Abstract
This article focuses on conducting global testing for association between a binary trait and a set of rare variants (RVs), although its application can be much broader to other types of traits, common variants (CVs), and gene set or pathway analysis. We show that many of the existing tests have deteriorating performance in the presence of many nonassociated RVs: their power can dramatically drop as the proportion of nonassociated RVs in the group to be tested increases. We propose a class of so-called sum of powered score (SPU) tests, each of which is based on the score vector from a general regression model and hence can deal with different types of traits and adjust for covariates, e.g., principal components accounting for population stratification. The SPU tests generalize the sum test, a representative burden test based on pooling or collapsing genotypes of RVs, and a sum of squared score (SSU) test that is closely related to several other powerful variance component tests; a previous study (Basu and Pan 2011) has demonstrated good performance of one, but not both, of the Sum and SSU tests in many situations. The SPU tests are versatile in the sense that one of them is often powerful, although its identity varies with the unknown true association parameters. We propose an adaptive SPU (aSPU) test to approximate the most powerful SPU test for a given scenario, consequently maintaining high power and being highly adaptive across various scenarios. We conducted extensive simulations to show superior performance of the aSPU test over several state-of-the-art association tests in the presence of many nonassociated RVs. Finally we applied the SPU and aSPU tests to the GAW17 mini-exome sequence data to compare its practical performance with some existing tests, demonstrating their potential usefulness.
Keywords: adaptive SPU (aSPU) test, genome-wide association study (GWAS), score statistic, sequencing data, sum of powered score (SPU) test, sum of squared score (SSU) test, sum test
THE recent advances in sequencing technologies have made it feasible to conduct global testing for association between complex traits and rare variants (RVs) (Bansal et al. 2010). The most popular approach in genome-wide association studies (GWASs) is to test on each single nucleotide variant (SNV) one by one and then select the SNVs meeting a stringent significance level after adjusting for multiple testing. However, such a strategy may be low powered due to the weak signal contained within each individual RV for its extremely low minor allele frequency (MAF). Hence, developing new association tests tailored to RVs has been an active research area in the past few years. Due to low MAFs of RVs, to achieve practically meaningful power, the majority of existing approaches focus on testing on a group of RVs, rather than on each individual RV (Capanu et al. 2011); the main idea is to boost power through aggregating information across multiple RVs in an analysis unit, such as a gene (e.g., Morgenthaler and Thilly 2007; Li and Leal 2008; Madsen and Browning 2009; Liu and Leal 2010; Han and Pan 2010; Hoffmann et al. 2010; Li et al. 2010; Price et al. 2010; Zhang et al. 2010; Zhu et al. 2010; Luo et al. 2011; Neale et al. 2011; Ionita-Laza et al. 2011; Feng et al. 2011; Pan and Shen 2011; Basu and Pan 2011; Gordon et al. 2011; Wu et al. 2011; Fan et al. 2013). As theoretically shown (Cox and Hinkley 1974) and demonstrated in our simulations, there is no uniformly most-power test for this purpose, which means that, depending on the unknown truth, including specific association effect directions and sizes, a given and fixed test may or may not be powerful. Hence, there have been intensive efforts in developing adaptive tests for RVs (e.g., Pan and Shen 2011; Lin and Tang 2011; Zhang et al. 2011; Lee et al. 2012; Chen et al. 2012; Derkach et al. 2013; Sun et al. 2013). However, due to their limited extents of adaptivity (e.g., with a predetermined and fixed set of the weights on RVs), these adaptive tests are still not flexible (or adaptive) enough with loss of power in some situations. A main motivation in this article is to develop a broader family of association tests such that at least one of them is powerful for a given situation. We develop such a family of tests, called the sum of powered score (SPU) tests, which generalize the sum (of score) test (Sum) and the sum of squared score (SSU) test (Pan 2009). The Sum test is a representative of the burden tests based on genotype pooling or collapsing (Morgenthaler and Thilly 2007; Li and Leal 2008; Madsen and Browning 2009), whereas the SSU test is closely related to kernel machine regression [and its implementation for RVs, SKAT (Sequence Kernel Association Test)] (Wu et al. 2010, 2011), C-alpha test (Neale et al. 2011), and an empirical Bayes test for high-dimensional data (Goeman et al. 2006); see Pan 2011 and Basu and Pan (2011). In many simulation setups, one, but not both, of the Sum test and SSU test has been shown to be powerful (Basu and Pan 2011). For example, with different association directions of causal RVs, the Sum test suffers from a loss of power, while the SSU test performs much better. However, we emphasize that, in analysis of multiple RVs, there exist nonassociated RVs. For example, in cancer research it has been observed that the vast majority of RVs do not appear to confer risk (Capanu et al. 2011). Hence, it is important to assess the performance of a test in the presence of nonassociated RVs in the group of the RVs to be tested. In fact, as to be shown, the performance of the Sum test deteriorates rapidly as the number of nonassociated RVs increases, whereas the SSU test is more robust but nevertheless may gradually become less competitive. It seems that the performance of various tests has not been fully investigated for the case with many nonassociated RVs, including some new adaptive tests, such as a kernel-based adaptive clustering (KBAC) test (Liu and Leal 2010), a P-value weighted sum test (PWST) (Zhang et al. 2011), an estimated regression coefficient (EREC) test (Lin and Tang 2011), an adaptive SSU (aSSU) test (Pan and Shen 2011), and an optimized SKAT (SKAT-O) test (Lee et al. 2012). As to be shown, it turns out that these tests suffer more from substantial power loss and are no longer competitive in the presence of a high proportion of nonassociated RVs. In contrast, regardless of the number of nonassociated RVs, at least one of our proposed SPU tests may remain relatively more powerful. Since the identity of the most powerful SPU test also changes with the unknown true association pattern with causal and noncausal RVs, we propose a simple yet highly adaptive SPU (aSPU) test to maintain high power across a wide range of scenarios.Our proposed aSPU test is often much more powerful than many existing adaptive tests in the presence of many nonassociated RVs.
We conducted extensive simulation studies to compare our newly proposed tests with several state-of-the-art tests, such as the PWST, EREC, aSSU, and SKAT-O tests, which all appeared after the publication of and thus were not compared in Basu and Pan (2011). As an active research topic, quite a few association tests for RVs have been proposed in the past 2 or 3 years. However, unfortunately, most of them have not been fully compared to each other, especially in the presence of many nonassociated RVs, which is expected to be a norm instead of an exception in analysis of RVs. Hence, as a second aim of this article, we assess the performance of these existing tests along with our newly proposed tests, offering some insights into their potential in practical use. Our study can be regarded as an update and follow-up to Basu and Pan (2011).
Methods
Data and notation
Our proposed methods are based on general regression models and thus can be applied to binary, quantitative, and survival responses or traits in the framework of generalized linear models and Cox proportional hazards model while adjusting for covariates, such as environmental variables and principal components accounting for population stratification. To be concrete, we consider only the case–control study design with a binary response/trait and no covariates; more general cases can be similarly approached, as shown in our example. Suppose that for subject i = 1, …, n, Yi = 0 or 1 is a binary response or trait, e.g., an indicator of disease, and Xi = (Xi1, …, Xik)′ is a group of predictors of interest, such as k RVs from a candidate gene or region. We use additive coding for each RV; that is, Xij is the count of the minor allele at RV j for subject i. Consider a logistic regression model:
| (1) |
We want to test the null hypothesis H0: β = (β1, …, βk)′ = 0; that is, there is no association between any RVs and the trait under H0.
Many of the existing tests and our new tests are based on the score vector U = (U1, …, Uk)′ for β in the logistic regression model (1) and V = Cov(U|H0) (Pan 2009; Basu and Pan 2011; Lin and Tang 2011; Wu et al. 2011; Lee et al. 2012),
where and are the sample means of Yi’s and Xi’s, respectively. An advantage of using a score-based test is that the closed form of the score vector is available and only a null model (i.e., the model under H0) needs to be fitted, and thus is computationally much faster, sometimes even only feasible, as compared to the corresponding Wald or likelihood-ratio test, for which a more general and complicated model has to be fitted and may not even converge (e.g., when k > n). Furthermore, it is noted that the score vector U in the joint model (1) is the same as UM = (UM,1, …, UM,k)′, with UM,j being the score statistic for βM,j in the marginal model
| (2) |
where subscript M denotes parameters from a marginal model. In contrast, for example, in general, the maximum-likelihood estimates (MLEs) and differ, and our experience suggests that the Wald test based on the marginal models is more powerful than that based on the joint model. The most popular single variant-based analysis corresponds to a minimum P-value (UminP) test combining univariate score tests for the marginal models,
where Vjj = Var(Uj|H0) is the jth diagonal element of V. To adjust for multiple testing, one could apply the conservative Bonferroni adjustment or better, as implemented here, based on the asymptotic null distribution of U ∼ N(0, V) under H0, use numerical integrations (or simulations) to obtain an asymptotically exact P-value for the UminP test (Conneely and Boehnke 2007).
