Abstract
eIF5A is an essential and evolutionary conserved translation elongation factor, which has recently been proposed to be required for the translation of proteins with consecutive prolines. The binding of eIF5A to ribosomes occurs upon its activation by hypusination, a modification that requires spermidine, an essential factor for mammalian fertility that also promotes yeast mating. We show that in response to pheromone, hypusinated eIF5A is required for shmoo formation, localization of polarisome components, induction of cell fusion proteins, and actin assembly in yeast. We also show that eIF5A is required for the translation of Bni1, a proline-rich formin involved in polarized growth during shmoo formation. Our data indicate that translation of the polyproline motifs in Bni1 is eIF5A dependent and this translation dependency is lost upon deletion of the polyprolines. Moreover, an exogenous increase in Bni1 protein levels partially restores the defect in shmoo formation seen in eIF5A mutants. Overall, our results identify eIF5A as a novel and essential regulator of yeast mating through formin translation. Since eIF5A and polyproline formins are conserved across species, our results also suggest that eIF5A-dependent translation of formins could regulate polarized growth in such processes as fertility and cancer in higher eukaryotes.
Keywords: eIF5A, polarized growth, mating, formins, translation
THE essential eIF5A protein is highly conserved from archaea to mammals, and has a distant ortholog, EF-P, in eubacteria (Park et al. 2010). Interestingly, eIF5A is the only known protein to undergo a unique and essential post-translational modification to form the hypusine residue, and this modification is necessary for its interaction with the 80S ribosome (Jao and Chen 2006; Zanelli et al. 2006). eIF5A was initially described to be involved in the formation of the first peptide bond in translation initiation (Benne and Hershey 1978). Later reports showed that eIF5A acts in translation elongation as its depletion resulted in the accumulation of polysomes in ribosome run-off experiments, yielding polysome profiles similar to those of elongation factor mutants. Additionally, depletion of eIF5A increased the transit time of ribosomes along mRNAs (Gregio et al. 2009; Saini et al. 2009; Li et al. 2010).
Recently, it has been shown that bacterial EF-P is essential for translation of proteins with consecutive proline residues (Doerfel et al. 2013; Ude et al. 2013), and during the course of our studies, this function was also proven for eIF5A in Saccharomyces cerevisiae (Gutierrez et al. 2013). In vitro experiments showed that eIF5A relieves the stall of ribosomes during translation of at least three consecutive prolines, and depletion of eIF5A in yeast cells strongly reduced the translation of polyproline (polyPro) proteins (Gutierrez et al. 2013). However, the connection between the molecular function, physiological roles, and possible targets of eIF5A is still missing.
Two or more isoforms of eIF5A have been identified in all eukaryotes (Park et al. 2010). In humans, two hypusinated isoforms exist; eIF5A-1 is constitutively expressed and is abundant in proliferating cells and some human cancer tissues. In contrast, the expression of eIF5A-2 is normally very low and tissue specific and is mainly found in testis, brain, and certain cancer cell lines. The connection between eIF5A and cancer development is further supported by studies indicating that overexpression of eIF5A-2 is associated with metastasis in several cancer types (Caraglia et al. 2013). The knockout of eIF5A in mice causes early embryonic lethality, indicating a role in mammalian development (Nishimura et al. 2012). In C. elegans, mutation of eIF5A produces slow larval growth, structural abnormalities, and sterile adults (Hanazawa et al. 2004). In Saccharomyces cerevisiae, eIF5A is encoded by two homologous genes, TIF51A and TIF51B. TIF51A is essential and mainly expressed under aerobic conditions, whereas the nonessential TIF51B is only expressed under anaerobic conditions (Schnier et al. 1991; Wohl et al. 1993). The synthesis of the hypusine residue of eIF5A takes place in two steps. The first step involves the enzyme deoxyhypusine synthase, encoded by the yeast essential gene DYS1, transferring an aminobutyl moiety from the polyamine spermidine to a specific lysine group in the N terminus of eIF5A. In a second step, the enzyme deoxyhypusine hydroxylase, encoded in yeast by the nonessential gene LIA1, yields the active hypusinated eIF5A protein through the addition of a hydroxyl group. It is remarkable that both enzymes, which act on just one protein, are evolutionary conserved (Park et al. 2010).
The physiological role of eIF5A has begun to be visualized from several functional studies. Depletion of eIF5A in yeast and inhibition of eIF5A hypusination in higher eukaryotes both cause cell cycle arrest at G1 phase (Hanauske-Abel et al. 1994; Kang and Hershey 1994). Additionally, eIF5A has been identified as a factor involved in the control of mRNA decay, although it is still unclear whether this function is direct or indirect (Zuk and Jacobson 1998; Valentini et al. 2002; Schrader et al. 2006). In yeast temperature-sensitive mutants, the overexpression of factors involved in proper actin organization can suppress the temperature-sensitive phenotype of eIF5A mutants (Valentini et al. 2002; Zanelli and Valentini 2005). It was also documented that eIF5A temperature-sensitive mutants showed changes in actin dynamics at restrictive temperature (Chatterjee et al. 2006). A functional association between eIF5A and cell wall integrity has been established from the studies showing that growth defects of eIF5A and deoxyhypusine synthase (DYS1) temperature-sensitive mutants are partially suppressed in the presence of the osmotic stabilizer sorbitol and that DYS1 mutant is sensitive to cell-wall-perturbing compounds (Valentini et al. 2002; Galvao et al. 2013). Additionally, the overexpression of several components of the PKC pathway also restores growth of eIF5A mutants (Valentini et al. 2002).
Polyamines (putrescine, spermidine, and spermine) are essential for life and their mechanism of action in cells has been thoroughly investigated, although their exact mode of action is far from being resolved. In S. cerevisiae, the metabolic synthesis of spermidine requires the proteins encoded by the genes SPE1, -2 and -3. Under spermidine limiting conditions, yeast spe2 mutants show morphological abnormalities and most of intracellular spermidine is used for the hypusine modification of eIF5A, suggesting that activation of eIF5A is probably the most important function of spermidine in yeast cells (Chattopadhyay et al. 2008). In mammalian cells, spermidine regulates translation at initiation and elongation steps by unknown mechanisms that are partially dependent on eIF5A. These data suggest that the role of polyamines in translation can account for their essentiality in cellular proliferation (Landau et al. 2010).
