Abstract
Typically pathogens deploy virulence effectors to disable defense. Plants defeat effectors with resistance proteins that guard effector targets. Here we show that a pathogen exploits a resistance protein by activating it to confer susceptibility. Interactions of victorin, an effector produced by the necrotrophic fungus Cochliobolus victoriae, TRX-h5, a defense-associated thioredoxin, and LOV1, an Arabidopsis susceptibility protein, recapitulate the guard mechanism of plant defense. In LOV1’s absence, victorin inhibits TRX-h5 resulting in compromised defense but not disease by C. victoriae. In LOV1’s presence, victorin binding to TRX-h5 activates LOV1 and elicits a resistance-like response that confers disease susceptibility. We propose victorin is or mimics a conventional pathogen virulence effector that was defeated by LOV1 and confers virulence to C. victoriae solely because it incites defense.
Disease susceptibility and resistance are normally considered opposite plant responses to pathogen challenge. However for disease caused by the fungus Cochliobolus victoriae, susceptibility and the host resistance response appear to be one and the same (1). Most pathogens gain virulence by expressing effectors that target proteins integral to host defense. The guard model posits that plants defeat pathogen virulence by guarding effector targets with resistance (R) proteins in a process called effector-triggered immunity or R-gene resistance (2,3). The largest class of R proteins consists of nucleotide-binding-leucine rich repeat (NB-LRR) proteins related to innate immune response proteins in animals (2,3). The Arabidopsis thaliana gene LOV1 encodes a typical NB-LRR but is unique because it confers sensitivity to the fungal toxin victorin, and thus susceptibility (S) rather than resistance to Cochliobolus victoriae (1). Although LOV1 conditions disease susceptibility, it initiates a defense-like response and requires structural features identical to those of resistance-associated NB-LRRs (1,4). Additionally, LOV1 is widespread and conserved in Arabidopsis, implying that it is maintained for resistance to an unidentified pathogen (4). In support of this presumption is the original description of C. victoriae as causal to Victoria blight of oats. Victoria blight affects oats bred for single-gene (Pc2) resistance to the crown rust pathogen, Puccinia coronata (5). The locus conferring C. victoriae susceptibility, Vb, and Pc2 were never genetically resolved and are surmised to be one and the same (5). Hence, multiple lines of evidence associate susceptibility to C. victoriae with disease resistance, but mechanistic proof of this association is lacking. We propose susceptibility to C. victoriae conforms to the guard model of plant defense. We find victorin exhibits characteristics of a canonical virulence effector by targeting TRX-h5, a thioredoxin required for redox control of NPR1 (6). As a key regulator of local and systemic acquired resistance (7), NPR1 presents a conspicuous effector target. We also find that LOV1 is activated (causes cell death) when TRX-h5 binds victorin. However, activation of this NB-LRR guard (LOV1) leads to disease susceptibility instead of resistance, presumably by facilitating C. victoriae’s necrotrophic exploitation of the associated host cell death (1). Thus, victorin is an atypical virulence effector because it confers virulence by evoking rather than suppressing defense.
Thioredoxins (TRXs) regulate the redox homeostasis of cells by functioning as protein disulfide oxidoreductases. TRXs contain two active-site cysteines that form an oxidized disulfide or exist as free sulfhydryls that reduce and regulate the activity of target proteins. Of the eight Arabidopsis h-type TRXs, only TRX-h5 is genetically required for victorin sensitivity (8) and induced by biotic stress (9). While TRX-h5 mutants are completely victorin-insensitive, over-expression of TRX-h3, the constitutive leaf TRX-h, can partially rescue TRX-h5 mutants (8).
Victorin sensitivity in Arabidopsis requires the TRX-h5 protein but not its enzymatic activity, as mutation of the TRX-h5 essential, active-site C42 or both thioredoxin reductases does not compromise victorin-induced cell death. Mutation of TRX-h5 at active-site C39 does abolish victorin-induced cell death (8). Because TRX-h5 but not its enzymatic activity is required for LOV1 function, and because NB-LRR proteins are known to survey host proteins for modification, we evaluated TRX-h5 for victorin-induced modifications (10). We found TRX-h5 exhibits an ~1 kilodalton (kD) shift in electrophoretic mobility when treated with victorin in vivo (Fig 1). Victorin (MW~ 1 kD) induced a similar shift to all thioredoxins tested (Fig. S1), suggesting victorin is generally reactive to thioredoxins. Furthermore, biotin-labeled victorin co-immunoprecipitated with shifted TRX-h5, indicating victorin binds TRX-h5 (Fig 1B). A covalent association of victorin with TRX-h5 was confirmed by mass spectrometry (S2). The ability of TRX-h5C42S but not TRX-h5C39S to bind victorin correlated with the respective abilities of these proteins to support victorin-induced cell death (Fig. 1A, S3), suggesting victorin binding is essential for cell death and occurs at C39. We provide several lines of evidence that this is the case. First, victorin’s aldehyde moiety is essential for both toxicity (11) and binding to TRX-h5 (Fig. 1C). Secondly, victorin binds TRX-h5 under reducing conditions (free sulphydryls), but not non-reducing conditions or in the presence of the sulfhydryl blocker N-ethyl-malamide (Fig 1D). Finally, mutation of the only cysteine in TRX-h5 (C10) other than the active-site cysteines does not affect victorin binding or induced cell death (Fig. S3). Collectively these data suggest victorin binds TRX-h5 at the active site C39 which is required for LOV1 activation.