A brief review of some existing tests
Basu and Pan (2011) compared the performance of many existing association tests for RVs. Their major conclusion is that if there is (nearly) a common association strength for causal RVs with no or few nonassociated RVs, then the burden tests, such as the Sum test (Pan 2009), were most powerful; otherwise, the SSU test (Pan 2009) and its close relatives (Pan 2011), kernel machine regression (KMR or SKAT) (Wu et al. 2010, 2011) and C-alpha test (Neale et al. 2011) performed best. The Sum test is based on a working assumption that in the joint logistic regression model (1), we have a common association parameter between the k RVs and the trait, say β1 = … = βk = βc. Then we need only to test a null hypothesis with a single parameter H0: βc = 0, corresponding to fitting a simple logistic regression model:
| (3) |
On the other hand, the SSU test is a variance-component test: assuming that β1, …, βk in model (1) are independent random effects with mean 0 and variance τ2, it can be derived as a score test on a null hypothesis with a single parameter H0: τ2 = 0. Specifically, both the Sum test and SSU test are based on the score vector U,
from which it is clear that the Sum test, as other burden tests, such as the CMC test (Li and Leal 2008) and the weighted Sum test (Madsen and Browning 2009), will lose its power if the causal RVs have different association directions, leading to different signs of Uj’s and thus a small test statistic TSum, failing to reject H0. In contrast, since the components of U is squared in the SSU test (and KMR and C-alpha test), the SSU test and its close relatives do not lose power with different association directions due to the sum over , instead of over Uj as in the Sum test. A more general score-based statistic can be written as
where ζ = (ζ1, …, ζk)′ is a vector of weights for the k RVs (Lin and Tang 2011). For example, if ζj = −1 or 1 depending on whether and its P-value < 0.1, then TG is the adaptive Sum (aSum) test of Han and Pan (2010). Two new tests that were not reviewed in Basu and Pan (2011) are also special cases of the above general test TG. First, if ζj = 2(pj − 0.5), where pj is the P-value for a one-sided Wald test for Hj,0: βM,j = 0 vs. Hj,1: βM,j < 0 with a test statistic , then TG is the PWST of Zhang et al. (2011). Second, if , then the TG test is the EREC test of Lin and Tang (2011); it is noted that, due to the instability of estimating βM,j for a RV, Lin and Tang (2011) proposed shrinking toward a constant d or −d, with d = 1 for binary traits. Each of the above three adaptive tests accommodates different association directions by using the signs of ’s, thus overcoming a main shortcoming of the Sum test, retaining high power in the presence of different association directions. Nevertheless, with RVs, for the same reason that motivates pooling or collapsing RVs in most association tests proposed so far, only limited information is contained in each RV, implying that all the above weighting schemes may not work well in some situations, as elaborated later.
In addition to differing association directions for causal RVs, a more common issue is the existence of many nonassociated RVs among the group of RVs to be tested. In particular, with many nonassociated RVs, as shown by Basu and Pan (2011), the burden tests, including the Sum test, lose their power quickly, while the SSU test and its close relatives perform much better. On the other hand, intuitively, if we can exclude nonassociated RVs in constructing a test statistic, it may help improve the power. Along this line, Pan and Shen (2011) proposed a class of adaptive Neyman-type tests (Neyman 1937), including an adaptive Sum (aSum+) and an adaptive SSU (aSSU) test, which are based on RV selection, instead of weighting. Specifically, first, one orders the components of the score vector U in a descending order based on the magnitudes of Uj and , respectively, for the aSum+ and aSSU tests. Second, suppose that the P-values for the Sum and SSU tests based on the first j components of the ordered U are PSum,j and PSSU,j respectively; then the test statistics for the two adaptive tests are
and the final P-values are obtained by permutations or simulations. In short, the aSum+ and aSSU tests work by selecting the first few components of a reordered score vector U that are most informative (with smallest P-values) while possibly ignoring other components of U for nonassociated or weakly associated RVs. In particular, the aSum+ test accounts for possibly different association directions by using only those positively associated RVs; however, it may suffer from power loss due to its ignoring those negatively associated RVs. To improve over the aSum+ test, Pan et al. (2011) proposed an adaptive Sum test based on two directional searches, denoted as aSum2d, to use both positively and negatively associated SNVs, and found its improved power in detecting gene–gene interactions for CVs. Specifically, we first reorder the components of U in a descending order of Uj as for the aSum+ test and suppose that the P-value of the Sum test applied to the last j components of the reordered U is PSum,−j; then the aSum2d test statistic is
We can then use permutations or simulations to obtain P-values for TaSum2d (and TaSum– if needed). If desired, one can also just use TaSum– to test for only negatively associated RVs.
Another adaptive test, called KBAC, was proposed by Liu and Leal (2010). A unique feature of the KBAC test is to detect not only the main effects of, but also possible interactions among, RVs. For the latter purpose, rather than weighting on each individual RV, it uses a kernel-based weight on each unique pattern (or combination) of the genotypes across the k RVs. It upweights a genotype pattern that appears more frequently in cases (i.e., with a higher risk of disease) and then contrasts the frequencies of genotype patterns between the case and control groups by taking a weighted sum of their frequency differences. As pointed out by Basu and Pan (2011), there are two potential limitations. First, since its test statistic is a weighted sum of the frequency differences between the case and control groups, the presence of opposite association directions may contribute to both positive and negative frequency differences, leading to a small test statistic and thus loss of power. Second, as the number of nonassociated RVs increases, there will be a larger number of unique genotype patterns and thus a smaller number of subjects with each genotype pattern, leading to loss of power. These two points are confirmed later.
A new class of tests and a data-adaptive test
Our basic observation is that, depending on the unknown pattern of association effects of the group of RVs to be tested, different tests may be more powerful; in spite of the generality of the TG test, its performance critically depends on the choice of the weights, and any fixed choice may or may not be most suitable. Hence our primary goal is to construct a class of versatile tests such that for a given scenario, at least one of the tests is powerful. Then we combine these tests to obtain a data-adaptive test that will maintain high power across a wide range of scenarios. For this purpose, we choose weight ζj as informative and as simple as possible. Since most existing association tests use the score vector U, suggesting that most information is already contained in U, we would simply use U to construct weights. In particular, since we have U ∼ N(0, V) under H0, we know that a large |Uj| offers strong evidence to reject H0,j: βj = 0. Specifically, we choose for an integer γ ≥ 1, leading to a SPU test:
| (4) |
With various values of γ ≥ 1, we obtain a class of the SPU tests. The SPU tests cover the Sum and SSU tests as two special cases with a corresponding γ = 1 and γ = 2, respectively. Importantly, as γ increases, the SPU(γ) test puts more weights on the larger components of U while gradually ignoring the remaining components. An extreme case is that, as an even number γ → ∞, we have
Since the SPU tests are based on resampling methods to calculate their P-values, they are invariant to any monotone transformation of their test statistics, such as (⋅)1/γ. That is, we can equivalently define , which uses only the largest component of |U|. More generally, as we increase the value of γ, we put higher and higher weights on the larger components of U, effectively realizing RV selection. On the other hand, an even integer of γ automatically eliminates the effects of different signs of Uj’s, avoiding power loss of the Sum test in the presence of different association directions. However, an odd integer of γ might be more suitable, as in the SPU(1) or Sum test, when the associations are all in the same direction.
We know that under H0, the score vector U has an asymptotic Normal distribution N(0, V). Hence, in theory, we can derive the asymptotic distribution of TSPU(γ), which, however, may not be easy to calculate. As an alternative, we recourse to permutations (Churchill and Doerge 1994). Specifically, we permute the original set of traits Y to obtain a new set of traits Y(b), based on which we calculate the score vector U(b) and the null statistic ; after b = 1, …, B permutations, we calculate the P-value as . We used B = 200 in our simulations for a nominal significance level at 5%.
In the presence of covariates, we propose generalizing the above permutation scheme. Specifically, first we regress Y on the covariates to fit a null model under H0 to obtain and residual ; second, we permute the set of the residuals r = {ri|i = 1, …, n} to obtain a permuted set r(b); third, we calculate the new score vector based on the permuted residuals as and the corresponding null statistic and after repeating the above steps for b = 1, …, B, we calculate the P-value as .
Alternatively, we also propose using the parametric bootstrap: we first fit a null model under H0 to obtain and then simulate a new set of traits for b = 1, …, B; we calculate the test statistic based on each set of simulated Y(b) and calculate the P-value as in the permutation method.
Since the power of a SPU(γ) test depends on the choice of γ while the optimal choice of γ depends on the unknown true association pattern of the RVs to be tested, it would be desirable to data-adaptively choose the value of γ. For this purpose, we propose an aSPU test that simply combines the P-values of multiple SPU tests (with various values of γ), although other combining methods are also possible (Pan et al. 2010; Cheung et al. 2012). Suppose that we have some candidate values of γ in Γ, e.g., Γ = {1, 2, 3, …, 8, 15, 16, 31, 32, ∞} as used in our later simulations, and suppose that the P-value of the SPU(γ) test is PSPU(γ); then our combining procedure is to take the minimum P-value:
Of course, TaSPU is no longer a genuine P-value; we use the permutation or parametric bootstrap to estimate its P-value. It may appear that a double permutation or bootstrap procedure is needed, but indeed not necessary. For example, if we use the permutation method, first, we permute the original set of traits to obtain Y(b) and the corresponding score vector U(b) for b = 1, 2, …, B. We then calculate the corresponding SPU test statistics and their P-values . Thus, we have , and the final P-value of the aSPU test is .