Spermidine is also indispensable for fertilization efficiency in eukaryotes, from humans to yeast (Lefevre et al. 2011). During sexual reproduction of S. cerevisiae, haploid cells grow a mating projection, the shmoo, toward a pheromone gradient produced by the partner cell. Shmoo formation requires spermidine (Bauer et al. 2013) and allows the fusion of yeast cells of different sexual type to form a diploid zygote. Yeast pheromone response involves the activation of a conserved mitogen-activated protein kinase (MAPK) module through its recruitment by the Ste5 scaffold protein to the pheromone receptor-associated G-protein. The activated MAPK Fus3 modulates the expression of mating-specific genes, promotes cell-cycle arrest, and controls polarized morphogenesis (reviewed in Slaughter et al. 2009; Merlini et al. 2013). One of the targets of Fus3 is the formin Bni1 (Matheos et al. 2004). Like other eukaryotic formins, Bni1 functions in actin cable polymerization (Evangelista et al. 1997; Evangelista et al. 2002; Pruyne et al. 2002; Sagot et al. 2002a,b) and contains several conserved protein domains, which allow its proper localization at sites of polarized growth and its interaction with proteins of the polarisome, such as Spa2 (Fujiwara et al. 1998; Liu et al. 2012). Upon pheromone detection, phosphorylation of Bni1 by Fus3 promotes its localization to the shmoo tip, and thus resulting in the polarized assembly of actin cables (Matheos et al. 2004). Actin plays an essential and complex role in shmoo formation since Bni1-induced actin cables are required for stable recruitment of the Ste5 scaffold at the shmoo, yielding polarized recruitment and full activation of MAPK Fus3 (Qi and Elion 2005). Spermidine is not only crucial for yeast mating but also essential for effective fertilization in Caenorhabditis elegans, suggesting that its role in fusion of haploid gametes is conserved in higher eukaryotes (Bauer et al. 2013).
We investigated the connection between spermidine-hypusinated eIF5A and polarized growth during shmoo formation in S. cerevisiae. Our results show a mechanistic connection between eIF5A and translation of Bni1 formin to allow organization of actin cables to form the shmoo. Eukaryotic formins, like Bni1, contain polyPro segments in their sequences. Our data suggest that eIF5A has an essential role in fertility and polarized cell growth in eukaryotes through the translation of polyPro formins.
Materials and Methods
Yeast strains and growth conditions
The strains and plasmids used in this study are listed in Supporting Information, Table S1 and Table S2, respectively, and primers are listed in Table S3. All strains were generated in the S. cerevisiae haploid strain BY4741 (MATa his3∆0 leu2∆0 met15∆0 ura3∆0) and grown in liquid YPD medium (1% yeast extract, 2% peptone, 2% glucose). A PCR-based genomic tagging technique was employed to tag genomic full-length BNI1 or BNI1 harboring the deletion of amino acids 1240–1954 at the C terminus (BNI1ΔCt) with 6HA. The plasmid PA171 was used as a template for PCR reactions using primers BNI1-1 and BNI1-2 for HA tagging of full-length BNI1, and BNI1-2 and BNI1-5 for C-terminus-truncated BNI1 (Table S3). The resulting cassettes were transformed in tif51A-1 and wild-type strains and transformants were selected in synthetic complete medium lacking histidine. To generate the strains containing a genomic 6HA tag of a BNI1 mutant harboring the deletion of polyPro stretches (amino acids 1239–1307) (BNI1ΔPro), the C terminus BNI1-HA sequence was amplified from genomic DNA of the strain PAY723 using primers BNI1–2 and BNI1–6. The use of the primer BNI1–6 resulted in the deletion of nucleotides 3715–3921 in BNI1, which encode for the polyPro stretches of the Bni1 protein. The resulting PCR product was transformed in tif51A-1 and wild-type cells, and transformants were selected in synthetic complete medium lacking histidine.
For experiments with null mutants, strains were grown at 30° to the mid-log phase. For experiments with temperature-sensitive mutants, strains were maintained at the permissive temperature of 25° and transferred to the nonpermissive temperature of 37° during mid-log phase. Otherwise indicated, all pheromone treatments were done with 10 μg/ml of α-factor (Sigma) for 2 hr.
To overexpress Bni1 protein, a multicopy plasmid (PA291) containing a GAL promoter fused to BNI1-4HA (pGAL-BNI-HA) was used (kindly provided by David Pellman) (Kono et al. 2012) (Table S2). Wild type, tif51A-1, tif51A-3, and bni1Δ were transformed with PA291, and transformants were selected in synthetic complete media lacking uracil. To induce Bni1 overexpression, cells were grown in liquid synthetic complete media lacking uracil and containing 2% galactose as the only carbon source (Sgal −ura).
Shmooing efficiency and Spa2 localization
Strains were grown in experimental temperatures and treated with α-factor for 2 hr. Cells were visualized by microscopy and ∼300 cells were analyzed per sample from at least two independent experiments. To specifically visualize the sublocalization of Spa2, an overexpression plasmid containing SPA2-GFP fusion was used (Arkowitz and Lowe 1997). Localization of Spa2-GFP was monitored by visual inspection in at least 100 cells from two independent experiments.
F-actin staining
For the visualization of F-actin, cells were fixed with 4% formaldehyde for 1 hr at experimental temperatures and then collected by centrifugation. Pellets were incubated overnight with 9% Alexa Fluor 488-phalloidin (Molecular Probes) and resuspended in PBS to be visualized by fluorescence microscopy.
Microscopy
All microscopy were carried out at room temperature using the Axioskop 2 fluorescence microscope (Zeiss), and images were taken using an AxioCam MRm (Zeiss). For fluorescent microscopy, the power unit AttoArc HBO 100W (Zeiss) was used to visualize the indicated fluorophores.
