Inc), and larvae were humanely killed in high-dose tricaine (1.6 mg/mL), followed by lysis in PCR extraction buffer.24 Deletion mutations were detected by PCR and agarose gel electrophoresis (Figure 1E bottom). Primers were designed using Primer325 and oligonucleotides ordered from Integrated DNA Technologies. All oligonucleotide sequences are listed in supplemental Tables 1-2 (see supplemental Data available on the Blood Web site).
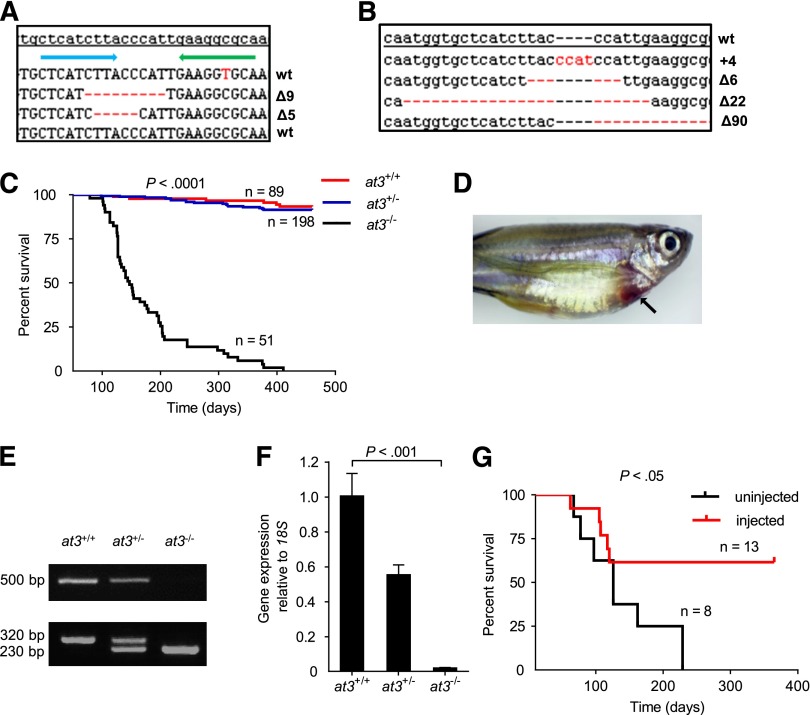
Figure 1.
Targeted disruption of at3 using genome editing nucleases results in adult lethality. (A) ZFN-induced deletions in somatic cells. Of 16 pooled morphologically normal embryos from 2 individual experiments, 3 of 192 sequences were identified with Δ5 (2x) and Δ9 (1x) in exon 5. The top line is the at3 reference genome sequence from an unrelated individual. The blue and green arrows indicate 5′ and 3′ ZFN-binding sites, respectively. Deletions are indicated by red dashes. (B) Sequences of ZFN-induced mutations transmitted through the germline. Red dashes and letters indicate deletions and insertions, respectively. (C) Survival curve of a total of 3 clutches derived from at3Δ90 heterozygous incrosses shows loss of homozygotes. Offspring were genotyped at 2 to 4 months of age and tracked over 1 year. The results demonstrate a highly significant difference in the survival of homozygous mutants by log-rank (Mantel-Cox) analysis (P < .0001). (D) Externally visible secondary hemorrhage in an at3Δ90 homozygous mutant is indicated by an arrow. This fish was identified while alive, anesthetized, and the image was captured using a Leica MZ16FA microscope and a Canon EOS60D camera. (E-F) RT-PCR analysis of at3Δ90 mutants demonstrates reduction of at3 expression in heterozygotes and absence of homozygous mutant mRNA. (E) Top: Total mRNA was prepared from adult fish (4 mpf) followed by qualitative RT-PCR. Bottom: Genotyping of at3Δ90 mutants. (F) Total mRNA was prepared from pooled larvae (3 dpf, n = 15 for each genotype) followed by quantitative real-time PCR. Error bars represent standard deviation and statistical significance was determined by t test. (G) at3Δ22 homozygous mutants were injected with a ubi regulated at3 cDNA expression vector and followed for 1 year.
Construction of at3 in vivo expression vectors
Vectors pubi-zat3-EGFP and pubi-hAT3-EGFP were constructed by inserting complementary DNA (cDNA) sequences under control of the ubi promoter in frame with enhanced green fluorescent protein (EGFP) in pDestTol2pA2_ubi:EGFP26 (supplemental Figure 1). Zebrafish at3 was amplified from total adult cDNA using NcoI tagged primers, and cloned into NcoI digested pDestTol2pA2_ubi:EGFP. Human AT3 was amplified from liver cDNA using primers designed for sequence- and ligation-independent cloning, and cloned into NcoI digested pDestTol2pA2_ubi:EGFP as described.27
Zebrafish and human at3 substitutions were introduced into pubi-zat3-EGFP and pubi-hAT3-EGFP, respectively, as described with some modifications.28 Mutagenic primers amplified the plasmids using Phusion DNA polymerase (New England Biolabs) as follows: 98°C for 3 minutes; 4 cycles of 98°C for 10 seconds, 55°C for 30 seconds, 72°C for 8 minutes; 12 cycles of 98°C for 10 seconds, 60°C for 30 seconds, 72°C 8 minutes; 72°C for 10 minutes. PCR products were column purified, digested with DpnI, and transformed into TOP10 cells, and mutations confirmed by sequencing. Amino acid numbering of human AT3 begins at +1 after the propeptide cleavage site (amino acid 33 from the initiation methionine), reflecting absence of the 32-aa propeptide. The N-terminal amino acids of zebrafish At3 are not conserved with the mammalian AT3 propeptide (supplemental Figure 2), therefore, +1 for the former is from the initiation methionine.
One-cell injection of at3 expression vectors
Circular at3 expression plasmids (25 ng/µL) and transposase mRNA (25 ng/µL)29 were coinjected into 1-cell stage embryos from at3∆90 or at3∆22 heterozygous incrosses. Fluorescent embryos were raised to adulthood or selected for laser-mediated endothelial injury.
In vitro binding studies
Zebrafish at3 cDNA was cloned into the BamHI site of pcDNA3.1V5/HIS (Invitrogen) to produce pcDNA3.1-zat3. HEK 293 cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in 5% CO2. The cells were transiently transfected with pcDNA3.1-zat3 using polyethylenimine (Sigma-Aldrich), incubated in serum-free DMEM for 36 hours, and lysed in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, and 0.5% Nonidet P40. Recombinant protein was purified using anti-V5 beads (Invitrogen), mixed with 10 µg of human thrombin (Sigma-Aldrich), incubated at 4°C for 2 hours, and washed 4 times with lysis buffer. Cell lysates and beads were boiled and separated on a 4% to 20% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel (Bio-Rad). Western blotting was performed using anti-human thrombin antibody (Affinity Biologicals) as described in the next section.
Adult blood collection and western blotting
Adult fish blood was collected in EDTA-coated microcapillary tubes (Drew Scientific Inc) as described (see supplemental Methods for a detailed description).30,31 Plasma was obtained by centrifugation at 1000g for 10 minutes at room temperature, and stored at −80°C. Two microliters from pooled wild-type or at3∆90/∆90 fish were boiled for 5 minutes in sample buffer (Bio-Rad) and β-mercaptoethanol (Sigma-Aldrich), resolved on 4% to 20% SDS-PAGE gels, and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked in 5% nonfat milk in 1% Triton in Tris-buffered saline at 4°C overnight and incubated with zebrafish anti-fibrinogen antibody for 1 hour at room temperature, washed, incubated with horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology), washed again, and developed with chemiluminescent substrate (SuperSignal West Femto; Thermo-Scientific), and scanned with FluorChem (ProteinSimple).