We note the practicality of permutation- or other resampling-based methods for P-value calculations. First, due to extremely low MAFs of some RVs, it is always dubious whether asymptotic results are applicable. Second, it is computationally feasible to use permutation-based tests for genome-wide searches. In practice, we can first use a smaller B, say B = 1000, to scan a genome and then gradually and repeatedly increase B for a few groups of RVs that pass an initial significance criterion (e.g., P-value < 5/B) in the previous step; in this way, contrary to otherwise claimed, it is indeed feasible to apply a permutation-based test to genome scans and obtain highly significant results (if any). We have applied permutation- or simulation-based aSPU test to genome-wide scans to yield P-values < 10−6.
Finally we comment on the choice of Γ. The following considerations guide the choice of the integers γ ≥ 1 in Γ. First, to cover the burden and variance-component tests, which have been shown empirically to perform well under some situations (Basu and Pan 2011), we would include γ = 1 and γ = 2 in Γ. Second, depending on whether the phenotype-RV association directions vary, we may need to use either even or odd integers γ’s to yield high power; if unsure, then it is suggested to use both odd and even integers γ’s. Third, depending on how sparse the association signals are, one may use smaller or larger γ’s. For example, the more the RVs to be tested and the fewer associated RVs to be expected, then larger γ’s would be desirable. We have found that often Γ = {1, 2, 3, …, 8, ∞} suffices. For demonstration, we have included γ = ∞, which is not necessary, but may be beneficial when testing on CVs. In the following, we also show the results of the SPU(γ) tests for γ ∈ Γ; we have two purposes. The first is to show varying operating characteristics of the various SPU tests. For example, we show higher power of SPU(3) or SPU(4) than that of SPU(1) and SPU(2), demonstrating the power gain of using some SPU(γ) test with γ > 2. Second, we show that often SPU(8) gives results almost the same as those of SPU(∞), suggesting no need to use other larger γ’s. In practice, we suggest using the aSPU test that combines the strengths (and possibly weaknesses) of various SPU tests; the aSPU test can be regarded as a rigorous (and almost exact) means for multiple testing adjustment with the use of several SPU tests, while the results of the SPU tests may shed light on the underlying genetic architecture. For example, if a large γ gives the most significant P-value, it may indicate a high degree of signal sparsity; if some odd γ’s yield more significant results than even γ’s, then most or all of the large associations are in the same direction. More elaborately, as shown below, an analysis of a SPU test can also imply the relative contribution of each SNV to the aggregated association (if any).
SNV selection
A limitation of most global tests is their inability for variant selection: even if the global null hypothesis is rejected, they may not give any information on which RVs are (or are not) likely to be associated with disease. We note that the aSPU test can be used to rank the importance of the RVs. First, we estimate the optimal value of γ < ∞, chosen by the aSPU test. Second, we assess the relative contribution of each RV r to the aSPU test as . Third, we rank the RVs based on their Cr values, and we can select the top k1 RVs such that the sum of their relative contributions with α1 = 0.8, say; the choice of α1 determines the tradeoff between increasing true positives and increasing false positives. Generally, we can use Cr to prioritize and generate hypotheses on the selection of causal RVs.
Further comments and extensions
Below we briefly comment on the advantages of the aSPU test over several other adaptive tests. First, since the power of any univariate test for a single RV may be low (which is exactly the reason why we combine information across multiple RVs, e.g., through pooling or collapsing), the P-value of such a test may not be informative; the aSum test and PWST based on such P-values may not perform well. Second, we note that the adaptive Neyman-type tests, such as the aSSU test, are based on the idea of variable or RV selection, while the SPU tests are more based on weighted averaging of variables or RVs. As discussed extensively in the model selection literature (Yuan and Yang 2005; Shen and Huang 2006) and in a genetic application (Newton et al. 2007), if signals are strong enough, then model selection is expected to perform better; otherwise, model averaging is preferred. In our current context, again due to extremely low MAFs of RVs, no matter how strong its association strength, information contained within each individual RV is only quite limited. Thus we expect that the model averaging-based SPU tests to outperform the model selection-based aSSU or other adaptive Neyman-type tests. Third, we note that the EREC test is related to the SPU tests. As shown in Pan (2009), we have
where Diag(V) is a diagonal matrix with its jth diagonal element as Vjj. Hence, if is much larger than d, then , which is roughly proportional to U (if the MAFs of the RVs are in a close range), suggesting that the EREC test will be similar to the SPU(2), i.e., SSU test. On the other hand, if is small relative to d, then , implying that the EREC test will behave similarly to the SPU(1) (or Sum) test. Generally, we expect that the EREC test behaves between the SPU(1) and SPU(2) tests.
Furthermore, several approaches (Lee et al. 2012; Derkach et al. 2013; Sun et al. 2013), including SKAT-O, have been proposed to combine a burden test like SPU(1) and a variance-component test like SPU(2). In contrast, our proposed aSPU test is based on combining a broader set of tests including but beyond SPU(1) and SPU(2), hence is more flexible and adaptive. In the presence of many nonassociated RVs, the weight ζ = 1 or ζ = U may not suffice: we may need weights Uγ − 1 with γ > 2. In other words, with many nonassociated RVs, the power of the EREC or SKAT-O [or similar tests combining SPU(1) and SPU(2)] can be much lower than a SPU(γ) test with a large γ and lower than the aSPU test. In addition, since the aSum, PWST, and EREC tests use the marginal estimates , which have to be obtained iteratively during each permutation, whereas the score vector U is much easier to obtain, the SPU tests are much faster.
In the presence of external or prior biological information, as for other tests, it may be helpful to incorporate some external weights (differing from ζ discussed earlier) into the SPU tests. Given some external weights wj, we can have a weighted SPU (wSPU) test as
while all other aspects, including the construction of an adaptive wSPU test, remain the same as before. For example, if it is believed that causal RVs tend to have lower MAFs, as advocated by Madsen and Browning (2009), one can use a wj inversely proportional to the MAF of the jth RV. In this way, by suitably weighting both CVs and RVs, it is possible to use the adaptive wSPU test for a joint analysis of CVs and RVs (Ionita-Laza et al. 2013). Alternatively, wj can be a predicted likelihood of the jth RV’s being functional or deleterious based on some computational algorithms (Wei et al. 2011). As other tests, the performance of the wSPU tests depends on how informative or correct the external weights are, while the choice of the external weights may not always be clear; hence we skip further discussions on the use of such external weights.
We have proposed using permutations (or the parametric bootstrap) to calculate the P-values for the SPU and aSPU tests. If the asymptotic normality of the score vector is expected to approximately hold, e.g., in analysis of CVs, we may replace the permutation or bootstrap with simulation-based methods, which will be much faster (Zou et al. 2004; Seaman and Muller-Myhsok 2005).
Results
Simulation setups
We conducted extensive simulation studies to evaluate and compare the performance of various tests. We simulated genotypes as in Wang and Elston (2007). First, a latent vector Z = (Z1, …, Zk)′ was generated from a multivariate Normal distribution N(0, R), where R had a first-order autoregressive (AR1) covariance structure with its (i, j)th element Rij = Corr(Zi, Zj) = ρ|i − j|; we used ρ = 0 and ρ = 0.9 to generate independent and correlated RVs, respectively. Second, the latent vector Z was dichotomized to yield a haplotype with some specified MAFs, each of which was randomly selected from a uniform distribution between 0.001 and 0.01 during each simulation. Third, the above two steps were repeated to generate two independent haplotypes, which were then combined to obtain genotype Xi = (Xi1, …, Xik)′ for subject i. Fourth, for a nonnull case we randomly chose k1 causal RVs with their corresponding βj ≠ 0 while all other βj = 0; for a null case, all βj = 0. Fifth, the disease status Yi of subject i was generated from the logistic regression model (1). We used β0 = −log(0.05/0.95) for a 5% background disease probability; that is, Pr(Yi = 1|Xi = 0) = 0.05. Finally, as in a case–control study, we sampled n/2 cases and n/2 controls in each data set.
We considered a few setups with combinations of various values of ρ = 0 or 0.9, k1 = 8 or 1, and n = 1000. We varied the number of nonassociated RVs k − k1 between 0 and 128, and a range of possible values of βj ≠ 0 to cover from a common association effect to varying association strengths or directions and from a single to multiple causal RVs.
Throughout the simulations, the test significance level was fixed at α = 0.05. The results were based on 1000 independent replicates for each setup. We compared the performance of the SPU tests with several state-of-the-art adaptive tests not reviewed in Basu and Pan (2011), including one based on a Bayesian hierarchical GLM (BhGLM) (Yi et al. 2011). As a benchmark, we also included the UminP test that tests on each individual RV separately and then combine them by taking their minimum P-value.
Simulation results
To save space, we focus on a few cases with correlated RVs (i.e., neighboring RVs were in linkage disequilibrium). It is not only more general to consider correlated RVs (or covariates), but also in agreement with real sequence data as generated from the 1000 Genomes Project (Zhang et al. 2013). The extensive simulation results with independent RVs and other association parameters were similar to those presented below and thus are relegated to supporting information, File S1.