Western blot and quantitative RT-PCR
Western blot analyses were performed as previously described (Miguel et al. 2013). Antibodies used were anti-HA 12C5A (Roche), anti-Hxk2 (a kind gift from F. Rández-Gil, Instituto de Agroquímica y Tecnología de los Alimentos, Valencia), and anti-eIF5A antibody (kindly provided by S. R. Valentini, Säo Paulo State University, Säo Paulo, Brazil) (Valentini et al. 2002). Band intensities of immunoblots were detected and quantified with the luminescent image analyzer ImageQuant LAS 4000 mini (GE Healthcare). Reverse transcription and quantitative PCR (RT-qPCR) analyses, performed as described in Garre et al. (2013), were used to quantify BNI1-HA, BNI1ΔCt-HA, and BNI1ΔPro-HA mRNA amount (primers BNI1–3 and BNI1–4, Table S3), and HXK2 mRNA amount (primers HXK2-1 and HXK2-2, Table S3) was determined as a control. Transcript amount was normalized against endogenous ACT1 expression using established primers ACT1-1 and ACT1-2 (Table S3). Translation efficiencies of BNI1-HA, BNI1ΔCt-HA, BNI1ΔPro-HA, and HXK2 were estimated as the protein/mRNA ratio, using the Western and RT-qPCR data (normalized against ACT1 expression) obtained from cell samples of the same experiment. BNI1-HA, BNI1ΔCt-HA, and BNI1ΔPro-HA translation efficiencies are expressed relative to HXK2 translation efficiency and represented as a fraction against 25° for each strain.
β-Galactosidase assay
To investigate the induction of cell fusion genes in response to pheromone, the membrane protein FUS1 was expressed in a fusion plasmid containing the reporter gene LacZ (Table S2). As a positive control, osmotic stress-induced gene STL1 coupled with LacZ was used (Table S2). Biological triplicates were grown at 25° to the mid-log phase and transferred to the nonpermissive temperature of 37° for 4 hr and then incubated with 10 μg/ml of α-factor for 1 or 2 hr. For STL1 induction, control cells were stressed with 0.6 M KCl for 45 min. β-Galactosidase activity was determined from three biological triplicates as previously described (Garre et al. 2012).
Results
Pheromone-induced shmoo formation is inhibited in eIF5A mutants
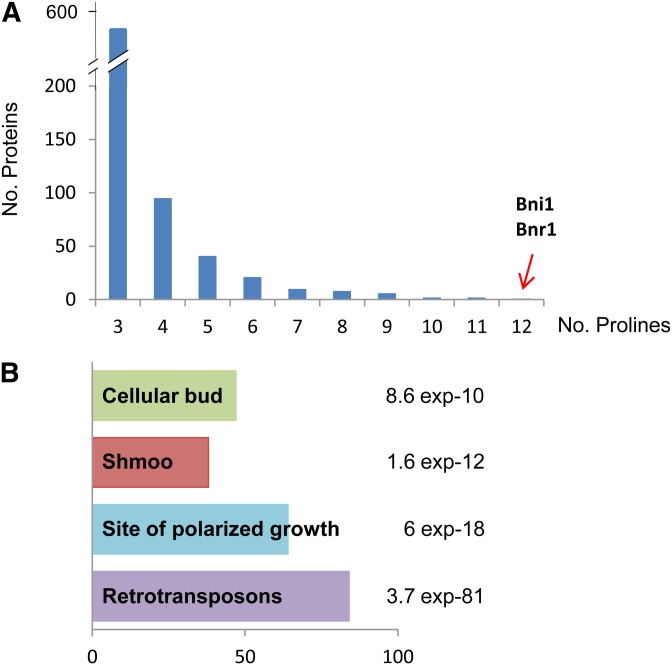
Recently, it was discovered that the bacterial eIF5A-ortholog factor EF-P is essential for the translation of proteins with consecutive prolines (polyPro proteins) (Doerfel et al. 2013; Ude et al. 2013). Furthermore, S. cerevisiae mutant in the spermidine biosynthesis SPE2 gene is unable to mate in low-spermidine media (Bauer et al. 2013), and given the role of spermidine in eIF5A activation (Park et al. 2010), we proposed that hypusinated eIF5A may be involved in fertility through the translation of polyPro proteins with roles in fertilization. To test this hypothesis, we first investigated the number and functional roles of the polyPro proteins in S. cerevisiae. As shown in Figure 1A, 549 yeast proteins were found to contain at least one motif of three consecutive proline residues and the number of proteins is much reduced for those with four or more consecutive prolines. A search in the Gene Ontology database (GO) revealed four significant overrepresented categories, including “site of polarized growth” and “mating projection (shmoo)” (Figure 1B). Only two yeast proteins, Bnr1 and Bni1, contain 10 or more consecutive prolines. These two proteins are evolutionary conserved formins, regulating actin cytoskeleton during budding (Imamura et al. 1997; Pruyne et al. 2004), with Bni1 also involved in shmoo formation during mating (Evangelista et al. 1997). From these data, we investigated whether eIF5A is involved in shmoo formation through the regulation of Bni1.
Figure 1.
Number and functional categories of S. cerevisiae proteins containing three or more consecutive prolines. (A) Number of yeast proteins containing polyPro stretches with the indicated consecutive prolines. Formins Bni1 and Bnr1 are the only yeast proteins with polyproline stretches with 12 or more prolines. (B) GO functional categories significantly overrepresented in the 549 yeast genes containing a polyPro stretch with at least three consecutive prolines. Bars represent the number of genes found in each category and the P-value is indicated for each category.