In situ hybridization
RNA in situ hybridization was carried out as described.32 Probes were generated from total zebrafish cDNA using primers with SP6 and T7 overhangs, followed by in vitro transcription with digoxygenin-labeled nucleotides.
Thrombin injection
Four milliunits of bovine thrombin (Sigma-Aldrich) were retro-orbitally infused33 into 4 dpf larvae from at3∆90/+ intercrosses. Phenotypes were collected 2 minutes postinjection by observers blinded to genotype. After phenotyping, larvae were lysed and genotyped (see supplemental Methods for a detailed description).
Laser-mediated endothelial injury
Endothelial injury was executed using a pulsed nitrogen dye laser and focusing system (MicroPoint; Andor Technology) as described.14,17 Larvae (3-4 dpf) were anesthetized in tricaine, embedded in 0.8% low-melt agarose on glass cover slips, and visualized on an inverted microscope (Olympus IX71, 40× objective). The posterior cardinal vein (PCV) endothelium was targeted and laser-ablated at the fifth somite distal from the anal pore at power level 18 using 100 pulses. The time to occlusion was recorded up to 2 minutes. Larvae were recovered from agarose and genotyped.
Fibrinogen injection and ε-aminocaproic acid treatment
Offspring from at3∆90/+ and at3∆90/∆90 crosses were either infused with 50 ng (2 nL) human fibrinogen or bovine serum albumin (Sigma-Aldrich) in saline at 3 dpf, or preincubated with 0.1M ε-aminocaproic acid at 2 dpf, and laser injury performed at 60 minutes postinjection or 24 hours postincubation, respectively, followed by genotyping.
FITC-fibrinogen labeling and infusion
Human fibrinogen was labeled with fluorescein isothiocyanate (FITC; Thermo-Scientific), PD-10 column purified, and dye-to-protein molar ratio determined by manufacturer’s instructions. Eight to 20 ng of a 1:1 molar ratio of FITC:fibrinogen was infused at 3 dpf.
Embryos were incubated in 50 µg/mL warfarin (Sigma-Aldrich) for 48 hours prior to FITC-fibrinogen infusion, or larvae infused with 1 mg/mL tissue plasminogen activator (tPA; Alteplase) via retro-orbital injection. At 3 dpf, larvae were infused with FITC-fibrinogen and evaluated 1 hour postinjection or postinfusion with tPA, and images were acquired with a charge-coupled device camera. Fluorescence intensity was determined either qualitatively by a blinded observer or quantitatively using Scion/ImageJ imaging software.34 Corrected total cell fluorescence was integrated density after subtraction of background (area selected × mean background fluorescence).35
RNA isolation, generation of cDNA, and reverse transcription PCR
Larvae (3 dpf) were anesthetized in tricaine and biopsied as described36 for genotyping. Larvae were maintained in RNAlater (Ambion) and 15 of each genotype pooled for RNA extraction. Total RNA was isolated using the Purelink RNA isolation kit (Ambion), and reverse transcribed with Superscript Synthesis (Invitrogen). cDNAs were amplified (MyiQ; Bio-Rad) using SYBR Green Master Mix (Applied Biosystems) at 95°C for 10 minutes, 40 cycles of 95°C for 30 seconds, 55°C for 30 seconds. Each was analyzed in triplicate and normalized to 18S ribosomal RNA, and expression presented as fold changes (ΔΔ cycle threshold [ΔΔCt]).37
Histologic examination
Humanely killed adult zebrafish were fixed in 4% buffered formalin overnight at room temperature, bisected in the midsagittal plane, embedded in paraffin, and sagittal sections produced at least 50 µm apart. Sections (3-4 µm) were hematoxylin and eosin stained.
Statistical analysis
Analyses were performed using χ2, Mann-Whitney U, Fisher exact, or 2-tailed Student t tests. Log-rank testing of survival curves was performed using Prism (GraphPad Software). For statistical comparisons between individual transgenic rescue experiments, results from homozygous mutants were normalized to the median of their heterozygous clutchmates. P values were calculated with the Mann-Whitney U test by comparing the normalized data, followed by Bonferroni correction.
Results
Targeted disruption of at3 using genome editing nucleases results in adult lethality in association with massive intracardiac thrombosis
We used OPEN19,22,23 to produce 4 ZFN pairs designed to cleave exon 5 of the zebrafish at3 genomic locus (zebrafish nomenclature guidelines dictate use of “at3” for the gene, “At3” for protein38). RNA encoding each ZFN pair was injected into the cytoplasm of single-cell embryos, with 1 pair producing somatic mutations (Figure 1A) at a frequency of 1.6% (3 of 192 sequenced clones from 2 pools of 8 embryos). The remaining embryos were raised to adulthood, and 46 of these F0 fish were mated to wild-type fish to confirm germline transmission. We identified 4 founders with the following mutations: 4-bp insertion (at3+4), 6-bp deletion (at3∆6), 22-bp deletion (at3∆22), and 90-bp deletion (at3∆90) (Figure 1B). at3+4, at3∆22, and at3∆90 produced frameshifts in exon 5 and are predicted to result in null alleles (the 90-bp deletion removes the splice donor site) (supplemental Table 3). at3∆22 and at3∆90 heterozygotes were incrossed, with similar results for both mutants. Genotyping between 2 and 24 dpf revealed the expected Mendelian distribution (Table 1) with no visible phenotypes. However, beginning at ∼2 months postfertilization (mpf), we observed a significant loss of homozygotes (Table 1), with ∼80% dying between 2 and 7 mpf (Figure 1C). Approximately 5% of at3−/− mutants had externally visible hemorrhage (Figure 1D). Reverse transcription PCR (RT-PCR) indicated reduction of at3 expression in heterozygotes, and absence of expression in homozygous mutants, presumably due to nonsense-mediated decay39 (Figure 1E-F).
Table 1.
Genotyping results of heterozygous incrosses from mutant line at3Δ90
| Line | Age | +/+ | +/− | −/− | P (χ2) |
|---|---|---|---|---|---|
| Δ90 | 12 dpf | 18 | 49 | 23 | 0.53 |
| 24 dpf | 20 | 43 | 21 | 0.96 | |
| 2 mpf | 55 | 94 | 24 | 0.002 | |
| 3 mpf | 57 | 101 | 23 | 5.0E-04 | |
| 4 mpf | 90 | 209 | 22 | 2.4E-13 |
To rule out potential off-target ZFN mutations, we injected at3∆22 incrosses with an at3 cDNA under control of the zebrafish ubiquitin (ubi) promoter (ubi-at3)26. After 1 year, homozygotes displayed significantly greater survival compared with uninjected at3−/− fish (Figure 1G).