First, all the tests maintained well-controlled type I error rates (Table 1). Second, we consider the case with nonzero βj’s randomly drawn from a uniform distribution U(1, 2), representing the association pattern with varying association strengths but all in the same direction (Table 2). Among the SPU tests, the SPU(1), SPU(2), SPU(4), or SPU(6) were the respective winners with no, a medium, and a large number of nonassociated RVs. This is in agreement with our analysis earlier that an increasing proportion of nonassociated RVs requires a larger value of γ in the SPU(γ) test to weed off the effects of nonassociated RVs. In particular, the quickly deteriorating performance of the SPU(1) (i.e., Sum) test was striking. Compared to some more powerful SPU tests, the UminP test was low powered because the UminP test used information from only the most significantly associated RV while ignoring other associated RVs. It is noted that a SPU(γ) test with 8 ≤ γ < ∞ was only slightly more powerful than SPU(∞), suggesting no need to use γ > 8.
Table 1. Empirical type I error rates of various tests for the cases with a group of eight nonassociated RVs and another group of nonassociated RVs.
| No. of nonassociated RVs | |||||||
|---|---|---|---|---|---|---|---|
| Test | 0 | 8 | 16 | 32 | 64 | 96 | 128 |
| UminP | 0.025 | 0.020 | 0.017 | 0.018 | 0.015 | 0.007 | 0.011 |
| SPU(1) | 0.051 | 0.055 | 0.044 | 0.049 | 0.048 | 0.041 | 0.051 |
| SPU(2) | 0.048 | 0.055 | 0.034 | 0.029 | 0.039 | 0.037 | 0.035 |
| SPU(3) | 0.049 | 0.051 | 0.040 | 0.041 | 0.032 | 0.038 | 0.037 |
| SPU(4) | 0.051 | 0.046 | 0.033 | 0.040 | 0.025 | 0.037 | 0.029 |
| SPU(5) | 0.053 | 0.048 | 0.039 | 0.050 | 0.026 | 0.043 | 0.030 |
| SPU(6) | 0.056 | 0.046 | 0.042 | 0.048 | 0.024 | 0.039 | 0.022 |
| SPU(7) | 0.057 | 0.044 | 0.040 | 0.046 | 0.028 | 0.038 | 0.023 |
| SPU(8) | 0.054 | 0.042 | 0.040 | 0.044 | 0.022 | 0.036 | 0.023 |
| SPU(16) | 0.055 | 0.041 | 0.041 | 0.047 | 0.029 | 0.037 | 0.025 |
| SPU(32) | 0.053 | 0.042 | 0.043 | 0.047 | 0.030 | 0.039 | 0.025 |
| SPU(∞) | 0.053 | 0.042 | 0.042 | 0.047 | 0.031 | 0.039 | 0.025 |
| aSPU | 0.055 | 0.044 | 0.041 | 0.048 | 0.038 | 0.047 | 0.038 |
| aSum+ | 0.045 | 0.058 | 0.045 | 0.053 | 0.048 | 0.051 | 0.046 |
| aSum2d | 0.062 | 0.054 | 0.042 | 0.049 | 0.052 | 0.043 | 0.045 |
| aSSU | 0.066 | 0.047 | 0.045 | 0.045 | 0.046 | 0.048 | 0.052 |
| KBAC | 0.049 | 0.061 | 0.035 | 0.049 | 0.047 | 0.057 | 0.051 |
| aSum | 0.066 | 0.056 | 0.033 | 0.048 | 0.048 | 0.055 | 0.042 |
| PWST | 0.061 | 0.047 | 0.031 | 0.049 | 0.040 | 0.040 | 0.042 |
| EREC | 0.056 | 0.058 | 0.044 | 0.046 | 0.050 | 0.045 | 0.046 |
| BhGLM | 0.044 | 0.061 | 0.042 | 0.042 | 0.043 | 0.039 | 0.048 |
| SKAT | 0.057 | 0.064 | 0.042 | 0.046 | 0.050 | 0.046 | 0.049 |
| SKAT-O | 0.058 | 0.065 | 0.047 | 0.040 | 0.051 | 0.047 | 0.052 |
The RVs within each group were correlated, but there was no between-group correlation; all results were based on 1000 simulation replicates.
Table 2. Empirical power of various tests for the cases with a group of eight causal RVs with ORs randomly drawn from U(1,2), and another group of nonassociated RVs.
| No. of nonassociated RVs | |||||||
|---|---|---|---|---|---|---|---|
| Test | 0 | 8 | 16 | 32 | 64 | 96 | 128 |
| UminP | 0.874 | 0.812 | 0.768 | 0.733 | 0.659 | 0.619 | 0.586 |
| SPU(1) | 0.939 | 0.852 | 0.746 | 0.577 | 0.411 | 0.300 | 0.244 |
| SPU(2) | 0.926 | 0.908 | 0.904 | 0.872 | 0.832 | 0.801 | 0.769 |
| SPU(3) | 0.917 | 0.903 | 0.893 | 0.870 | 0.829 | 0.802 | 0.786 |
| SPU(4) | 0.909 | 0.896 | 0.890 | 0.882 | 0.854 | 0.840 | 0.835 |
| SPU(5) | 0.902 | 0.894 | 0.879 | 0.875 | 0.843 | 0.834 | 0.834 |
| SPU(6) | 0.901 | 0.882 | 0.872 | 0.873 | 0.843 | 0.835 | 0.835 |
| SPU(7) | 0.899 | 0.881 | 0.868 | 0.869 | 0.836 | 0.830 | 0.828 |
| SPU(8) | 0.898 | 0.876 | 0.863 | 0.864 | 0.833 | 0.834 | 0.826 |
| SPU(16) | 0.885 | 0.860 | 0.848 | 0.852 | 0.821 | 0.814 | 0.814 |
| SPU(32) | 0.878 | 0.855 | 0.844 | 0.850 | 0.817 | 0.807 | 0.810 |
| SPU(∞) | 0.877 | 0.852 | 0.844 | 0.846 | 0.814 | 0.801 | 0.806 |
| aSPU | 0.923 | 0.898 | 0.894 | 0.869 | 0.842 | 0.829 | 0.811 |
| aSum+ | 0.940 | 0.893 | 0.866 | 0.801 | 0.677 | 0.607 | 0.532 |
| aSum2d | 0.921 | 0.857 | 0.805 | 0.719 | 0.618 | 0.504 | 0.453 |
| aSSU | 0.900 | 0.867 | 0.847 | 0.819 | 0.767 | 0.695 | 0.658 |
| KBAC | 0.903 | 0.785 | 0.722 | 0.613 | 0.466 | 0.331 | 0.283 |
| aSum | 0.948 | 0.892 | 0.855 | 0.756 | 0.636 | 0.480 | 0.407 |
| PWST | 0.823 | 0.729 | 0.698 | 0.613 | 0.508 | 0.400 | 0.380 |
| EREC | 0.943 | 0.901 | 0.887 | 0.833 | 0.738 | 0.656 | 0.579 |
| BhGLM | 0.934 | 0.863 | 0.779 | 0.619 | 0.434 | 0.299 | 0.248 |
| SKAT | 0.927 | 0.914 | 0.906 | 0.870 | 0.823 | 0.800 | 0.749 |
| SKAT-O | 0.940 | 0.915 | 0.899 | 0.858 | 0.799 | 0.767 | 0.696 |
All results were based on 1000 simulation replicates. The highest powered nonadaptive and adaptive tests are italic.
Among the adaptive tests, the aSPU test was the overall winner; its performance in the presence of many nonassociated RVs was most impressive: for example, with 126 nonassociated RVs, the power of the aSPU test was 0.811, much higher than 0.749 of SKAT (with a linear kernel used throughout), 0.658 of aSSU, 0.567 of EREC, 0.532 of aSum+, 0.380 of PWST, 0.331 of KBAC, and 0.248 of BhGLM. It is noted that the power of the aSPU test was always close to that of the most powerful SPU test, whose identity however changed with the setup. Although all other adaptive tests performed well with no or few nonassociated RVs, they failed to do so otherwise. In particular, the aSSU test was much less powerful than the SSU test with many nonassociated RVs, presumably due to the difficulty in RV selection with relatively weak signals with each individual RV.
Third, for the case with both varying association directions and effect sizes of the causal RVs (Table 3), among the SPU tests, as the number of nonassociated RVs increased, SPU(2), SPU(4), SPU(6), and SPU(8) became most powerful, respectively, and as expected, the SPU(1) (i.e., Sum) test was the least powerful. For example, with 128 null RVs, the power of SPU(1) and that of SPU(2) were only 0.070 and 0.261, respectively, much lower than 0.370 of SPU(7) and SPU(8); accordingly, the power of aSPU was 0.329, much higher than 0.235 of SKAT and 0.195 of SKAT-O. It was also confirmed that there was no need to use a SPU(γ) test with γ > 8 to gain power. Among the adaptive tests, the aSPU test was the winner, although the PWST and SKAT were most powerful with no or only few nonassociated RVs, but quickly lost their edge as the number of nonassociated RVs increased. The aSSU test performed second best after the aSPU test, presumably due to easier RV selection with larger effect sizes of some causal RVs. The BhGLM, aSum, and KBAC tests did not perform well in this case. Similar results with higher significance levels α and with covariates were obtained as shown in File S1.
