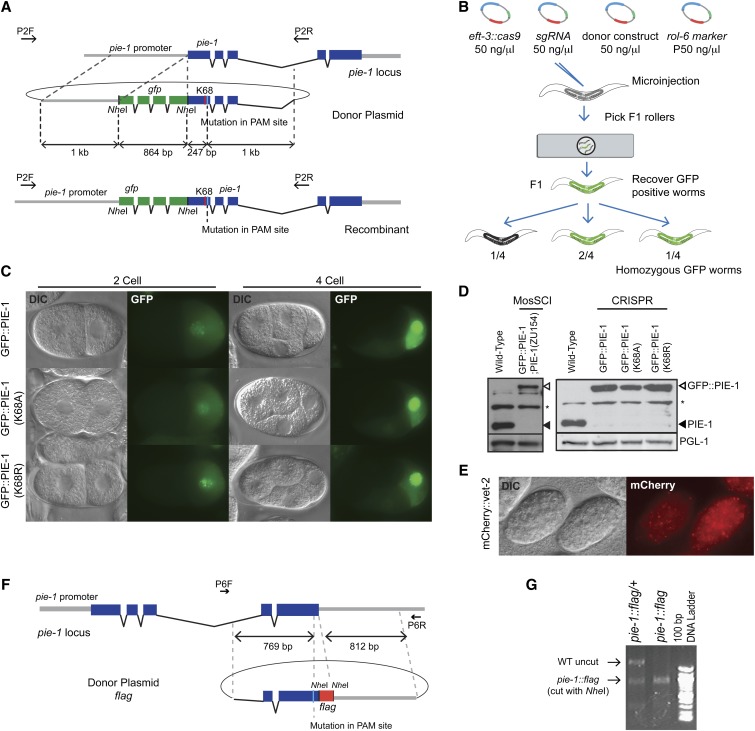
Figure 3.
HR-mediated knock-in to generate fusion genes at endogenous loci. (A) Schematic of the Cas9/sgRNA target site and the donor plasmid for gfp::pie-1 knock-ins. The donor plasmid contains the gfp coding sequence inserted immediately after the start codon of pie-1, 1 kb of homology flanking the CRISPR-Cas9 cleavage site, and a silent mutation in the PAM site. (B) Strategy to screen for gfp knock-in lines. We placed three F1 rollers at a time on a 2% agar pad and screened for GFP expression using epifluorescence microscopy. GFP-expressing worms were individually recovered and allowed to make F2 progeny for 1 day before being lysed for PCR and DNA sequence analysis. We confirmed Mendelian inheritance of gfp knock-in alleles among F2 progeny. (C) GFP::PIE-1 expression in the germline of two- to four-cell embryos of gfp::pie-1 knock-in strains. (D) Immunoblot analysis showing PIE-1 expression levels in wild-type animals, MosSCI-mediated gfp::pie-1 knock-in animals, and CRISPR-Cas9-mediated gfp knock-in animals. A MosSCI strain of gfp::pie-1; pie-1(zu154) was obtained by crossing gfp::pie-1 (LGII) with the pie-1(zu154) (LGIII) null mutant. (E) mCherry expression in late embryos of the mCherry::vet-2 knock-in strain. (F) Schematic of Cas9/sgRNA target sequence, PAM site, and donor plasmid for pie-1::flag knock-in. The PAM is located in the last exon of pie-1. The donor plasmid includes flag coding sequence immediately before the pie-1 stop codon and ∼800-bp homology arms flanking the target site. (G) PCR and restriction analysis of an HR event. PCR products were generated using the primers indicated in F, and the products were digested with NheI. The pie-1::flag gene conversion introduces an NheI RFLP that is observed in F1 heterozygous and F2 homozygous pie-1::flag animals.