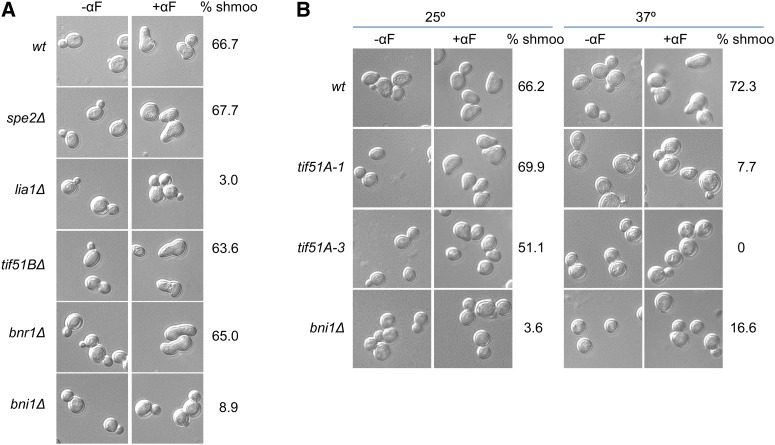
With this aim, we first evaluated the ability to form shmoo projections under pheromone treatment of mutants in genes SPE2 (biosynthesis of spermidine), LIA1 (deoxyhypusine hydroxylase of eIF5A), mutants in the formins BNR1 and BNI1, and mutant in the nonessential eIF5A gene, TIF51B. The lia1Δ mutant was unable to form shmoos in the presence of α-factor, whereas, as expected in a rich media where spermidine is available, spe2Δ showed a percentage of shmoo formation similar to the wild type (Figure 2A). Since the only known target of Lia1 is eIF5A, these results suggest that hypusination of eIF5A is necessary for shmoo formation and that perhaps the vital requirement of spermidine relies on eIF5A activation by hypusination. Furthermore, tif51BΔ was able to shmoo, and, as it has been described, the formin Bni1, but not Bnr1, is essential for shmooing (Figure 2A).
Figure 2.
Yeast shmoo formation requires hypusinated eIF5A. (A and B) Representative DIC images of wild type and mutants with or without treatment of 10 μg/ml α-factor for 2 hr. Null mutants were maintained in 30° (A), and temperature-sensitive mutants were maintained at 25° or transferred to 37° for 4 hr (B). Quantification of percentage of cells containing shmoos in samples treated with α-factor are indicated. Approximately 300 cells were manually counted for each sample from at least two independent experiments.
Next, since eIF5A is an essential protein in yeast, we used eIF5A temperature-sensitive strains previously described (Valentini et al. 2002; Li et al. 2011) to evaluate the formation of mating projections. Mutants tif51A-1 and tif51A-3 contain a single (Pro83 to Ser) and double (Cys39 to Tyr, Gly118 to Asp) mutation, respectively, in eIF5A protein. Both mutants showed a drop in viability after 8 hr of incubation at 37°, with a more severe growth defect in tif51A-3 (Figure S1, A and B). Accordingly, eIF5A protein was mostly depleted after 4 hr of incubation at 37° in both mutants (Figure S1, C and D). Treatment with 10 μg/ml α-factor induced shmoo formation in eIF5A mutants and wild type at permissive temperature (25°) (Figure 2B). However, at restrictive temperature (37°), tif51A-1 yielded very low shmooing (7.7%) and tif51A-3 was unable to shmoo, and most cells in both strains appear ellipsoidal. As previously described, bni1 mutant showed a low percentage of shmoo formation at both temperatures (Figure 2B). As positive controls in shmoo formation at 37°, we also used two other temperature-sensitive mutants in essential genes (top2-1 mutation in topoisomerase II and xpo1-1 mutation in exportine Crm1/Xpo1) and observed that, in both, the percentage of shmoo formation was similar at permissive and restrictive temperatures (Figure S2). Taken together, these results indicate that hypusinated eIF5A is required for successful shmoo formation.
eIF5A is required to shmoo localize Spa2, induce FUS1 gene, and assemble actin cables during pheromone stimulation
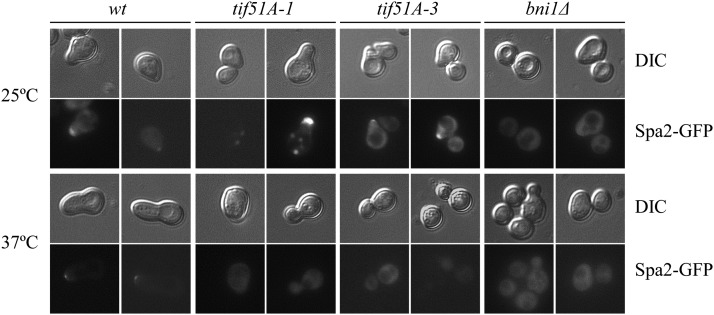
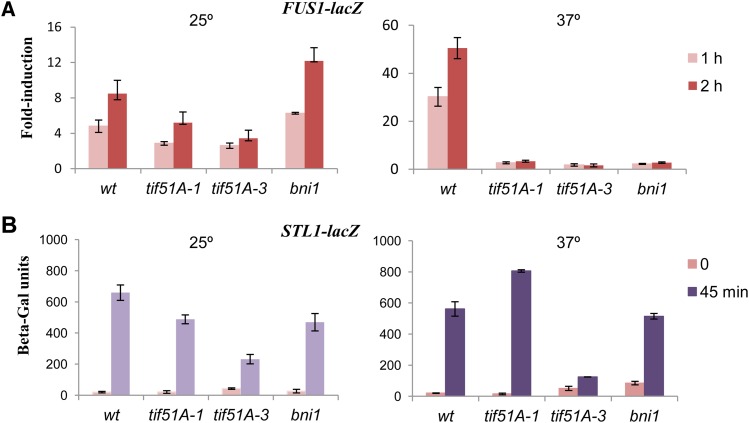
We further investigated the basis for the defect in shmoo formation in eIF5A mutants by visualizing the localization of Spa2, a component of the polarisome that localizes at the shmoo tip during mating through its interaction with Bni1 (Fujiwara et al. 1998; Liu et al. 2012). Upon pheromone treatment at 25°, wild-type and eIF5A mutant cells showed Spa2-GFP localization at the shmoo tip, meanwhile Spa2 was delocalized in bni1Δ (Figure 3). Inactivation of eIF5A by incubation at 37° inhibited polarized localization of Spa2 in tif51A-1 and tif51A-3 mutants (Figure 3). Furthermore, to determine the full activation of the MAP kinase Fus3 with α-factor, we examined the expression of the FUS1 gene, which encodes a pheromone-induced transmembrane protein involved in cell fusion (Trueheart and Fink 1989). A FUS1-lacZ fusion was induced under pheromone treatment in wild type and to a lower level in tif51A mutants at permissive temperature. However, no FUS1-lacZ induction was observed in eIF5A mutants at restrictive temperature (Figure 4A). In bni1Δ, FUS1 gene was not induced at 37°, and surprisingly, FUS1 induction was observed at 25° (Figure 4A). In contrast to the observed defects in FUS1 induction, tif51A mutants were able to induce an osmotic stress-responsive gene (STL1) fused to LacZ during osmotic stress treatment, indicating that, at experimental conditions, tif51A mutants maintain the ability to induce signaling pathways in response to external stimuli (Figure 4B).