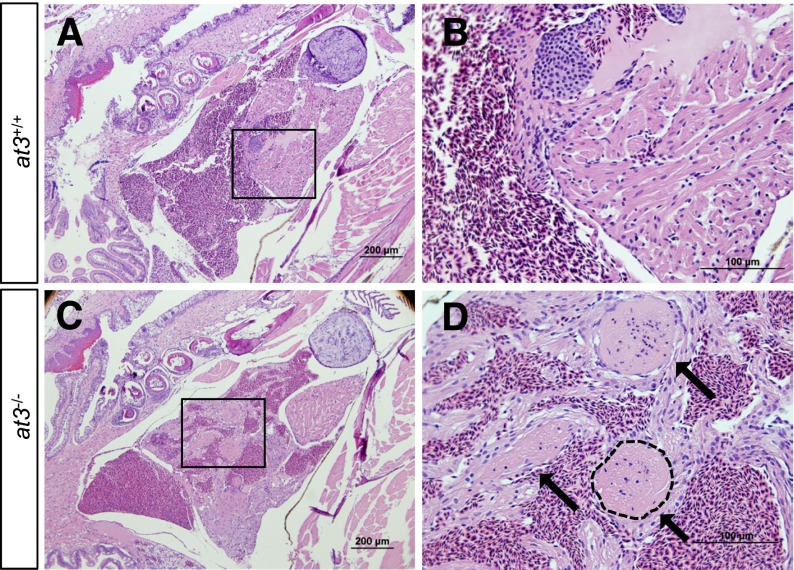
Histologic examination of surviving at3−/− fish at 2 to 3 mpf revealed large intracardiac thrombi in all homozygous mutants examined (n = 10), but not in wild-type (n = 10) or heterozygous mutants (n = 10) (Figure 2A-D). Notably, evaluation of a ubi-at3 rescued homozygous at3∆22 mutant found no evidence of thrombosis at 16 mpf (supplemental Figure 3).
Figure 2.
Loss of At3 results in adult intracardiac thrombosis. (A-D) at3−/− fish die in association with large intracardiac thrombi. H&E-stained cardiac sections from wild-type clutchmates (A-B) and at3Δ90 homozygous mutants (C-D). The boxed regions in panels A and C are shown at higher magnification in panels B and D, respectively. Sections were photographed with an Olympus BX-51 upright light microscope using an Olympus DP-70 digital camera. Three large thrombi present in panels C and D are indicated by arrows in panel D. One thrombus is outlined. Scale bar 200 µm (A,C), 100 µm (B,D). H&E, hematoxylin and eosin.
at3 homozygous mutants display an increased time to occlusion in an induced model of venous thrombosis
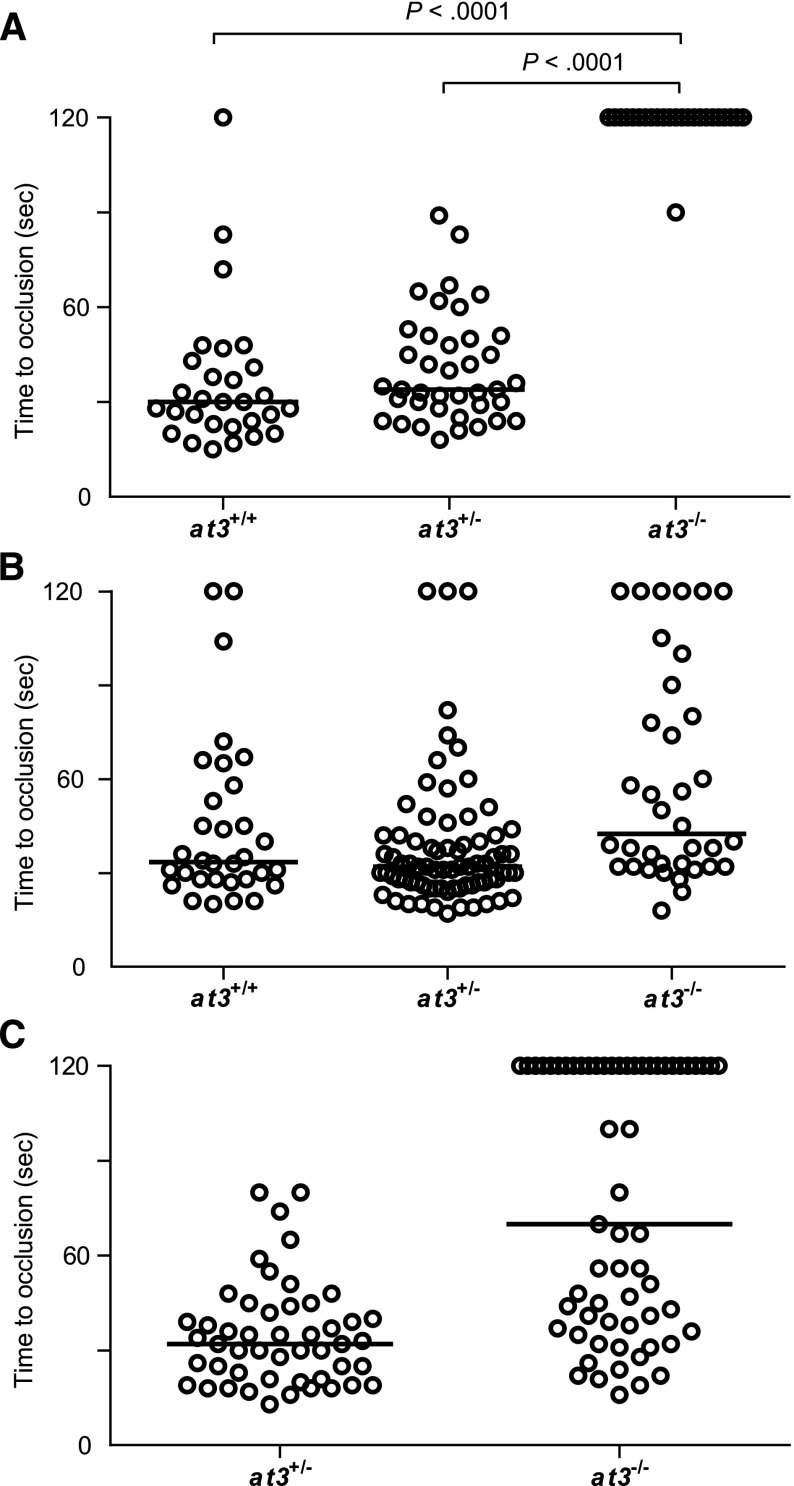
Complete deficiency of AT3 in mice and humans results in lethal thrombosis in utero.1,5 Although loss of zebrafish At3 resulted in apparently lethal thrombosis in adults, embryos and larvae appeared grossly normal with no overt evidence of pathologic thrombosis. We performed laser injury14,17 at 3 to 4 dpf on larvae derived from at3+/− intercrosses, targeting the posterior cardinal vein (PCV) (orthologous to the mammalian inferior vena cava; supplemental Figure 4 and supplemental Video). As expected, wild-type and at3+/− larvae exhibited vessel occlusion within 2 minutes, but at3−/− mutants failed to occlude within that time period (Figure 3A). To confirm the role of At3 deficiency in this bleeding phenotype, we injected ubi regulated human and zebrafish at3 cDNAs into single-cell embryos of wild-type and at3−/− clutchmates, followed by laser injury at 3 dpf. Fifty-two and 83% of human and zebrafish transgene-injected embryos, respectively, demonstrated a time to occlusion of <2 minutes (P < .0001 by the Mann-Whitney U test, Figure 3B-C).