Table 3. Empirical power of various tests for the cases with a group of eight causal RVs with ORs = (3, 1/3, 2, 2, 2, 1/2, 1/2, 1/2) and another group of nonassociated RVs.
| No. of nonassociated RVs | |||||||
|---|---|---|---|---|---|---|---|
| Test | 0 | 8 | 16 | 32 | 64 | 96 | 128 |
| UminP | 0.507 | 0.379 | 0.324 | 0.288 | 0.208 | 0.197 | 0.157 |
| SPU(1) | 0.341 | 0.227 | 0.159 | 0.111 | 0.074 | 0.070 | 0.070 |
| SPU(2) | 0.631 | 0.542 | 0.485 | 0.435 | 0.332 | 0.279 | 0.261 |
| SPU(3) | 0.563 | 0.493 | 0.451 | 0.413 | 0.303 | 0.323 | 0.274 |
| SPU(4) | 0.625 | 0.540 | 0.508 | 0.478 | 0.402 | 0.386 | 0.351 |
| SPU(5) | 0.601 | 0.514 | 0.480 | 0.465 | 0.374 | 0.383 | 0.358 |
| SPU(6) | 0.619 | 0.529 | 0.504 | 0.490 | 0.398 | 0.401 | 0.367 |
| SPU(7) | 0.600 | 0.521 | 0.485 | 0.480 | 0.386 | 0.399 | 0.370 |
| SPU(8) | 0.610 | 0.530 | 0.494 | 0.485 | 0.402 | 0.404 | 0.370 |
| SPU(16) | 0.595 | 0.524 | 0.488 | 0.470 | 0.390 | 0.395 | 0.356 |
| SPU(32) | 0.595 | 0.523 | 0.487 | 0.469 | 0.388 | 0.389 | 0.353 |
| SPU(∞) | 0.592 | 0.520 | 0.484 | 0.467 | 0.386 | 0.387 | 0.355 |
| aSPU | 0.589 | 0.511 | 0.467 | 0.461 | 0.366 | 0.361 | 0.329 |
| aSum+ | 0.596 | 0.497 | 0.419 | 0.370 | 0.268 | 0.245 | 0.214 |
| aSum2d | 0.560 | 0.450 | 0.391 | 0.326 | 0.220 | 0.193 | 0.169 |
| aSSU | 0.598 | 0.511 | 0.464 | 0.424 | 0.337 | 0.311 | 0.296 |
| KBAC | 0.525 | 0.392 | 0.327 | 0.254 | 0.181 | 0.135 | 0.129 |
| aSum | 0.549 | 0.415 | 0.299 | 0.232 | 0.138 | 0.145 | 0.127 |
| PWST | 0.675 | 0.554 | 0.460 | 0.354 | 0.267 | 0.197 | 0.196 |
| EREC | 0.545 | 0.448 | 0.365 | 0.298 | 0.189 | 0.176 | 0.154 |
| BhGLM | 0.434 | 0.290 | 0.186 | 0.123 | 0.082 | 0.072 | 0.071 |
| SKAT | 0.650 | 0.557 | 0.474 | 0.425 | 0.318 | 0.272 | 0.235 |
| SKAT-O | 0.622 | 0.495 | 0.406 | 0.378 | 0.260 | 0.226 | 0.195 |
All results were based on 1000 simulation replicates. The highest powered nonadaptive and adaptive tests are italic.
Data example
We applied the methods to the mini-exome sequence data provided by the Genetic Analysis Workshop (GAW) 17 (Almasy et al. 2011). The exome sequence data contain 24,487 SNVs in 3205 genes from 697 unrelated subjects. Our analyses focused on RVs with MAFs no larger than 1%; after removing those more frequent SNVs, we had 2476 genes containing at least 1 RV, with a total of 18,131 RVs. We conducted gene-based analyses.
The phenotypes were generated by GAW17 organizers based on some disease liability models with covariates. In particular, biological knowledge on pathways, especially the vascular endothelial growth factor (VEGF) pathway, and on predicted deleterious coding variants was utilized to design a realistic simulation model. Fixing the sequence data for the 697 subjects, 200 independent sets of a binary phenotype were generated. In addition to some causal SNVs, three risk factors, age, gender, and smoking status, were possibly associated with the binary phenotype. An advantage of using the GAW17 data is the opportunity to assess statistical power of any given method due to the known causal SNVs and the availability of replicated phenotypes. Hence, in addition to conducting a genome-wide scan, we applied the methods to each causal gene with all 200 sets of the binary phenotype to estimate the power.
A genome-wide scan:
To demonstrate the practical use of the proposed methods, we first conducted a genome-wide scan on all the 2476 genes with the first set of the binary phenotype. A logistic regression model was fitted to each gene with the three covariates,
for i = 1, … 697, where Xi1, …, Xik are the k RVs in the gene to be tested and Yi is the binary phenotype. In the presence of the covariates, we used the parametric bootstrap to calculate the P-values for the SPU, aSPU, SKAT, and SKAT-O tests.
Throughout the data analysis, we used the following “step-up” procedure to determine the number of bootstraps (or permutations), B. We started with B = 103 amd then gradually increased B: if an estimated P-value was <5/B, we increased B to 10 times of its current value to reestimate the P-value, and the process was repeated until no estimated P-value was <5/B. For the genome scan on the GWA17 data, we tried B = 103, 104, and up to 105 to obtain P-values. On a multicore computer with 100 cores, it took about 0.21 hr to test the 2476 genes based on B = 103, 0.05 hr to test the 50 genes (with their P-values < 0.005 in the previous round) with B = 104, and 0.12 hr for testing the 5 genes based on B = 105. If a single-core computer was used, a conservative estimate of the time to be taken would be the above time multiplied by 100, which would take <2 days. If we doubled the sample size, it took about three times the original time. Note that our code was completely in R and not yet optimized; implementing the core part in C or another compiled language is planned and expected for at least a 10-fold speedup. Hence, to be more accurate, we can replace the used threshold 5/B with a larger value such as 50/B.
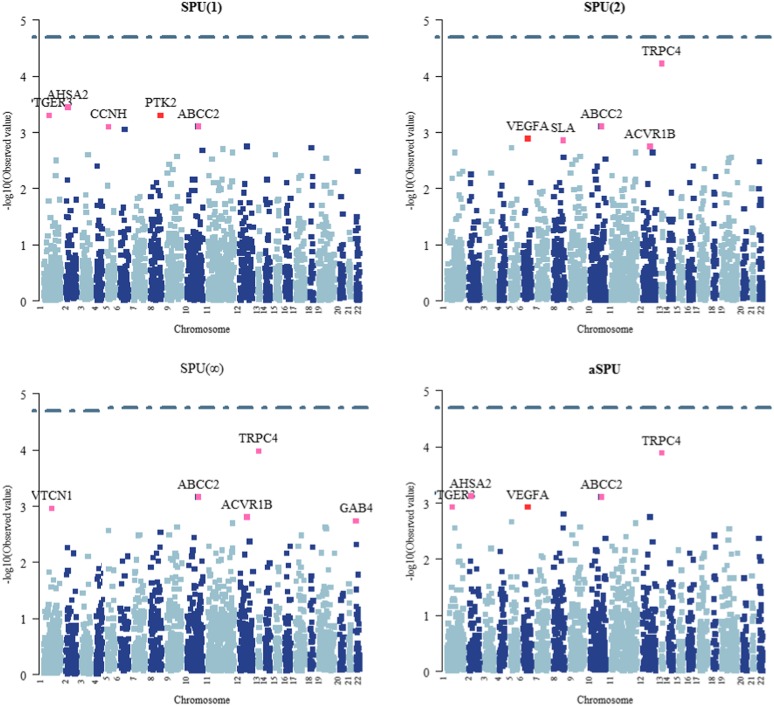
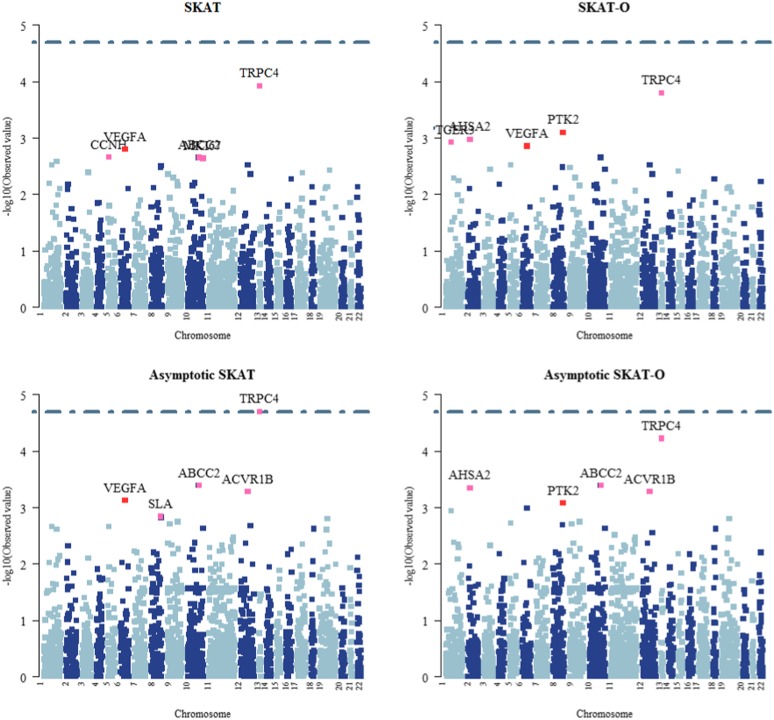
Figure 1 shows the Manhattan plots for the tests. Since we were testing on 2476 genes, at the usual family-wide significance level of 0.05 and with a Bonferroni adjustment, we would use a gene-wise significance level of 0.05/2476 = 2.02 × 105, which would suggest using a bootstrap replication number up to 106 to 107.