Figure 3.
Localization of the polarisome protein Spa2 at the shmoo tip in response to pheromone requires eIF5A. Representative GFP and their corresponding DIC images of wild type, bni1Δ, tif51A-1, and tif51A-3 mutants overexpressing Spa2-GFP. Cells were maintained at 25° or transferred to 37° for 4 hr. Cells were examined after treatment with 10 μg/ml α-factor for 2 hr.
Figure 4.
Expression of FUS1-lacZ in response to pheromone requires eIF5A. (A and B) Expression of FUS1-LacZ (A) or STL1-LacZ fusion (B) in wild type and mutants. Cells were grown at 25° or 4 hr at 37° and then treated with 10 μg/ml α-factor for either 1 or 2 hr (A) or stressed with 0.6 M KCl for 45 min (B). Error bars represent the standard error of the mean of three or more biological replicates.
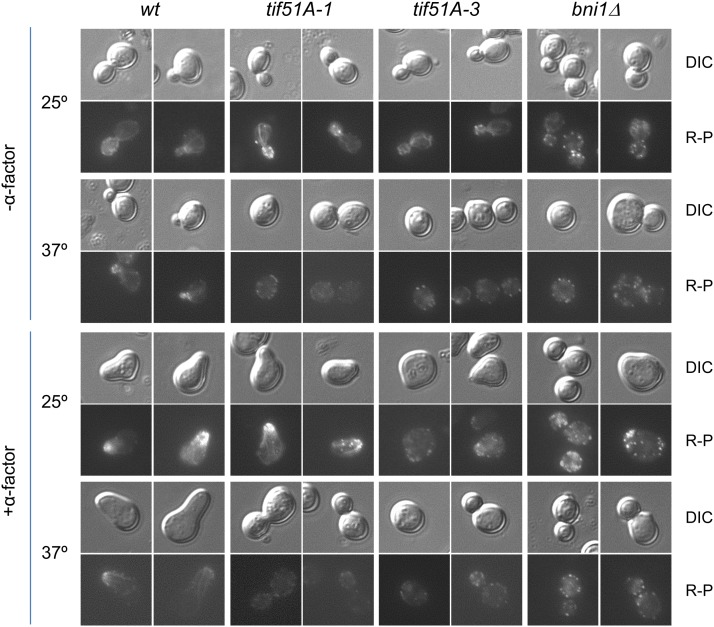
It has been shown that depletion of eIF5A causes changes in the actin cytoskeleton by unknown mechanisms (Chatterjee et al. 2006; Zanelli and Valentini 2007). We therefore investigated the requirements of eIF5A for pheromone-induced polarization of actin cables. Phalloidine staining of actin before and after treatment with pheromone was visualized by fluorescent microscopy (Figure 5). At 25°, in the absence of α-factor, eIF5A mutants showed a normal pattern of actin with patches and cables (Figure 5). Addition of α-factor at permissive temperature induced the formation of actin cables from the shmoo in tif51A-1 and wild-type cells, and actin was visualized mainly as patches in tif51A-3. However, at 37° with α-factor, both eIF5A mutants were unable to form actin cables, but contained delocalized actin patches. Consistent with previous reports (Matheos et al. 2004), actin was observed as intense delocalized patches in all conditions in bni1 mutant. Therefore, eIF5A is necessary for the formation of actin cables upon pheromone stimulation.
Figure 5.
Polymerization and localization of actin cables require eIF5A during shmoo formation. Representative fluorescence and their corresponding DIC images of wild type, tif51A-1, tif51A-3, and bni1Δ showing F-actin staining by phalloidin at 25° or after incubation for 4 hr at 37°, in the presence or absence of treatment with 10 μg/ml α-factor for 2 hr.
Taken together, these results indicate that eIF5A controls different outcomes of pheromone signaling, including induction of pheromone-responsive genes and localization of components of the polarisome at the shmoo. Given the essential role of actin in the induction of cell polarization during mating (Slaughter et al. 2009), these downstream effects could be explained by a direct role for eIF5A in actin polymerization.