Figure 3.
Loss of At3 prevents endothelial injury induced venous occlusion. The endothelium was targeted and injured at 3 dpf, and the time to occlusion measured in seconds with a maximum of 120. All data shown are from Δ90 mutants. (A) Time to occlusion of larvae derived from at3+/− incrosses at 3 dpf (at3−/−, n = 16). Results were similar for at3Δ22 mutants. (B) Time to occlusion of larvae (3 dpf) derived from at3+/− incross progeny injected at the 1-cell stage with the zebrafish at3 cDNA transgene (at3−/−, n = 36) is significantly different from homozygotes in panel A, P < .0001. (C) Time to occlusion of larvae (3 dpf) derived from at3+/− × at3−/− progeny injected at the 1-cell stage with the human at3 cDNA transgene (at3−/−, n = 61) is significantly different from homozygotes in panel A, P < .0001. Circles represent individual larvae. Data are from at least 3 independent experiments. Statistical significance was determined by the Mann-Whitney U test. Horizontal bars represent the median time to occlusion.
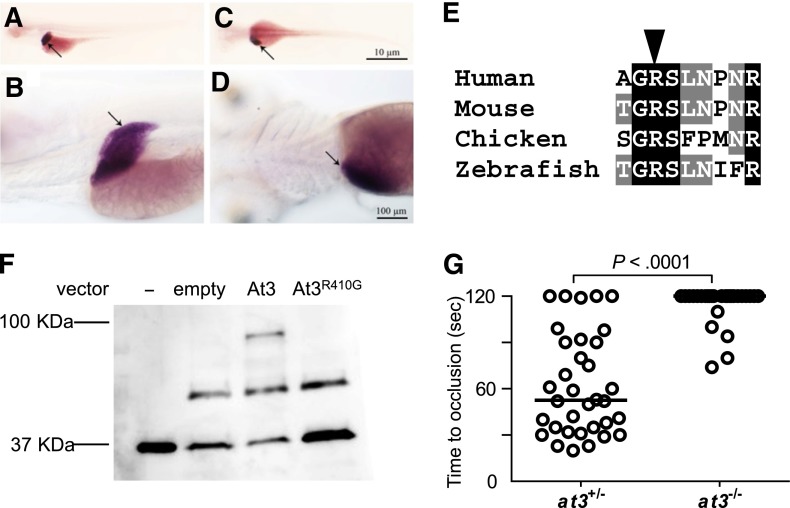
The unexpected prolongation of clotting time in at3−/− larvae suggested the possibility of differential localization of zebrafish At3 compared with its mammalian ortholog. However, in situ hybridization at 5 dpf revealed strong mRNA expression localized predominantly to the liver, consistent with mouse and human AT340,41 (Figure 4A-D). Minor expression of At3 has been noted in the notochord and neural tube of embryonic day 10.5 mice,42 whereas early zebrafish embryos demonstrate weak yolk syncytial layer expression from 16 through 48 hours postfertilization.43 Alignment with human AT3 revealed that the RCL was highly conserved across species (supplemental Figure 2). The orthologous P1 arginine was identified (Figure 4E), substituted with glycine (zebrafish Arg410Gly, orthologous to human Arg393), and assessed for binding to human thrombin in vitro (Figure 4F). Although wild-type At3 exhibited binding to thrombin and formed a putative thrombin-antithrombin complex, this interaction was absent in reactions with At3R410G (Figure 4F). To prove that the mutant phenotype was dependent on known serine protease activity, an At3R410G expressing cDNA was injected into at3−/− embryos. Consistent with the in vitro–binding studies, At3R410G failed to rescue the at3−/− larval bleeding phenotype (Figure 4G). Taken together, these data strongly suggest that the observed at3−/− larval phenotype is due to loss of anticoagulant activity.
Figure 4.
Conservation of zebrafish At3 expression and function. (A-D) In situ hybridization for at3 mRNA demonstrates liver-specific expression in 5 dpf larvae. Larvae were photographed with an Olympus BX-51 upright light microscope using an Olympus DP-70 digital camera. (A-B) Left lateral view at low and high magnification, respectively. (C-D) Dorsal view at low and high magnification, respectively. Arrow indicates the liver. Scale bar, 10 µm (C), 100 µm (D). In situ hybridization with a sense strand probe did not show any signal (not shown). (E) Alignment of the region flanking the AT3 P1 arginine (arrowhead). (F) Western blot analysis (using anti-human thrombin antibody) on recombinant At3 pulled down with human thrombin after expression in HEK 293 cells. Thrombin input only (−), pCDNA3.1 without insert (empty), pCDNA3.1zat3 (At3), and pCDNA3.1zat3R410G (At3R410G). The intermediate band present at ∼50 kDa is an unknown protein present in the cell lysate and recognized by the antibody, but unrelated to expression of At3. (G) Time to occlusion of larvae (3 dpf) derived from at3+/− × at3−/− progeny injected at the 1-cell stage with the zebrafish At3R410G expressing cDNA transgene regulated by the ubi promoter (at3−/−, n = 34). Horizontal bars represent the median of time to occlusion.
Hypofibrinogenemia in at3 homozygous mutants explains the increased time to occlusion in larvae
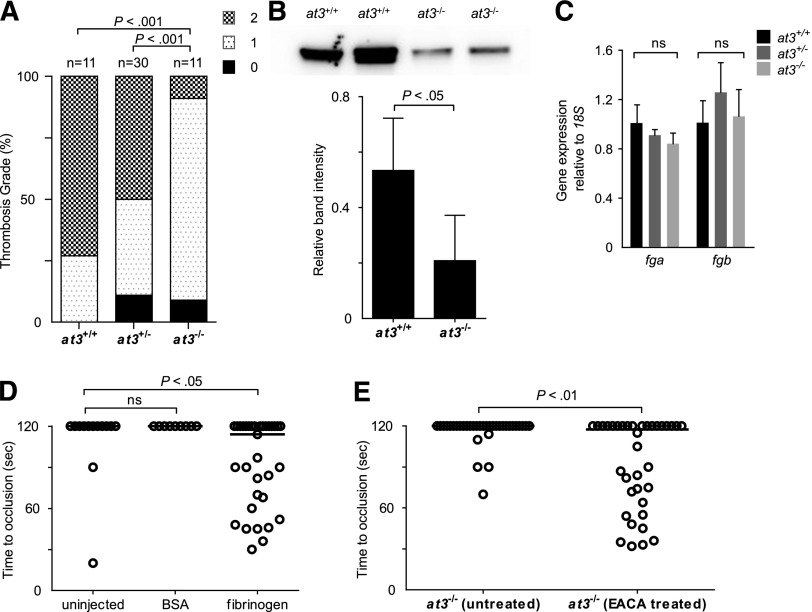
To explain the paradoxical bleeding of at3−/− larvae, we hypothesized that loss of At3 function results in unregulated procoagulant activity, leading to consumption of fibrinogen, and lack of occlusion after endothelial injury. We infused activated bovine thrombin into larvae to evaluate relative concentrations of clottable fibrinogen at 3 to 4 dpf.33 We observed a significant decrease in the number of larvae with severe vascular obstruction (9% of injected at3−/− vs 73% and 50% of at3+/+ and at3+/−, respectively) (Figure 5A), data consistent with lower levels of fibrinogen in at3−/− mutants. Furthermore, quantification of fibrinogen antigen in adult plasma revealed a 68% decrease in at3−/− relative to at3+/+ (P < .05, Figure 5B).