Figure 1.
Manhattan plots for the GAW17 data.
None of the tests detected any significant genes. Nonetheless, we highlighted the top five most significant genes based on each test to show their differing operating characteristics. In Figure 1, it is clear that the three representative SPU tests gave overlapping but different sets of the top genes. It is interesting to note that the aSPU test combined the results of the SPU tests.
As a comparison, Figure 1 also shows the results for the parametric bootstrap- and asymptotics-based SKAT and SKAT-O tests. First, we note the differences among the top genes between the resampling- and asymptotics-based tests, although their overall trends were similar, implying that one has to be cautious in using asymptotics-based tests for analysis of RVs. Second, we note the difference between the results of SKAT and SKAT-O; the latter had some similarity to that of the SPU(1) test, as expected. Most importantly, although similar to some extent, the top ranked genes by the aSPU, SKAT, and SKAT-O tests were still different, suggesting the potential usefulness of the aSPU test as a complement to SKAT and SKAT-O. We also note that most of the genes contained no more than 30 RVs; otherwise, the difference between the aSPU and SKAT or SKAT-O could be larger, as suggested in our simulations.
Power comparison:
With the 200 replicated sets of the binary phenotype, the GAW17 data offer a unique opportunity to estimate the power of any test when applied to real sequence data. Due to the small sample size and relatively small effect sizes of the causal RVs, there was low power to detect any causal RVs in the GAW17 data. Accordingly, we used a less stringent gene-wise significance level of 0.05 (i.e., without multiple testing adjustment) and estimated the power of a test as the sample proportion of its rejecting the null hypothesis among the 200 sets of the replicated phenotypes. We considered all 35 causal genes (with at least one causal RV) and tested on each separately. The main results are shown in Table 4.
Table 4. Estimated power for some causal genes with the GAW17 data.
| Chr | Gene | No. of RVs | No. of causal | SPU(1) | SPU(2) | SPU(3) | SPU(4) | SPU(5) | SPU(6) | SPU(7) | SPU(8) | SPU(∞) | aSPU | SKAT | SKAT-O |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PIK3C2B | 60 | 23 | 0.565 | 0.445 | 0.650 | 0.400 | 0.395 | 0.355 | 0.360 | 0.350 | 0.340 | 0.600 | 0.435 | 0.560 |
| 6 | VNN1 | 6 | 1 | 0.185 | 0.230 | 0.315 | 0.235 | 0.380 | 0.320 | 0.350 | 0.325 | 0.140 | 0.270 | 0.215 | 0.185 |
| 3 | BCHE | 28 | 13 | 0.110 | 0.190 | 0.215 | 0.185 | 0.175 | 0.165 | 0.165 | 0.165 | 0.160 | 0.210 | 0.195 | 0.175 |
| 8 | LPL | 15 | 2 | 0.090 | 0.130 | 0.135 | 0.110 | 0.115 | 0.110 | 0.115 | 0.115 | 0.110 | 0.135 | 0.125 | 0.125 |
| 10 | SIRT1 | 23 | 9 | 0.095 | 0.105 | 0.110 | 0.065 | 0.070 | 0.060 | 0.055 | 0.055 | 0.015 | 0.105 | 0.090 | 0.100 |
| 14 | SOS2 | 7 | 2 | 0.100 | 0.270 | 0.285 | 0.265 | 0.275 | 0.265 | 0.275 | 0.265 | 0.245 | 0.220 | 0.255 | 0.200 |
| 19 | RRAS | 5 | 2 | 0.235 | 0.140 | 0.155 | 0.145 | 0.155 | 0.150 | 0.155 | 0.150 | 0.100 | 0.180 | 0.135 | 0.200 |
| 8 | PLAT | 25 | 8 | 0.225 | 0.135 | 0.145 | 0.110 | 0.105 | 0.100 | 0.100 | 0.100 | 0.070 | 0.155 | 0.130 | 0.195 |
| 9 | VLDLR | 23 | 8 | 0.080 | 0.120 | 0.125 | 0.110 | 0.120 | 0.105 | 0.115 | 0.105 | 0.075 | 0.090 | 0.125 | 0.090 |
| 17 | SREBF1 | 21 | 10 | 0.050 | 0.085 | 0.090 | 0.105 | 0.100 | 0.105 | 0.100 | 0.105 | 0.100 | 0.085 | 0.090 | 0.070 |
| 4 | KDR | 14 | 8 | 0.365 | 0.350 | 0.160 | 0.105 | 0.105 | 0.100 | 0.100 | 0.100 | 0.020 | 0.280 | 0.365 | 0.390 |
| 13 | FLT1 | 25 | 8 | 0.125 | 0.160 | 0.170 | 0.150 | 0.160 | 0.155 | 0.155 | 0.160 | 0.065 | 0.125 | 0.150 | 0.165 |
| 14 | HSP90AA1 | 20 | 3 | 0.050 | 0.275 | 0.180 | 0.195 | 0.170 | 0.140 | 0.110 | 0.120 | 0.030 | 0.155 | 0.335 | 0.250 |
We excluded the genes for which the maximum power of all the tests was <10% and partitioned the remaining causal genes into three groups based on whether the aSPU test was more powerful than both SKAT and SKAT-O, or between them or less than them. It is clear that for some genes, the aSPU test was more powerful than SKAT and SKAT-O. For gene PIK3C2B, due to the connections between SPU(1) and burden tests and between SPU(2) and SKAT, given that the SPU(1) was more powerful than SPU(2), it is not surprising to see that SKAT-O (which combines SKAT and a burden test) was more powerful than SKAT; furthermore, perhaps due to the relatively high signal sparsity, the SPU(3) turned out to be the most powerful among the SPU tests, leading to the aSPU test being more powerful than both SKAT and SKAT-O. A similar but different situation was found with gene VNN1. That the SPU(1) test was less powerful than SPU(2) might explain why SKAT was more powerful than SKAT-O; however, interestingly, since the SPU(5) test was most powerful among the SPU tests while SPU(4)–SPU(8) were also relatively high powered, we found the aSPU test more powerful than both SKAT and SKAT-O. On the other hand, when SPU(1) or SPU(2) was (nearly) more powerful than other SPU tests, the aSPU test combining all the SPU tests (with only one or two high powered but more low-powered ones) lost edge to either SKAT or SKAT-O, but not both, as for gene RRAS. In some situations, as for gene KDR, since both SPU(1) and SPU(2) were far more superior than other SPU tests, the aSPU test ended up being less powerful than both SKAT and SKAT-O. In summary, we found that, as expected, there was no uniform winner among the aSPU, SKAT, and SKAT-O tests; under some situations, the aSPU test could be more powerful than both SKAT and SKAT-O.
Since all the causal genes contained only a relatively small number of noncausal RVs, and all causal RVs were deleterious (Almasy et al. 2011), most often the SPU(3) test was most powerful among the SPU tests. In contrast, perhaps as expected, the SPU(∞) was almost always least powerful. Importantly, we note that the power of the SPU(3) test could be much larger than SPU(1) and SPU(2), representatives of the burden tests and variance-component tests. For example, for gene PIK3C2B with 23 causal RVs among a total of 60 RVs, the power of SPU(3) was 0.650, much larger than 0.565 and 0.445 for SPU(1) and SPU(2), respectively. It is also confirmed that, even with the same association direction but also with nonassociated RVs, the SPU(1) test might lose power, as for gene VNN1 and BCHE. These points reinforce what was observed in simulation studies: we may need to use SPU tests beyond SPU(1) and SPU(2) to yield high power, supporting the use of the aSPU test in some applications.
We briefly mention two limitations of the GAW17 data. First, the number of RVs in any gene was often relatively small in this mini-exome sequence data set; hence the clear advantage of SPU(γ) tests with γ > 3 did not show up, which in turn limited the potential advantage of the aSPU test. However, with the increasing availability of whole-genome sequence data, we expect a much larger number of RVs in or near a gene, for which we may see a more dramatic advantage of the aSPU test. Second, since all the causal SNVs were deleterious, it favored SKAT-O, which might not perform as well otherwise (as shown in simulations; see Table 3).