eIF5A is required for the translation of the polyproline formin Bni1
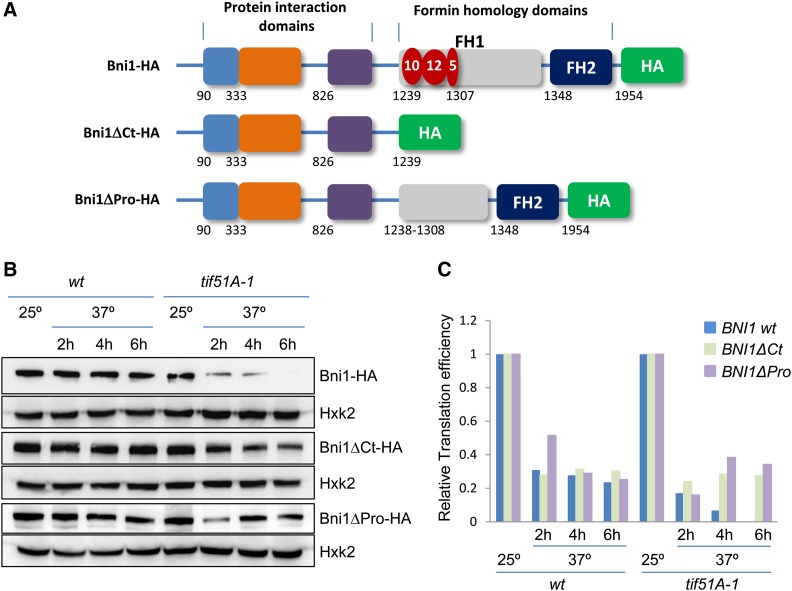
Cells depleted in eIF5A show similar defects in polarization during mating as the bni1 mutant. The formin Bni1 is a large protein containing three segments of 10, 12, and 5 consecutive proline residues in the formin homology domain 1 (FH1), the domain required for assembly of actin filaments (Figure 6A) (Sagot et al. 2002b). Since it has been shown that the bacterial ortholog of eIF5A, EF-P, enhances the synthesis of proteins containing polyPro stretches (Doerfel et al. 2013; Ude et al. 2013), we hypothesized that yeast eIF5A may be involved in the translation of the polyPro motifs of Bni1 and that low levels of Bni1 in eIF5A mutants is the basis for their inability to shmoo. To test this, we obtained two genomic HA-tagged versions of BNI1 in wild type and tif51A-1; a full-length BNI1 with HA after the coding region (BNI1-HA) and a truncated BNI1 with HA fused before the first polyPro stretch (BNI1ΔCt-HA) (Figure 6A). The shift of wild-type cells from 25° to 37° slightly reduced the protein levels of Bni1-HA and Bni1ΔCt-HA. However, Bni1-HA level was importantly reduced in tif51A-1 cells after 2 hr of incubation at 37° and was almost undetectable after 6 hr. Conversely, we only observed a partial reduction in Bni1ΔCt-HA protein levels in tif51A-1 after the shift to 37°, which was sustained up to 6 hr (Figure 6B). We analyzed BNI1-HA and BNI1ΔCt-HA mRNA expression in the same cell cultures by RT-qPCR and calculated the protein/mRNA ratios as an indication of translation efficiency, which was compared to the translation efficiency of a control gene, HXK2, encoding hexokinase 2, a protein with no polyPro motifs. After temperature shift, tif51A-1 showed an important decrease in the translation efficiency of full-length BNI1, reaching almost zero after 6 hr. In comparison, the translation efficiency of C-terminus truncated BNI1 lacking the polyPro motifs was sustained from 2 to 6 hr upon eIF5A depletion (Figure 6C). These results suggested that eIF5A controls the expression of the Bni1 formin and is required for the translation of the polyPro motifs. To further demonstrate that the polyPro motifs are responsible for the BNI1 translation dependency on eIF5A, we obtained an additional bni1 mutant, in which only a short genomic region of the FH1 domain containing the polyPro stretches was deleted, maintaining intact the rest of the BNI1 C terminus (BNI1ΔPro-HA) (Figure 6A). In tif51A-1, the translation efficiency of Bni1ΔPro also reduced at 37° but was sustained at 4 and 6 hr of incubation at restrictive temperature, with similar efficiencies observed in the wild-type strain (Figure 6, B and C). This result, showing eIF5A independency of this BNI1 version with the polyPro stretches deleted, strongly supports the conclusion that eIF5A is involved in the translation of the proline-rich motifs of Bni1.
Figure 6.
Translation of the proline stretches of the formin BNI1 requires eIF5A. (A) Schematic diagrams showing C-terminal HA genomic tagging of full-length (BNI1-HA), C-terminal truncated (Δ1240–1954, BNI1ΔCt-HA), and proline-deleted (Δ1239–1307, BNI1ΔPro-HA) Bni1. Formin homology (FH) and protein interaction domains are indicated, as well as the polyPro stretches with the number of consecutive prolines represented in red. The numbers below the boxes indicate the first amino acid of each domain or stretch. (B) Immunoblots with antibodies against HA and hexokinase 2 protein (Hxk2) to show expression of full-length Bni1, C-terminal truncated Bni1, proline-deleted Bni1 and Hxk2, in wild type and tif51A-1 at 25° or 37° at the indicated times. (C) Translation efficiency of BNI1-HA, BNI1ΔCt-HA, and BNI1ΔPro-HA relative to translation efficiency of HXK2 in wild type and tif51A-1. Protein/mRNA ratios for Bni1, Bni1ΔCt-HA, Bni1ΔPro-HA, and Hxk2 were calculated by Western (B) and quantitative PCR from the same samples. Translation efficiencies of BNI1-HA, BNI1ΔCt-HA, and BNI1ΔPro-HA are calculated relative to translation efficiency of HXK2 and represented as a fraction against 25° for each strain.
It is noteworthy to mention that the above results could have an alternative interpretation, where the differences in protein amount seen between Bni1 wild type and Bni1 mutants are due to different protein degradation rates, and it could be that eIF5A is involved in the stabilization of only the wild-type Bni1 protein. Although we cannot exclude this possibility, the fact that the specific elimination of the Bni1 polyPro stretches renders the protein eIF5A independent, deems the interpretation that the eIF5A elongation factor is involved in Bni1 translation rather than in the regulation of protein stability to be more reasonable. Bni1 protein stability has been studied in a previous work in which it was demonstrated that local cell wall/membrane damage results in the PKC-regulated proteosomal degradation of Bni1 (Kono et al. 2012). Moreover, it was described that the Bni1 N-terminal region (amino acids 1–642), which does not contain the polyPro motifs, is responsible for this regulated degradation that can be provoked by heat shock at 39° (Kono et al. 2012). Perhaps, our observation that Bni1 protein levels slightly drop in all strains during the shift from 25° to 37° (Figure 6B), could be explained by a regulated Bni1 degradation during a mild heat shock.