Figure 1.
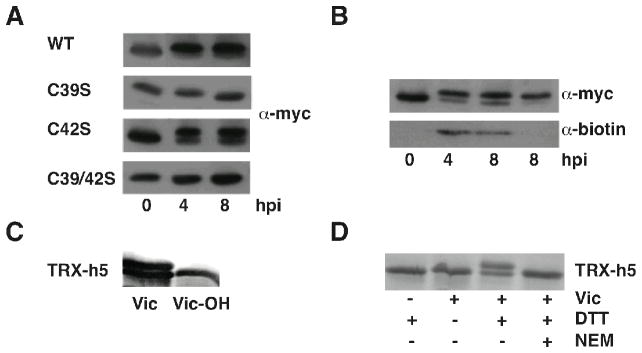
Victorin binds to TRX-h5. Immunoblot of wildtype and mutant, myc-tagged TRX-h5 proteins from Arabidopsis leaves hours after treatment (hpi) with (A) victorin, or (B) biotinylated-victorin. Proteins were detected with α-myc (A) or immunoprecipitated with α-myc and detected with α-biotin and α-myc (B). Victorin binding is seen as an ~ 1 kD increase in the apparent mass of TRX-h5. (C) Purified, His-tagged TRX-h5 incubated in vitro with 1mM DTT and 10 μM native victorin or victorin in which the aldehyde moiety has been reduced to a primary alcohol (Vic-OH). Protein detected by silver staining. (D) Purified, His-tagged TRX-h5 incubated in vitro with or without 10 μM victorin, 1 mM DTT, and 1mM NEM, and detected by Coomassie staining.
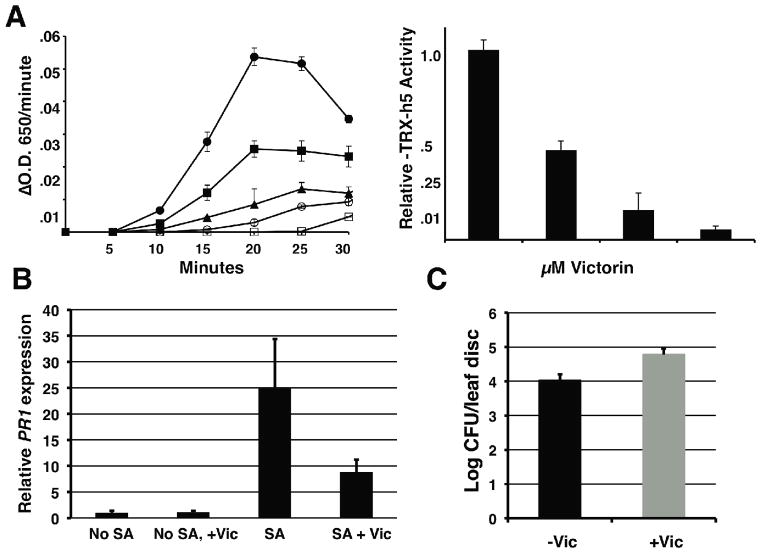
Activation of LOV1 upon victorin binding to TRX-h5 implies TRX-h5 is guarded because it plays a role in defense. One defense function of TRX-h5 is to regulate the redox state of NPR1 (6). NPR1 normally exists in the cytosol as a thiol-bound oligomer. Upon pathogen challenge or treatment with salicylic acid, TRX-h5 is upregulated (6,9). TRX-h5 then reduces NPR1 oligomers resulting in monomer release, translocation to the nucleus, and regulation of defense gene expression associated with both local and systemic resistance (6). Because victorin binds to the TRX-h5 active-site, it should inhibit its activity. We measured TRX-h5 actvity in vitro using a reduction-of-insulin assay (10) and found victorin dramatically inhibited TRX-h5 activity (Fig. 2A). Accordingly, victorin inhibition of TRX-h5 activity in planta (in the absence of LOV1) should compromise defense-gene expression conferred by NPR1. We found seedlings (lov1/lov1) treated with victorin exhibited three-fold lower salicylic-acid-induced PR1-expression (a reporter for NPR1 activation) than controls (Fig. 2B). This extent of PR1 repression is equivalent to that reported for TRX and thioredoxin reductase mutants (6). We also found victorin increased susceptibility to Pseudomonas syringae pv. maculicola (Psm) (Fig. 2C). This increase in disease susceptibility is equivalent to that displayed by an NPR1 mutant (6,7) (Fig. S4), statistically significant by T-test (P <0.001) and highly reproducible. In this capacity, and as implicated by the guard model, victorin acts as a canonical virulence effector through its inhibition of thioredoxin and subsequent repression of local resistance to Psm. Systemic application of victorin also inhibited salicylic-acid induced resistance (Fig. S5). Salicylic acid is associated with both local and systemic acquired resistance. However, because pathogens secrete effectors locally, the potential to inhibit systemic acquired resistance is difficult to place in a biological context. Interestingly, systemic acquired resistance discourages sequential infection of an individual plant and thus could provide formidable defense against pathogens that undergo polycyclic infection, such as rusts (12).