Figure 5.
Evidence for fibrinogen consumption in at3 homozygous mutant larvae and adults. All data shown are from Δ90 mutants. (A) Thrombin injection of larvae derived from at3+/− incrosses at 4 dpf. Bar graph represents percentage of each thrombosis grade. Grade 0, normal flow within the PCV and dorsal aorta; grade 1, partial obstruction of flow in the PCV and dorsal aorta due to thrombus formation; grade 2, complete obstruction of flow. Statistical significance was determined by Fisher exact test. (B) Top: Western blot analysis of fibrinogen in adult plasma isolated from at3+/+ and at3−/− clutchmates. Bottom: Quantification of fibrinogen in plasma from at3+/+ and at3−/− clutchmates from 3 separate experiments, n = 4 per genotype. Error bars represent standard deviation and statistical significance was determined by a paired t test. (C) Quantitative real-time PCR demonstrates no significant difference of fga and fgb mRNA expression in at3+/+ and at3−/− clutchmates (3 dpf, n = 15 per genotype). Error bars represent standard deviation and statistical significance was determined by a paired t test. (D) Rescue of time to laser-induced occlusion by human fibrinogen injection at 3 dpf. at3−/− mutant clutchmates were injected with human fibrinogen or BSA. Uninjected (n = 13), BSA injection (n = 9), and human fibrinogen injection (n = 33). (E) Effects of EACA (100 mM) pretreatment on at3−/− larvae. Untreated (n = 34), EACA treated (n = 36). Circles represent individual larvae. Statistical significance was determined by the Mann-Whitney U test. Horizontal bars represent the median time to occlusion. BSA, bovine serum albumin; EACA, ε-aminocaproic acid; ns, nonsignificant.
Examination of fibrinogen α (fga) and β (fgb) expression at 3 dpf demonstrated that there were no differences in mRNA accumulation (Figure 5C). Infusion with human fibrinogen prior to laser injury at 3 dpf rescued the bleeding phenotype in 52% of at3−/− larvae (Figure 5D, P < .05 by Mann-Whitney U test when compared with uninjected control). Treatment of larvae prior to laser injury with the fibrinolysis inhibitor ε-aminocaproic acid also rescued the at3−/− bleeding phenotype (Figure 5E, P < .01).
Hypofibrinogenemia in at3−/− mutants is secondary to DIC
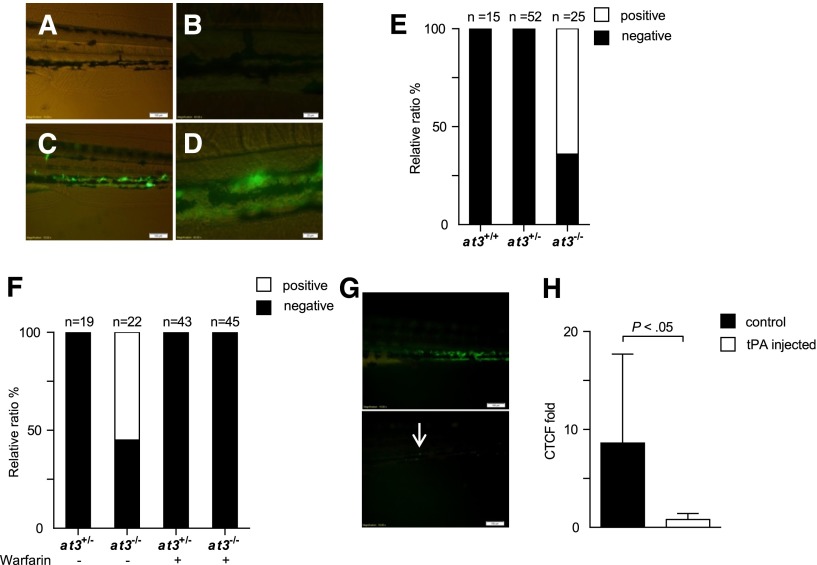
We hypothesized that the mechanism underlying the discrepant adult thrombotic and larval bleeding phenotypes in at3 mutants was DIC, with microscopic fibrin deposition. To identify the location of purported fibrin, human fibrinogen was labeled with FITC44 and infused into at3+/− intercross progeny. Widespread PCV fluorescence was observed in at3 homozygous mutant larvae after infusion, but absent in wild-type and heterozygous siblings (Figure 6A-E). Warfarin prevented laser-induced thrombosis in wild-type larvae (supplemental Figure 5), and pretreatment of at3−/− larvae eliminated fluorescence accumulation (Figure 6F). Coinjection of tPA partially prevented accumulation, as did postinjection (Figure 6G-H). Taken together, these data support the presence of DIC with consumption of fibrinogen in at3−/− mutants.
Figure 6.
at3−/− mutants display fibrin deposition, consistent with DIC. Fluorescent deposits were observed in the PCV in at3−/− mutant larvae after FITC-labeled fibrinogen injection. All data shown are from Δ90 mutants and were collected by an observer blinded to genotype and/or treatment. (A-B) Images of at3+/+ and (C-D) at3−/− larvae postinjection. Scale bars: (A,C), 100 µm; (B,D), 20 µm. (E) Percentage of larvae identified without (black) or with fluorescent PCV deposits (white), derived from at3+/− incrosses or at3+/− × at3−/−. Data are from 3 separate experiments. (F) Effects of warfarin pretreatment on at3−/− larvae prior to FITC-labeled fibrinogen injection demonstrate that fluorescence deposition is inhibited by anticoagulation. (G) Effects of tPA on at3−/− larvae post FITC-labeled fibrinogen injection. Presence of fluorescent venous deposits seen prior to injection of tPA (top) were significantly diminished post tPA injection (bottom). Scale bars: 100 µm. Arrow indicates minor residual fluorescence. (H) Quantification of CTCF of at3−/− larvae post tPA injection. Control, n = 7; tPA injected, n = 6. Error bars represent standard deviation and statistical significance was determined by an unpaired t test. Larvae were visualized on an inverted microscope Olympus IX71 with 40× objective and images were acquired with a charge-coupled device camera (Olympus DP72, cellSens software 1.3). CTCF, corrected total cell fluorescence.