Overexpression of Bni1 protein partially restores the shmoo formation defect of eIF5A mutants
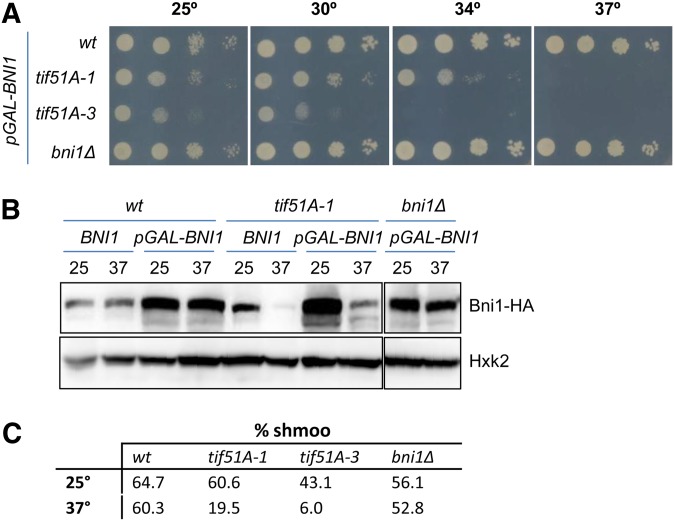
Finally, we explored the possibility of suppression of the eIF5A mutant shmoo defect by increasing the cellular levels of Bni1. To do so, we used a multicopy plasmid containing a BNI1-HA fusion under the control of the strong GAL promoter (Kono et al. 2012). Given the unessential nature of Bni1 to cell viability, the lack of this protein cannot explain the essentiality of eIF5A, and therefore, as expected, the growth defect at restrictive temperature of tif51A-1 and tif51A-3 mutant cells was not recovered when BNI1 was overexpressed in media containing galactose (Figure 7A). Upon galactose overexpression conditions, we observed a high increase in Bni1 protein levels in wild type and in tif51A-1 at permissive temperature with respect to the genomic endogenous Bni1 levels; however, Bni1 protein level increased only slightly in tif51A-1 at restrictive temperature (Figure 7B). This result further confirms that eIF5A is necessary for the production of Bni1 protein, even when overexpressing the BNI1 gene. Nevertheless, a slight increase in Bni1 protein was obtained in eIF5A mutants at restrictive temperature; therefore, we studied whether this increase could improve the shmoo formation of eIF5A mutants. As expected, a complete complementation of the shmoo formation defect in bni1 mutant was observed by inducing pGAL-BNI1. We also observed, at restrictive temperature, higher percentages of shmoo formation in tif51A-1 (19.5%) and tif51A-3 (6.9%) (Figure 7C) with respect to what was observed in these strains without BNI1 overexpression (7.7 and 0%, respectively; Figure 2B) This partial suppression of shmoo defect seen in eIF5A mutants by a slight increase of Bni1 protein levels further supports that the shmooing defect in these mutants is due to the low levels in Bni1 formin. Additionally, since increasing Bni1 protein levels does not produce a total restoration of shmoo formation ability in eIF5A mutants, it suggests that more Bni1 protein would be necessary for a full complementation or that other proteins involved in shmoo formation might be under the translational control of eIF5A.
Figure 7.
Overexpression of the formin BNI1 partially restores the shmoo formation defect of eIF5A mutants. (A) Overexpression of BNI1 does not restore the growth defect of eIF5A thermosensitive mutants at restrictive temperature. Growth of the indicated yeast strains containing a plasmid expressing BNI1 under the GAL promoter (pGAL-BNI1) was tested in Sgal −ura plates incubated at the indicated temperatures. (B) Immunoblots with antibodies against HA and hexokinase 2 protein (Hxk2) to show the expression of genomic Bni1 (BNI1) or the overexpression of Bni1 (pGAL-BNI1) at 25° or 37° for 4 hr. (C) Shmoo formation is partially restored in eIF5A mutants. The indicated strains containing a pGAL-BNI1 plasmid were grown at 25° in Sgal−ura media and then were maintained at 25° or transferred to 37° for 4 hr. Quantification of percentage of cells forming shmoos in samples treated with 10 μg/ml α-factor are indicated. Approximately 300 cells were manually counted for each sample from at least two independent experiments.
Discussion
The budding yeast is a model for studying the establishment of cellular asymmetry, in particular during the formation of a mating projection (shmoo) toward a pheromone gradient. It is remarkable that many of the genes that control cell polarity are conserved between yeast and more complex eukaryotes. Polarized growth requires the asymmetric organization of the cytoskeleton and the formation of actin cables nucleated by formins (Slaughter et al. 2009). S. cerevisiae formin Bni1 contains long stretches of consecutive prolines located in the formin homology domain 1, which has been conserved among eukaryotic formins, and is required for the formation of actin cables to form a shmoo in response to pheromone (Pruyne et al. 2002; Sagot et al. 2002b). In our study, we discovered that the translation factor eIF5A and the enzyme Lia1 required for its activation through hypusination are both essential for shmoo formation. Moreover, eIF5A mutants showed defects in all downstream events (polarized localization of the polarisome component Spa2, induction of the fusion gene FUS1, and actin polarization) of the pheromone response. Since Bni1 is activated by pheromone-responsive MAP kinase Fus3 and, concurrently, Bni1 is required for full activation of Fus3 by mediating its polarized localization to the shmoo (Matheos et al. 2004; Qi and Elion 2005), our data suggest that the defect in shmoo formation in eIF5A mutants is a direct consequence of dramatically low levels of Bni1 protein upon eIF5A depletion. Our results using three genomically tagged BNI1 versions, a full-length Bni1 whose translation is sensitive to eIF5A and two Bni1 proteins containing deletions of the polyPro stretches, by the deletion of the C terminus (Bni1ΔCt) or by a specific deletion of the three polyPro motifs (Bni1ΔPro), for which translation is eIF5A independent, strongly suggest that eIF5A is required for translation of these proline-rich sequences. Overall, our results connect, for the first time, eIF5A molecular function with a physiological role and a specific target.