Figure 2.
Victorin inhibits thioredoxin activity. (A, left side) TRX-h5-catalyzed reduction of insulin measured by Δ OD650 of insulin (0.1% w/v) in the presence of 1mM DTT (closed circle), or 1 mM DTT and victorin, 1 μM (closed square), 5 μM (triangle), or 10 μM (open circle). Open square is insulin reduction in the presence of 1mM DTT without TRX-h5. (A, right side) Relative specific activity of TRX-h5 in the presence of 0, 1, 5 or 10 μM victorin. Error bars represent standard error from triplicate assays. (B) Quantitative RT-PCR analysis of PR1 from leaves of three-week-old Arabidopsis (lov1, lov1) grown in hydroponic solution and transferred to water or 20 μg/ml victorin 48 hrs prior to spraying with water or 1mM salicylic acid (SA). RNA was isolated 24 hours after SA treatment. Standard error bars represent N=8, 4 biological × 2 technical replicates. Data were reproducible in separate experiments. (C) Colony forming units (CFUs) of Pseudomonas syringae pv. maculicola (Psm) from three-week-old Arabidopsis (lov1, lov1) leaves 2 days after treatment with Psm M6CΔE, 0.1 OD600. Leaves were infiltrated with water or 100 μg/mL victorin 2 hrs prior to treatment with Psm. Standard error bars represent N=6 biological replicates containing 3 leaf discs each.
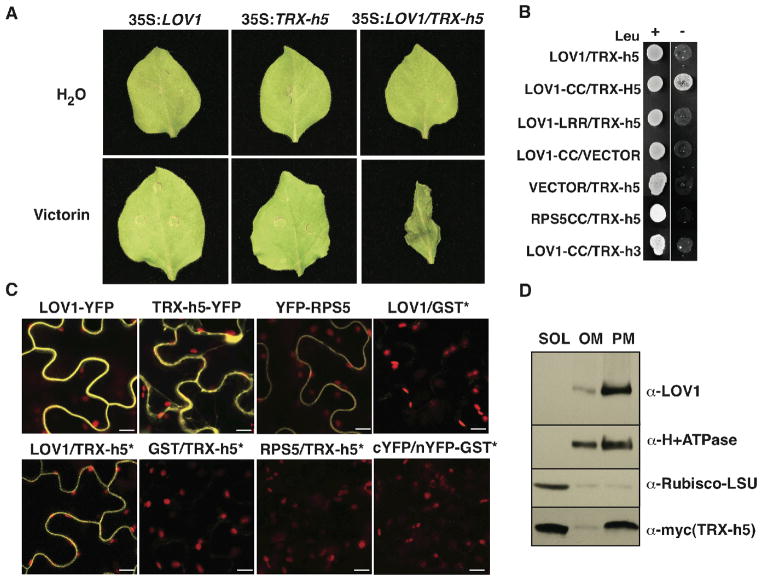
Heterologous expression of NB-LRR proteins in N benthamiana often results in NB-LRR autoactivation or activation in the presence of the cognate pathogen effector (3). Expression of only LOV1 in N. benthamiana did not result in cell death, even after treatment with victorin. However, co-expression of TRX-h5 and LOV1 conferred victorin sensitivity (Fig. 3A). Thus, LOV1 activation relies on TRX-h5, and their co-expression is both necessary and sufficient to confer victorin sensitivity. Furthermore, as in Arabidopsis (Fig. 1) (8), the ability of TRX-h5 to bind victorin is required for LOV1 activation (Fig. S3).