Evaluation of human AT3 mutations in vivo
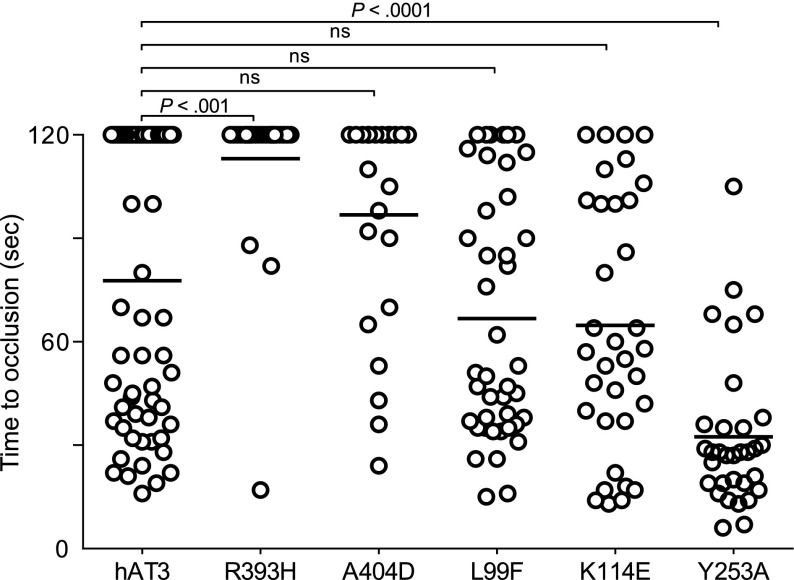
We examined several additional well-described human AT3 mutations associated with thrombosis, R393H,45-49 A404D,8 L99F,50,51 K114E,52,53, as well as a variant with greatly reduced anti-IXa and anti-Xa activity, Y253A54 (Figure 7). The P1 arginine mutant R393H was unable to reverse the DIC in at3−/− larvae. Surprisingly, the RCL mutant A404D and heparin-binding mutants L99F and K114E all demonstrated significant rescue, although the former was less potent than the latter two. Human AT3 Y253A also reversed the bleeding phenotype and was significantly more efficient when compared with human AT3 (P < .001).
Figure 7.
Effect of human AT3 substitutions in vivo. One-cell embryos were injected with individual wild-type or mutant human AT3 cDNAs, under regulation of the ubi promoter. At 3 dpf, the endothelium was targeted and the time to occlusion measured by a blinded observer. All larvae were derived from at3+/Δ90 incrosses or at3+/Δ90 × at3Δ90/Δ90. Data from at3Δ90/Δ90 mutants injected with wild-type human AT3 cDNA were compared with at3Δ90/Δ90 injected with mutant human AT3 cDNAs, R393H (at3Δ90/Δ90, n = 25), A404D (at3Δ90/Δ90, n = 23), L99F (at3Δ90/Δ90, n = 42), K114E (at3Δ90/Δ90, n = 14), Y253A (at3Δ90/Δ90, n = 32). For statistical analysis, data were normalized to heterozygous clutchmates and significance determined by the Mann-Whitney U test followed by Bonferroni correction (see “Methods”). Horizontal bars represent the median of time to occlusion. ns, nonsignificant.
Discussion
We report targeted mutagenesis of zebrafish at3 via ZFNs, one class of genome editing nucleases. Our data demonstrate that zebrafish survive complete loss of At3 with rampant DIC in the embryonic and larval periods, but apparently succumb to massive intracardiac thrombosis in adulthood. Concordant with our data, embryonic At3−/− mutant mice display a consumptive coagulopathy with secondary hemorrhage, although this causes in utero thrombosis and subsequent demise.5 It is notable that in both the complete mouse knockout and a heparin-binding mutant knockin (Arg48Cys),55 the heart is one of the most prominent sites of spontaneous thrombosis. The structure of the adult zebrafish heart has been described in detail,56 and overall cardiovascular development is highly comparable to mammalian and avian species. Taken together, our data support the contention that although the timing of ultimate lethal thrombosis is different between zebrafish and mammals, the underlying mechanisms of the coagulation cascade are conserved.
Our studies indicate that loss of At3 leads to dysregulation of coagulation, resulting in consumption of fibrinogen and prolonged clotting times in at3 homozygous mutant larvae. Although there were clearly visible spontaneous macrovascular thrombi in adult fish, injection of fluorescent fibrinogen identified disseminated microvascular thrombi in larvae, the latter consistent with clinical DIC. In patients, DIC is characterized by unregulated and excessive thrombin generation, resulting in fibrin formation with secondary plasminogen activation and fibrinolysis.57 Clinical bleeding and/or thrombosis develops depending on the balance between thrombin and plasmin generation. In at3−/− larvae, ε-aminocaproic acid, an inhibitor of plasmin, rescued the bleeding phenotype, demonstrating conservation of the fibrinolytic system. This also signifies possible contributions of fibrinolysis in this model, consistent with human DIC. These data demonstrate that loss of At3 results in unopposed procoagulant activity and possible secondary fibrinolytic activity, confirming a conserved role for zebrafish At3 as a key regulator of blood coagulation. Notably, extensive intracranial and subcutaneous hemorrhage was observed in mouse At3−/− embryos, without detectable accumulation of fibrinogen in those tissues.5 The authors posited that a consumptive coagulopathy depleted fibrinogen from the plasma, resulting in the observed bleeding, as we discerned in at3−/− mutants. Similarly, Proc knockout mice displayed intracranial hemorrhage in addition to thrombi in the heart and other organs, and clottable fibrinogen was undetectable.2 Recently, fga was targeted in zebrafish using ZFNs, and the authors demonstrated complete absence of fibrinogen.58 They did not observe specific bleeding in homozygous mutant larvae, but did note sporadic adult hemorrhage, both consistent with our data (Figure 1D). In contrast, they only detected a partial reduction in survival of homozygous mutants, supporting our data that suggest at3−/− mutants die of thrombosis rather than hemorrhage.
The observation that spontaneous, nonocclusive thrombosis due to DIC is so well tolerated in fish embryos and larvae raises important questions about the role of the coagulation system in vertebrate development. It is tempting to speculate that additional biological mechanisms may play a role, either through established clotting or other pathways. There may be protective species-specific factors for critical determinants of hemostatic balance, and the ability to perform high-throughput genetic screens in the zebrafish could lead to the identification of novel modifying factors.18 Another powerful application for zebrafish is small-molecule screening, which has led to the rapid clinical development of a novel therapeutic approach for cord blood transplant.59-61 Screens for novel anticoagulants or modulators of DIC might also have the potential to lead to innovative therapeutic modalities.
Alignment of human to zebrafish At3 reveals 55% identity and 70% similarity (supplemental Figure 2 and Kumar et al62). Because human AT3 rescued the prolonged time to occlusion in our endothelial injury assay, we performed semiquantitative evaluation of various human mutations and targeted substitutions. Substitution of the RCL P1 arginine (Arg393His) has been identified in several individuals and families with thrombophilia,45-49 and has been shown to alter AT3 function in vitro.63 Arg393His abolished AT3 activity, confirming the results of biochemical analysis as well as demonstrating the ability of this in vivo model to evaluate human coagulation factor variants. Ala404Asp, a substitution in the RCL with pleiotropic effects that alters interactions with both thrombin and heparin,8 demonstrated statistically significant rescue, although the average time to occlusion was longer than wild-type AT3. Leu99Phe,50,51 identified in patients with venous thromboembolism, demonstrated rescue consistent with wild-type AT3, as did another heparin-binding mutant Lys114Glu.52,53 The fact that heparin binding appears disposable was surprising, but consistent with human clinical data. Although mutations that abolish protease inhibition are sufficient to increase thrombosis risk in the heterozygous state, heparin-binding site mutations only manifest in homozygotes.64 Human Tyr253 is a critical residue of an AT3 IXa- and Xa-binding exosite.54 Surprisingly, AT3Tyr253Ala rescued the at3−/− coagulopathy with a greater potency than wild type. Although only semiquantitative, these data suggest that proper hemostatic balance primarily depends on thrombin inhibition. Also, AT3Tyr253Ala exhibits enhanced rates of thrombin inhibition in the presence of heparin,54 which could explain the increased activity. Taken together, these data provide novel insights into AT3 function in vivo. Our approach establishes a rapid and robust new paradigm for validating human genetic variants associated with coagulation disorders, as well as complements biochemical and structure/function studies.