We also performed a simulation study to confirm that our methods could control type I error rates satisfactorily with real sequence data at a higher significance level. We randomly selected three genes, RRAS, HIF3A, and PIK3C2B, with 5, 15, and 60 RVs, respectively. To mimic the GAW17 phenotype data, we randomly generated a binary phenotype Yi with Pr(Yi = 1) = 0.3 under H0 for each of the 697 subjects. We then tested H0 for possible association between the phenotype and each gene with 105 simulation replicates so that we could obtain more accurate type I error estimates for a higher significance level α. As shown in Table 5, our methods could satisfactorily control the type I error rate. We note that, for gene RRAS with only 5 RVs, many tests could be conservative with type I errors lower than the nominal level α, due to the highly discrete null distributions of the test statistics. Furthermore, due to its higher-level discreteness and extreme-value-type test statistic, the tail distribution of the SPU(∞) statistic had larger variability, which in turn could perturb that of the aSPU test. As shown in Table 4, since the SPU(∞) test was almost always lowest powered, we suggest excluding it when testing on a few RVs with a binary phenotype. Here, for the purpose of demonstration, we included the SPU(∞) test.
Table 5. Empirical type I error rates at the nominal significance level α based on 105 simulation replicates.
| Gene | α | SPU(1) | SPU(2) | SPU(3) | SPU(4) | SPU(5) | SPU(6) | SPU(7) | SPU(8) | SPU(∞) | aSPU | SKAT | SKAT-O |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RRAS | 0.01 | 0.00689 | 0.00974 | 0.00968 | 0.00963 | 0.00967 | 0.00965 | 0.00965 | 0.00968 | 0.00600 | 0.00791 | 0.00940 | 0.00908 |
| 0.001 | 0.00041 | 0.00099 | 0.00102 | 0.00100 | 0.00097 | 0.00100 | 0.00100 | 0.00100 | 0.00032 | 0.00063 | 0.00085 | 0.00072 | |
| HIF3A | 0.01 | 0.00902 | 0.00905 | 0.00922 | 0.00927 | 0.00924 | 0.00938 | 0.00931 | 0.00936 | 0.01019 | 0.00958 | 0.00980 | 0.00808 |
| 0.001 | 0.00106 | 0.00086 | 0.00099 | 0.00095 | 0.00101 | 0.00098 | 0.00102 | 0.00097 | 0.00126 | 0.00101 | 0.00088 | 0.00074 | |
| PIK3C2B | 0.01 | 0.00943 | 0.00901 | 0.00903 | 0.00894 | 0.00898 | 0.00915 | 0.00902 | 0.00917 | 0.00977 | 0.00933 | 0.00980 | 0.00995 |
| 0.001 | 0.00094 | 0.00081 | 0.00097 | 0.00095 | 0.00090 | 0.00093 | 0.00089 | 0.00095 | 0.00093 | 0.00116 | 0.00086 | 0.00106 |
SNV selection:
The aSPU test can be used for SNV selection. For comparison, we also included two methods: one was the UminP test as used in GWAS, and the other was Lasso-penalized logistic regression (Tibshirani 1996; Zhou et al. 2010). For UminP, we ranked the RVs based on their corresponding P-values. We used R package glmnet to fit Lasso-penalized logistic regression. There is a tuning parameter λ ≥ 0 in Lasso; as one reduces the value of λ, there will be more nonzero coefficient estimates , thus selecting more RVs to be included. In this way, we counted the number of nonzero and the corresponding number of true positives (i.e., the corresponding causal RVs). One problem with Lasso-penalized regression with RVs was that it was difficult to control the number of the nonzero coefficient estimates; as shown in Figure 2 for gene PIK3C2B, we could not obtain 40 or so nonzero coefficients, even with some labor-intensive fine tuning of λ.
Figure 2.
RV selection by aSPU and Lasso for a few causal genes with the GAW17 data.
The methods were compared on the basis of the number of true positives among a given number of their top-ranked RVs for a causal gene. We found that aSPU and Lasso performed similarly, but much better than UminP. Among the 15 causal genes, if we looked at the top six ranked RVs, the frequencies of selecting (0, 1, 2, 3, 4) true positives were the following: (10, 5, 0, 0, 0) by UminP, (3, 6, 4, 1, 1) by Lasso, and (4, 3, 6, 1, 1) by aSPU. Similar results were obtained for other numbers of top-ranked RVs. The bad performance of the UminP test could be due to the unstable variance estimate for a RV, which was too close to 0 (with a too small MAF) and thus dramatically inflated the corresponding test statistic. Figure 2 shows a few more examples in detail. It is confirmed that aSPU and Lasso performed similarly. Given the simplicity of the aSPU test and that Lasso may not yield some given numbers of nonzero coefficient estimates, the use of aSPU for ranking and selecting RVs seems to be promising.
Discussion
We have proposed and studied a class of SPU tests, which are versatile in the sense that in many scenarios at least one of the SPU tests has high power, although the identity of a more powerful SPU test may change with the varying scenario. The SPU tests are based on the score vector of a regression model, rendering it both computationally efficient and general to cover a wide range of applications with binary, quantitative, ordinal, and survival traits and possible covariates. In particular, compared to several other adaptive tests based on estimated (marginal) regression coefficients (e.g., Feng et al. 2011; Zhang et al. 2011; Lin and Tang 2011), the SPU tests, as any score-based test, requires only fitting a simplified model under the null hypothesis. In addition to its computational simplicity, a score-based test is more stable with RVs: for example, with a binary trait, the MLE of a regression coefficient for a RV does not exist if the minor allele appears in only cases or controls (but not both), leading to no convergence with an iterative algorithm to obtain the MLE. On the other hand, as shown earlier, the MLE of a (marginal) regression coefficient is approximately proportional to its score component, implying that, as expected, the score vector is as informative as a vector of the estimated regression coefficients while being computationally much simpler. Our major contribution is that, by recognizing the limitation of the existing adaptive tests with a fixed choice of weights, we allow many possible choices of weights indexed by a single parameter γ ≥ 1. There is a simple interpretation, and thus a guidance, on parameter γ: as the value of γ increases, we upweight the larger components of the absolute value of the score vector, |U|; that is, with a decreasing proportion of the causal RVs in the group to be tested, we expect a SPU(γ) test with a larger value of γ to be more powerful because its upweighting of the larger components of |U| essentially reduces or even eliminates noisy perturbations from many nonassociated RVs to the test statistic, thus maintaining high power in the presence of many nonassociated RVs. In addition, in the presence of both positive and negative association directions for causal RVs, an even number of γ will eliminate the canceling effect of positive and negative components of U. In particular, as compared with some new adaptive tests, such as KBAC, PWST, aSSU, EREC, and SKAT-O, our proposed aSPU test was more adaptive and performed much better in simulations when there were a large number of nonassociated RVs.
For its highly adaptive and versatile performance, the aSPU test has a wide spectrum of applications with other types of traits and/or other genetic variants. For example, our preliminary results showed promising performance of the aSPU test for polygenic testing on association between a binary trait and thousands of CVs (International Schizophrenia Consortium 2009). In principle, the aSPU test can be also applied to gene set or pathway analysis (Liu et al. 2008; Wang et al. 2010).
The relatively good performance of the SSU test and its close relatives, KMR or SKAT (Wu et al. 2010, 2011) and C-alpha test (Neale et al. 2011), was attributed to its treating the regression coefficients β as random effects and then testing on the variance component of the random effects (Basu and Pan 2011). Here, based on the formulation of the SPU tests, more general than the SSU test, we feel that it can be viewed from another angle: the good performance of the SSU test, or more generally, any SPU(γ) test, is due to the weighting of each score component Uj by itself or its power . Since Uj contains association information about RV j, such weights are both simple and informative: specifically, since U has a null distribution N(0, V), a larger component of |Uj| corresponds to stronger evidence of association between the jth RV and the trait and thus assigning a higher weight Uj or will help boost power by reducing the noises introduced to the test statistic with nonassociated RVs. However, depending on the unknown truth, such as the proportion of associated RVs and their specific association effects, a SPU(γ) test with a suitable γ will be more powerful than others. For example, in the presence of many nonassociated RVs, we would expect a larger γ to be more effective: a nonassociated RV is expected to have a smaller |Uj|, and thus almost a zero weight with , and may successfully eliminate the negative effects of many nonassociated RVs on testing. In particular, when the group of RVs to be tested contains a large proportion of nonassociated RVs, we found that a SPU(γ) test with γ > 2 could be much more powerful than the Sum and SSU tests, explaining when and why the aSPU test could outperform SKAT-O and other tests combining a burden test like SPU(1) and a variance component test like SPU(2) (Lee et al. 2012; Derkach et al. 2013; Sun et al. 2013). We also found that a SPU(γ) test with a large γ > 8 gave results similar to that of the SPU(∞) test in our simulations, suggesting that searching γ over 1 to 8 perhaps suffices in many applications.