Figure 3.
LOV1 guards TRX-h5. (A) Transient expression of LOV1 (left), TRX-h5 (middle), or LOV1 and TRX-h5 (right) in Nicotiana benthamiana infiltrated with water (upper) or 1μg/ml victorin (lower). (B) Yeast-two-hybrid assay of LOV1, TRX-h5, and control proteins on SD/-Trp/-His medium (left) and SD/-Leu/-Trp/-His medium (right). (C) Confocal fluorescence microscopy of N. benthamiana leaves transiently expressing proteins tagged with yellow fluorescent protein (YFP) or split-YFP*, from left to right (upper); 35S:LOV1-YFP; 35S:TRX-h5-YFP; 35S:YFP-RPS5; 35S:cYFP-LOV1+35S:nYFP-GST; (lower); 35S:cYFP-LOV1+35S:nYFP-TRX-h5; 35S:cYFP-GST+35S:nYFP-TRX-h5; 35S:cYFP-RPS5+35S:nYFP-TRX-h5; 35S:cYFP+35S:nYFP-GST. Red indicates chlorophyll autofluorescence and yellow, YFP fluorescence. Scale bars equal 10 μm. (D) Immunoblot of 10 μg of soluble (SOL), other membrane (OM), or plasma membrane (PM) protein fractions from two-phase extraction of N. benthamiana transiently expressing LOV1 and myc-TRX-h5.
Because LOV1 requires TRX-h5 co-expression and is activated when TRX-h5 (or enzymatically-inactive TRX-h5) binds victorin, we explored the possibility that LOV1 and TRX-h5 interact. Yeast two-hybrid analyses demonstrated TRX-h5 interacts strongly with the LOV1 CC domain, but not the LRR domain and weakly with full-length LOV1 (Fig. 3B). In contrast, TRX-h3 interacted only weakly with the LOV1 CC domain (Fig. 3B). A fluorescent fusion of LOV1 exclusively labeled the cell periphery (Fig. 3C) and subcellular fractionation revealed that LOV1 and TRX-h5 co-purify with the plasma membrane (Fig. 3D) even though TRX-h5 predominantly occupies the cytoplasm. TRX-h5 localization to the plasma membrane was also independently corroborated by proteome analyses (13). Finally, bimolecular fluorescence complementation supported an interaction of LOV1 and TRX-h5 at the plasma membrane (Fig. 3C). Co-immunoprecipitation of LOV1 and TRX-h5 was not successful, possibly because of conditions required for solubilizing LOV1. Nonetheless, the cumulative data indicate that LOV1 and TRX-h5 interact in some manner at the plasma membrane, consistent with the model that TRX-h5 is guarded by LOV1.
The guard model accounts for plants having immunity to a myriad of pathogens, while possessing a limited number of R genes (2,3). R gene limitation is possible because effector targets are limited and pathogens, however numerous, secrete functionally redundant virulence effectors. This implies that R genes across plant species evolve to guard common targets (14). We have observed victorin sensitivity in oats, Arabidopsis, barley, rice, Brachypodium (15) and bean (Fig. S6). Because victorin binds diverse thioredoxins (Fig. S1) and sensitivity is conditioned by a NB-LRR gene (LOV1) in Arabidopsis, inseparable from the Pc2 resistance gene in oats and mapped to a genomic region rich in NB-LRR genes in barley (15), data suggest that victorin sensitivity is evoked by a common mechanism across these species: by victorin binding to a thioredoxin that is guarded by a NB-LRR protein. Given this and the important defense functions for TRXs (6), it is possible that multiple pathogens target thioredoxins to enhance virulence (i.e. redundant virulence effectors). Significantly, C. victoriae does not cause disease on Arabidopsis in the absence of LOV1 or oat in the absence of Vb (5). This is important because it implies that victorin production did not evolve in C. victoriae to inhibit TRX-h5-conferred defense. Rather, C. victoriae employs victorin solely in its capacity as a defeated effector to exploit R-gene mediated defense for disease susceptibility. This suggests other defeated effectors could confer virulence if expressed by the appropriate pathogen.
Susceptibility to three other necrotrophic pathogens has been associated with R-like genes (16,17). Given the numbers of R genes in plant genomes and defeated virulence effectors collectively deployed by biotrophic pathogens, this study underpins the importance of understanding the limits to necrotroph exploitation of effector-triggered immunity (resistance-mediated susceptibility), so that future deployment of resistance does not lead to the emergence of new disease.