Acknowledgments
The authors thank Drs Shawn Xu and Xiaochun Yu for use of MicroPoint Laser Systems, the University of Michigan Sequencing Core, and Dr Jonathan McHugh for histology review. The authors would especially like to thank Dr David Ginsburg for support and critical reading of the manuscript.
This work was supported by American Heart Association #0675025N, a Hemostasis and Thrombosis Research Society Mentored Research Award, Eunice Kennedy Shriver National Institute of Child Health and Human Development HD028820 (J.A.S.), and National Institutes of Health R01 GM088040 (J.K.J.). Statistical analysis was supported by the Charles Woodson Fund for Clinical Research.
J.A.S. is the Diane and Larry Johnson Family Scholar of Pediatrics and Communicable Diseases.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Y.L. designed and performed research, analyzed data, and wrote the manuscript; C.A.K. designed and performed research, and analyzed data; M.L.M. and J.K.J. designed and performed research; C.E.R., P.T., M.C.H., A.H.V., T.R., and Z.H. performed research; R.M. analyzed data; S.T.O. designed research and analyzed data; and J.A.S. designed, performed, and supervised research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. The remaining authors declare no competing financial interests.
The current affiliation for M.L.M. is Editas Medicine, Cambridge, MA. The current affiliation for A.H.V. is Graduate Program in Molecular Biosciences, University of Chicago, Chicago, IL.
Correspondence: Jordan Shavit, Department of Pediatrics, University of Michigan, Room 8301 Medical Science Research Building III, 1150 W Medical Center Dr, Ann Arbor, MI 48109-5646; e-mail: jshavit@umich.edu.
References
- 1.Shavit JA, Ginsburg D. Hemophilias and Other Disorders of Hemostasis. In: Rimoin DL, Pyeritz RE, Korf B, editors. Emery and Rimoin’s Principles and Practice of Medical Genetics. 2013. pp. 1–33. [Google Scholar]
- 2.Jalbert LR, Rosen ED, Moons L, et al. Inactivation of the gene for anticoagulant protein C causes lethal perinatal consumptive coagulopathy in mice. J Clin Invest. 1998;102(8):1481–1488. doi: 10.1172/JCI3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burstyn-Cohen T, Heeb MJ, Lemke G. Lack of protein S in mice causes embryonic lethal coagulopathy and vascular dysgenesis. J Clin Invest. 2009;119(10):2942–2953. doi: 10.1172/JCI39325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saller F, Brisset AC, Tchaikovski SN, et al. Generation and phenotypic analysis of protein S-deficient mice. Blood. 2009;114(11):2307–2314. doi: 10.1182/blood-2009-03-209031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishiguro K, Kojima T, Kadomatsu K, et al. Complete antithrombin deficiency in mice results in embryonic lethality. J Clin Invest. 2000;106(7):873–878. doi: 10.1172/JCI10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huntington JA. Serpin structure, function and dysfunction. J Thromb Haemost. 2011;9(suppl 1):26–34. doi: 10.1111/j.1538-7836.2011.04360.x. [DOI] [PubMed] [Google Scholar]
- 7.Olson ST, Gettins PG. Regulation of proteases by protein inhibitors of the serpin superfamily. Prog Mol Biol Transl Sci. 2011;99:185–240. doi: 10.1016/B978-0-12-385504-6.00005-1. [DOI] [PubMed] [Google Scholar]
- 8.Luxembourg B, Delev D, Geisen C, et al. Molecular basis of antithrombin deficiency. Thromb Haemost. 2011;105(4):635–646. doi: 10.1160/TH10-08-0538. [DOI] [PubMed] [Google Scholar]
- 9.Hanumanthaiah R, Day K, Jagadeeswaran P. Comprehensive analysis of blood coagulation pathways in teleostei: evolution of coagulation factor genes and identification of zebrafish factor VIIi. Blood Cells Mol Dis. 2002;29(1):57–68. doi: 10.1006/bcmd.2002.0534. [DOI] [PubMed] [Google Scholar]
- 10.Davidson CJ, Tuddenham EG, McVey JH. 450 million years of hemostasis. J Thromb Haemost. 2003;1(7):1487–1494. doi: 10.1046/j.1538-7836.2003.00334.x. [DOI] [PubMed] [Google Scholar]
- 11.Davidson CJ, Hirt RP, Lal K, et al. Molecular evolution of the vertebrate blood coagulation network. Thromb Haemost. 2003;89(3):420–428. [PubMed] [Google Scholar]
- 12.Jiang Y, Doolittle RF. The evolution of vertebrate blood coagulation as viewed from a comparison of puffer fish and sea squirt genomes. Proc Natl Acad Sci USA. 2003;100(13):7527–7532. doi: 10.1073/pnas.0932632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fish RJ, Vorjohann S, Béna F, Fort A, Neerman-Arbez M. Developmental expression and organisation of fibrinogen genes in the zebrafish. Thromb Haemost. 2012;107(1):158–166. doi: 10.1160/TH11-04-0221. [DOI] [PubMed] [Google Scholar]
- 14.Vo AH, Swaroop A, Liu Y, Norris ZG, Shavit JA. Loss of fibrinogen in zebrafish results in symptoms consistent with human hypofibrinogenemia. PLoS ONE. 2013;8(9):e74682. doi: 10.1371/journal.pone.0074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrillo M, Kim S, Rajpurohit SK, Kulkarni V, Jagadeeswaran P. Zebrafish von Willebrand factor. Blood Cells Mol Dis. 2010;45(4):326–333. doi: 10.1016/j.bcmd.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh A, Vo A, Twiss BK, et al. Characterization of zebrafish von Willebrand factor reveals conservation of domain structure, multimerization, and intracellular storage. Adv Hematol. 2012;2012:214209. [DOI] [PMC free article] [PubMed]
- 17.Jagadeeswaran P, Carrillo M, Radhakrishnan UP, Rajpurohit SK, Kim S. Laser-induced thrombosis in zebrafish. Methods Cell Biol. 2011;101:197–203. doi: 10.1016/B978-0-12-387036-0.00009-8. [DOI] [PubMed] [Google Scholar]
- 18.Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122(7):2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foley JE, Yeh JR, Maeder ML, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN). PLoS ONE. 2009;4(2):e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sander JD, Cade L, Khayter C, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29(8):697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeder ML, Thibodeau-Beganny S, Osiak A, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31(2):294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4(10):1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat Protoc. 2009;4(12):1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 26.Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138(1):169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li MZ, Elledge SJ. SLIC: a method for sequence- and ligation-independent cloning. Methods Mol Biol. 2012;852:51–59. doi: 10.1007/978-1-61779-564-0_5. [DOI] [PubMed] [Google Scholar]
- 28.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32(14):e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7(1):133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Jagadeeswaran P, Sheehan JP. Analysis of blood coagulation in the zebrafish. Blood Cells Mol Dis. 1999;25(3-4):239–249. doi: 10.1006/bcmd.1999.0249. [DOI] [PubMed] [Google Scholar]
- 31.Pedroso GL, Hammes TO, Escobar TD, Fracasso LB, Forgiarini LF, da Silveira TR. Blood collection for biochemical analysis in adult zebrafish. J Vis Exp. 2012;(63):e3865. doi: 10.3791/3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 33.Kalish Y, Ghosh A, Shavit JA, Ginsburg D. Conservation of hemostatic system component function between zebrafish and mammals. Blood. 2009;114(22):1228–1229. [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappell KM, Sinnott R, Taus P, Maxfield K, Scarbrough M, Whitehurst AW. Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Mol Cell Biol. 2012;32(20):4131–4140. doi: 10.1128/MCB.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson RN, Elworthy S, Ingham PW, van Eeden FJ. A method for high-throughput PCR-based genotyping of larval zebrafish tail biopsies. Biotechniques. 2013;55(6):314–316. doi: 10.2144/000114116. [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood R. Genetic nomenclature Guide: With Information on Websites. 1998th ed. Cambridge, UK: Elsevier Trends Journals; 1998. [Google Scholar]
- 39.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13(11):700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patnaik MM, Moll S. Inherited antithrombin deficiency: a review. Haemophilia. 2008;14(6):1229–1239. doi: 10.1111/j.1365-2516.2008.01830.x. [DOI] [PubMed] [Google Scholar]
- 41.Diez-Roux G, Banfi S, Sultan M, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9(1):e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capellini TD, Dunn MP, Passamaneck YJ, Selleri L, Di Gregorio A. Conservation of notochord gene expression across chordates: insights from the Leprecan gene family. Genesis. 2008;46(11):683–696. doi: 10.1002/dvg.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thisse C, Thisse B. High throughput expression analysis of ZF-models consortium clones. ZFIN direct data submission. 2005. http://zfin.org.