Since the SPU tests use a mechanism of weighting to minimize the effects of nonassociated RVs, they are analogous to model averaging in the literature of model selection, in contrast to the adaptive Neyman-type tests, such as the aSSU test (Pan and Shen 2011), which are more in the line of model or RV selection. In the current context of analysis of RVs, due to limited information contained within each individual RV, model averaging is expected to outperform model selection, as supported by our empirical results when comparing between the aSPU and aSSU tests. Since in general neither model selection nor model averaging can dominate the other (Yuan and Yang 2005; Shen and Huang 2006), there may be a merit in combining the two approaches in other applications. In addition, a few modifications or extensions are possible. First, in our current implementation of the aSPU test, we simply take the minimum P-value (minP) to combine multiple SPU tests; other combining methods may be equally applied and may be preferred in certain situations (Pan et al. 2010). Second, it appears straightforward to extend the SPU tests to the case with variable thresholds (Price et al. 2010), which is related to adaptive Neyman-type tests (Pan and Shen 2011). Third, we have not evaluated the performance of the SPU tests in the presence of interactions among RVs; in particular, it would be interesting to compare their performance with the KBAC test that was partly designed to detect interactions (Liu and Leal 2010). Finally, although we have pointed out some possible extensions of the aSPU test for (1) analysis of CVs, (2) joint analysis of both CVs and RVs, and (3) pathway analysis of CVs and/or RVs, these topics warrant further investigation in future research. R code is posted on our website at http://www.biostat.umn.edu/∼weip/prog.html.
Supplementary Material
Acknowledgments
We thank the reviewers for many helpful and constructive comments. This research was supported by National Institutes of Health grants R01GM081535, R01CA169122, R01HL065462, R01HL105397, and R01HL116720 and by the Minnesota Supercomputing Institute.
Footnotes
Communicating editor: N. Yi
Literature Cited
- Almasy, L., T. D. Dyer, J. M. Peralta, J. W. Kent, J. C. Charlesworth et al., 2011 Genetic Analysis Workshop 17: mini-exome simulation. BMC Proc. 5(Suppl. 9): S2. [DOI] [PMC free article] [PubMed]
- Bansal V., Libiger O., Torkamani A., Schork N. J., 2010. Statistical analysis strategies for association studies involving rare variants. Nat. Rev. Genet. 11: 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Pan W., 2011. Comparison of statistical tests for association with rare variants. Genet. Epidemiol. 35: 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capanu M., Concannon P., Haile R. W., Bernstein L., Malone K. E., et al. , 2011. Assessment of rare BRCA1 and BRCA2 variants of unknown significance using hierarchical modeling. Genet. Epidemiol. 35: 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. S., Hsu L., Gamazon E. R., Cox N. J., Nicolae D. L., 2012. An exponential combination procedure for set-based association tests in sequencing studies. Am. J. Hum. Genet. 91: 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y. H., Wang G., Leal S. M., Wang S., 2012. A fast and noise-resilient approach to detect rare-variant associations with deep sequencing data for complex disorders. Genet. Epidemiol. 36: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneely K. N., Boehnke M., 2007. So many correlated tests, so little time! Rapid adjustment of p values for multiple correlated tests. Am. J. Hum. Genet. 81: 1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. R., Hinkley D. V., 1974. Theoretical Statistics. Chapman & Hall, London. [Google Scholar]
- Derkach A., Lawless J. F., Sun L., 2013. Robust and powerful tests for rare variants using Fisher’s method to combine evidence of association from two or more complementary tests. Genet. Epidemiol. 37: 110–121. [DOI] [PubMed] [Google Scholar]
- Fan R., Wang Y., Mills J. L., Wilson A. F., Bailey-Wilson J. E., et al. , 2013. Functional linear models for association analysis of quantitative traits. Genet. Epidemiol. 37: 726–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T., Elston R. C., Zhu X., 2011. Detecting rare and common variants for complex traits: sibpair and odds ratio weighted sum statistics (SPWSS, ORWSS). Genet. Epidemiol. 35: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman J. J., van de Geer S., van Houwelingen H. C., 2006. Testing against a high dimensional alternative. J. R. Stat. Soc., B 68: 477–493. [Google Scholar]
- Gordon D., Finch S. J., De La Vega F., 2011. A new expectation-maximization statistical test for case-control association studies considering rare variants obtained by high-throughput sequencing. Hum. Hered. 71: 113–125. [DOI] [PubMed] [Google Scholar]
- Han F., Pan W., 2010. A data-adaptive sum test for disease association with multiple common or rare variants. Hum. Hered. 70: 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T. J., Marini N. J., Witte J. S., 2010. Comprehensive approach to analyzing rare genetic variants. PLoS ONE 5(11): e13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium , 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I., Buxbaum J. D., Laird N. M., Lange C., 2011. A new testing strategy to identify rare variants with either risk or protective effect on disease. PLoS Genet. 7(2): e1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I., Lee S., Makarov D., Buxbaum J. D., Lin X., 2013. Sequence kernel association tests for the combined effect of rare and common variants. Am. J. Hum. Genet. 92: 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Emond M. J., Bamshad M. J., Barnes K. C., Rieder M. J., et al. , 2012. Optimal unified approach for rare variant association testing with application to small sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 91: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Byrnes A. E., Li M., 2010. To identify associations with rare variants, Just WHaIT: weighted haplotype and imputation-based tests. Am. J. Hum. Genet. 87: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. Y., Tang Z. Z., 2011. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 89: 354–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Ghosh D., Lin X., 2008. Estimation and testing for the effect of a genetic pathway on a disease outcome using logistic kernel machine regression via logistic mixed models. BMC Bioinformatics 9: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. J., Leal S. M., 2010. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to GeneMain effects and interactions. PLoS Genet 6: e1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Boerwinkle E., Xiong M. M., 2011. Association studies for next-generation sequencing. Genome Res. 21: 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen B. E., Browning S. R., 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5(2): e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthaler S., Thilly W. G., 2007. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST). Mutat. Res. 615: 28–56. [DOI] [PubMed] [Google Scholar]
- Neale B. M., Rivas M. A., Voight B. F., Altshuler D., Devlin B., et al. , 2011. Testing for an unusual distribution of rare variants. PLoS Genet. 7(3): e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton M. A., Quintana F. A., den Boon J. A., Sengupta S., Ahlquist P., 2007. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann. Appl. Stat. 1: 85–106. [Google Scholar]
- Neyman J., 1937. Smooth test for goodness of fit. Skandinavisk Aktuarietidskrift 20: 149–199. [Google Scholar]
- Pan W., 2009. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genet. Epidemiol. 33: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., 2011. Relationship between genomic distance-based regression and kernel machine regression for multi-marker association testing. Genet. Epidemiol. 35: 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Shen X., 2011. Adaptive tests for association analysis of rare variants. Genet. Epidemiol. 35: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Han F., Shen X., 2010. Test selection with application to detecting disease association with multiple SNPs. Hum. Hered. 69: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Basu S., Shen X., 2011. Adaptive tests for detecting gene–gene and gene–environment interactions. Hum. Hered. 72: 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Kryukov G. V., de Bakker P. I. W., Purcell S. M., Staples J., et al. , 2010. Pooled association tests for rare variants in exon-resequenced studies. Am. J. Hum. Genet. 86: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman S. R., Muller-Myhsok B., 2005. Rapid simulation of P values for product methods and multiple-testing adjustment in association studies. Am. J. Hum. Genet. 76: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Huang H.-C., 2006. Optimal model assessment, selection, and combination. J. Am. Stat. Assoc. 101: 554–568. [Google Scholar]
- Sun J., Zheng Y., Hsu L., 2013. A unified mixed-effects model for rare-variant association in sequencing studies. Genet. Epidemiol. 37: 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R., 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- Wang K., Li M., Hakonarson H., 2010. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 11: 843–854. [DOI] [PubMed] [Google Scholar]
- Wang T., Elston R. C., 2007. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am. J. Hum. Genet. 80: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, P., X. Liu, and Y. X. Fu, (2011 Incorporating predicted functions of nonsynonymous variants into exome sequencing data: a comparative study. BMC Proc. 5(Suppl. 9): S20. [DOI] [PMC free article] [PubMed]
- Wu M. C., Kraft P., Epstein M. P., Taylor D. M., Chanock S. J., et al. , 2010. Powerful SNP-set analysis for case-control genome-wide association studies. Am. J. Hum. Genet. 86: 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. C., Lee S., Cai T., Li Y., Boehnke M., et al. , 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N., Liu N., Zhi D., Li J., 2011. Hierarchical generalized linear models for multiple groups of rare and common variants: jointly estimating group and individual-variants effects. PLoS Genet. 7: e1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Yang Y., 2005. Combining linear regression models: When and how? J. Am. Stat. Assoc. 100: 1202–1214. [Google Scholar]
- Zhang L., Pei Y. F., Li J., Papasian C. J., Deng H. W., 2010. Efficient utilization of rare variants for detection of disease-related genomic regions. PLoS ONE 5(12): e14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Irvin M. R., Arnett D. K., Province M. A., Borecki I., 2011. A data-driven method for identifying rare variants with heterogeneous trait effects. Genet. Epidemiol. 35: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guan W., Pan W., 2013. Adjustment for population stratification via principal components in association analysis of rare variants. Genet. Epidemiol. 37: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Sehl M. E., Sinsheimer J. S., Lange K., 2010. Association screening of common and rare genetic variants by penalized regression. Bioinformatics 26: 2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Feng T., Li Y., Lu Q., Elston R. C., 2010. Detecting rare variants for complex traits using family and unrelated data. Genet. Epidemiol. 34: 171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Fine J. P., Hu J., Lin D. Y., 2004. An efficient resampling method for assessing genome-wide statistical significance in mapping quantitative trait loci. Genetics 168: 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.