Supplementary Material
Acknowledgments
The authors thank Jeff Chang and Michael Behrenfeld for valuable discussion. This work was supported in part by grants from the USDA NRI/AFRI (2005-35319-15361 and 2007-01598) and the National Science Foundation (IOS-0724954). OSU’s mass spectrometry facility and core is in part supported by a grant from the NIH/NIEHS P30ES000210.
Footnotes
References
- 1.Lorang JM, Sweat TA, Wolpert TJ. Plant Disease Susceptibility Conferred by a Resistance Gene. Proc Natl Acad Sci. 2007;37:14861–14866. doi: 10.1073/pnas.0702572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.DeYoung BJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nature Immunology. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweat TA, Lorang JM, Bakker EG, Wolpert TJ. Characterization of natural and induced variation in the LOV1 gene, a CC-NB-LRR gene conferring victorin sensitivity and disease susceptibility in Arabidopsis. Molec Plant Microbe Interac. 2008;21:7–19. doi: 10.1094/MPMI-21-1-0007. [DOI] [PubMed] [Google Scholar]
- 5.Wolpert TJ, Dunkle LD, Ciuffetti LM. Host-Selective Toxins and Avirulence Determinants: What’s in a Name? Ann Rev Phytopath. 2002;40:251–285. doi: 10.1146/annurev.phyto.40.011402.114210. [DOI] [PubMed] [Google Scholar]
- 6.Tada Y, et al. Plant Immunity Requires Conformational Changes of NPR1 via S-Nitrosylation and Thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. The Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweat TA, Wolpert TJ. Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis thaliana. Plant Cell. 2007;19:673–687. doi: 10.1105/tpc.106.047563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP. The Arabidopsis cystolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 2004;134:1006–1016. doi: 10.1104/pp.103.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Information on materials and methods is available on Science Online.
- 11.Wolpert TJ, Macko V, Acklin W, Arigoni D. Molecular features affecting the biological activity of host-selective toxins from Cochliobolus victoriae. Plant Physiol. 1988;88:37–41. doi: 10.1104/pp.88.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mundt CC. Importance of autoinfection to the epidemiology of polycyclic foliar disease. Phytopathology. 2009;99:1116–1120. doi: 10.1094/PHYTO-99-10-1116. [DOI] [PubMed] [Google Scholar]
- 13.Marmagne A, et al. A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Molecular and Cellular Proteomics. 2007;6:1980–1996. doi: 10.1074/mcp.M700099-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Wroblewski T, et al. Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol. 2009;150:1733–1749. doi: 10.1104/pp.109.140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorang J, Cuesta-Marcos A, Hayes PM, Wolpert TJ. Identification and mapping of adult-onset sensitivity to victorin in barley. Mol Breeding. 2010;26:545–550. [Google Scholar]
- 16.Nagy ED, Bennetzen JL. Pathogen corruption and site-directed recombination at a plant resistance gene cluster. Genome Res. 2008;18:1918–1923. doi: 10.1101/gr.078766.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faris JD, et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci USA. 2010;107:13544–13549. doi: 10.1073/pnas.1004090107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Early KW, et al. Gateway-compatible vectors for functional plant genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 19.Martin K, et al. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009;59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- 20.Clough SJ, Bent AF. Floral Dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 21.Holgrem A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J of Biochem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 22.Larson C, Widell S, Kjelbom P. Preparation of high-purity plasma membranes. Methods in Enzymol. 1987;148:558–568. [Google Scholar]
- 23.Rohmer L, Kjemtrup S, Marchesini P, Dangl JL. Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol Microbiol. 2003;47:1545–1562. doi: 10.1046/j.1365-2958.2003.03402.x. [DOI] [PubMed] [Google Scholar]
- 24.Gibeaut D, Hulett J, Cramer GR, Seemann JR. Maximal biomass of Arabidopsis thaliana using a simple low maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 1997;115:317–319. doi: 10.1104/pp.115.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian M, et al. Arabidopsis Actin-Depolymerizing Factor AtADF4 Mediates Defense Signal Transduction Triggered by the Pseudomonas syringae Effector AvrPphB. Plant Physiol. 2009;150:815–824. doi: 10.1104/pp.109.137604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.pEG202 was a gift from Hironori Kaminaka and Jeff J. Dangl.
- 27.Rhoads TW, et al. Measuring copper and zinc superoxide dismutase from spinal cord tissue using electrospray mass spectrometry. Anal Biochem. 2011;415:52–58. doi: 10.1016/j.ab.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.