- 44.Gregory M, Hanumanthaiah R, Jagadeeswaran P. Genetic analysis of hemostasis and thrombosis using vascular occlusion. Blood Cells Mol Dis. 2002;29(3):286–295. doi: 10.1006/bcmd.2002.0568. [DOI] [PubMed] [Google Scholar]
- 45.Bauer KA, Ashenhurst JB, Chediak J, Rosenberg RD. Antithrombin “Chicago”: a functionally abnormal molecule with increased heparin affinity causing familial thrombophilia. Blood. 1983;62(6):1242–1250. [PubMed] [Google Scholar]
- 46.Owen MC, Beresford CH, Carrell RW. Antithrombin Glasgow, 393 Arg to His: a P1 reactive site variant with increased heparin affinity but no thrombin inhibitory activity. FEBS Lett. 1988;231(2):317–320. doi: 10.1016/0014-5793(88)80841-x. [DOI] [PubMed] [Google Scholar]
- 47.Erdjument H, Lane DA, Panico M, Di Marzo V, Morris HR. Single amino acid substitutions in the reactive site of antithrombin leading to thrombosis. Congenital substitution of arginine 393 to cysteine in antithrombin Northwick Park and to histidine in antithrombin Glasgow. J Biol Chem. 1988;263(12):5589–5593. [PubMed] [Google Scholar]
- 48.Erdjument H, Lane DA, Panico M, et al. Antithrombin Chicago, amino acid substitution of arginine 393 to histidine. Thromb Res. 1989;54(6):613–619. doi: 10.1016/0049-3848(89)90127-8. [DOI] [PubMed] [Google Scholar]
- 49.Lane DA, Erdjument H, Flynn A, et al. Antithrombin Sheffield: amino acid substitution at the reactive site (Arg393 to His) causing thrombosis. Br J Haematol. 1989;71(1):91–96. doi: 10.1111/j.1365-2141.1989.tb06280.x. [DOI] [PubMed] [Google Scholar]
- 50.Olds RJ, Lane DA, Boisclair M, Sas G, Bock SC, Thein SL. Antithrombin Budapest 3. An antithrombin variant with reduced heparin affinity resulting from the substitution L99F. FEBS Lett. 1992;300(3):241–246. doi: 10.1016/0014-5793(92)80854-a. [DOI] [PubMed] [Google Scholar]
- 51.Kuhle S, Lane DA, Jochmanns K, et al. Homozygous antithrombin deficiency type II (99 Leu to Phe mutation) and childhood thromboembolism. Thromb Haemost. 2001;86(4):1007–1011. [PubMed] [Google Scholar]
- 52.Mushunje A, Zhou A, Huntington JA, Conard J, Carrell RW. Antithrombin ‘DREUX’ (Lys 114Glu): a variant with complete loss of heparin affinity. Thromb Haemost. 2002;88(3):436–443. [PubMed] [Google Scholar]
- 53.Picard V, Susen S, Bellucci S, Aiach M, Alhenc-Gelas M. Two new antithrombin variants support a role for K114 and R13 in heparin binding. J Thromb Haemost. 2003;1(2):386–387. doi: 10.1046/j.1538-7836.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 54.Izaguirre G, Olson ST. Residues Tyr253 and Glu255 in strand 3 of beta-sheet C of antithrombin are key determinants of an exosite made accessible by heparin activation to promote rapid inhibition of factors Xa and IXa. J Biol Chem. 2006;281(19):13424–13432. doi: 10.1074/jbc.M600415200. [DOI] [PubMed] [Google Scholar]
- 55.Dewerchin M, Hérault JP, Wallays G, et al. Life-threatening thrombosis in mice with targeted Arg48-to-Cys mutation of the heparin-binding domain of antithrombin. Circ Res. 2003;93(11):1120–1126. doi: 10.1161/01.RES.0000103634.69868.4F. [DOI] [PubMed] [Google Scholar]
- 56.Hu N, Yost HJ, Clark EB. Cardiac morphology and blood pressure in the adult zebrafish. Anat Rec. 2001;264(1):1–12. doi: 10.1002/ar.1111. [DOI] [PubMed] [Google Scholar]
- 57.Liebman HA, Weitz IC. Disseminated intravascular coagulation. In: Hoffman R, Benz EJ, Shattil SJ, Furie B, Cohen HJ, McGlave P, editors. Hoffman Hematology: Basic Principles and Practice. Philadelphia, PA: Churchill Livingstone; 2005. [Google Scholar]
- 58.Fish RJ, Di Sanza C, Neerman-Arbez M. Targeted mutation of zebrafish fga models human congenital afibrinogenemia. Blood. 2014;123(14):2278–2281. doi: 10.1182/blood-2013-12-547182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.North TE, Goessling W, Walkley CR, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goessling W, Allen RS, Guan X, et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell. 2011;8(4):445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cutler C, Multani P, Robbins D, et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood. 2013;122(17):3074–3081. doi: 10.1182/blood-2013-05-503177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar A, Bhandari A, Sarde SJ, Goswami C. Sequence, phylogenetic and variant analyses of antithrombin III. Biochem Biophys Res Commun. 2013;440(4):714–724. doi: 10.1016/j.bbrc.2013.09.134. [DOI] [PubMed] [Google Scholar]
- 63.Chuang YJ, Swanson R, Raja SM, Bock SC, Olson ST. The antithrombin P1 residue is important for target proteinase specificity but not for heparin activation of the serpin. Characterization of P1 antithrombin variants with altered proteinase specificity but normal heparin activation. Biochemistry. 2001;40(22):6670–6679. doi: 10.1021/bi002933d. [DOI] [PubMed] [Google Scholar]
- 64.van Boven HH, Lane DA. Antithrombin and its inherited deficiency states. Semin Hematol. 1997;34(3):188–204. [PubMed] [Google Scholar]