It is worth mentioning that the defects in shmooing (Figure 2) and actin cable formation (Figure 5) are more severe in eIF5A mutants than in bni1 mutant. Additionally, the restoration of shmoo formation is only partial when increasing Bni1 protein levels in eIF5A mutants (Figure 7). These results can be explained by the putative role of eIF5A in the translation of other polyPro proteins involved in shmoo formation and actin organization. Indeed, the GO functional category of “Mating Projection” was significantly represented in the group of proteins with three or more consecutive prolines (Figure 1). Specifically, 38 proteins of the 119 in this GO functional category are polyPro proteins, and five of them contain at least six consecutive prolines. In respect to the observed actin cable formation and cytoskeleton organization defects in eIF5A mutants, five yeast proteins with these functions contain polyPro motifs with at least seven consecutive prolines, with one of them being the only other yeast formin, Bnr1. All of these polyPro proteins are putative targets of eIF5A, which should be further investigated.
The role of eIF5A in yeast mating could be extended to fertility in higher eukaryotes. Spermidine is essential for successful reproduction in eukaryotes (Bauer et al. 2013) and is also the substrate for eIF5A hypusination, a modification that is required for eIF5A binding to the ribosomes (Park et al. 2010). Given that reproduction and cell fusion during eukaryotic reproduction requires polarized growth, our results suggest that translation of polyPro formins, conserved across species from yeast to humans, could regulate this process.
Yeast and mammalian essential eIF5A proteins are 65% identical. Moreover, human eIF5A expressed in yeast cells becomes hypusinated and can functionally substitute yeast eIF5A in strains carrying eIF5A mutation (Schnier et al. 1991; Magdolen et al. 1994). Additionally, eukaryotic formins are large proteins with conserved C-terminal FH domains, which contain multiple proline-rich motifs that recruit actin monomers and nucleate actin filaments (Breitsprecher and Goode 2013). From our results and the existence of high sequence and functional homologies in eIF5A and formins, it can be conceptualized that other important roles of human eIF5A could be a consequence of formin-controlled translation. In humans, 15 different formins exist and are involved in actin organization during cell adhesion, cytokinesis, cell polarity or morphogenesis, and have essential roles in cell proliferation and migration in the context of human disease (Deward et al. 2010; Breitsprecher and Goode 2013; Favaro et al. 2013). As eIF5A has been reported as a novel oncogenic protein in many types of human tumors and its overexpression has been reported to be associated with metastasis in multiple cancer types (Caraglia et al. 2013; Wang et al. 2013), and, additionally, formins participate in cancer cell migration and invasion (Lizarraga et al. 2009; Kitzing et al. 2010; Zhu et al. 2011; Favaro et al. 2013), it would also be of great interest to explore the connection between eIF5A and formin translation in cancer and metastasis.
Supplementary Material
Acknowledgments
We thank M. E. Pérez-Martínez and I. Quilis for their help in viability and β-galactosidase experiments; J. E. Pérez-Ortín for critical reading of the manuscript; and F. Rández-Gil, G. Ammerer, J. Warringer, and S. R. Valentini for materials. We acknowledge funding from the Spanish MCINN (BFU2010-21975-C03-01), the Generalitat Valenciana (PROMETEO 2011/088 and ACOMP/2012/001), Universitat de València (UV-INV-AE13-139034), and support from European Union funds (FEDER). T.L. and B.B.-P. are recipients of a PROMETEO and a VALi+d predoctoral (ACIF2010/085) contract, respectively, from the Generalitat Valenciana.
Footnotes
Communicating editor: A. Hinnebusch
Literature Cited
- Arkowitz R. A., Lowe N., 1997. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J. Cell Biol. 138: 17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M. A., Carmona-Gutierrez D., Ruckenstuhl C., Reisenbichler A., Megalou E. V., et al. , 2013. Spermidine promotes mating and fertilization efficiency in model organisms. Cell Cycle 12: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R., Hershey J. W., 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253: 3078–3087. [PubMed] [Google Scholar]
- Breitsprecher D., Goode B. L., 2013. Formins at a glance. J. Cell Sci. 126: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraglia M., Park M. H., Wolff E. C., Marra M., Abbruzzese A., 2013. eIF5A isoforms and cancer: Two brothers for two functions? Amino Acids 44: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I., Gross S. R., Kinzy T. G., Chen K. Y., 2006. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Molecular genetics and genomics. MGG 275: 264–276. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M. K., Park M. H., Tabor H., 2008. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc. Natl. Acad. Sci. USA 105: 6554–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWard A. D., Eisenmann K. M., Matheson S. F., Alberts A. S., 2010. The role of formins in human disease. Biochim. Biophys. Acta 1803: 226–233. [DOI] [PubMed] [Google Scholar]
- Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., et al. , 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M. S., Chow C. J., Adames N., et al. , 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276: 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A., 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4: 260–269. [DOI] [PubMed] [Google Scholar]
- Favaro P., Traina F., Machado-Neto J. A., Lazarini M., Lopes M. R., et al. , 2013. FMNL1 promotes proliferation and migration of leukemia cells. J. Leukoc. Biol. 94: 503–512. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Tanaka K., Mino A., Kikyo M., Takahashi K., et al. , 1998. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9: 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao F. C., Rossi D., Silveira Wda S., Valentini S. R., Zanelli C. F., 2013. The deoxyhypusine synthase mutant dys1–1 reveals the association of eIF5A and Asc1 with cell wall integrity. PLoS ONE 8: e60140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garre E., Romero-Santacreu L., Barneo-Munoz M., Miguel A., Perez-Ortin J. E., et al. , 2013. Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress. PLoS ONE 8: e61240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garre E., Romero-Santacreu L., De Clercq N., Blasco-Angulo N., Sunnerhagen P., et al. , 2012. Yeast mRNA cap-binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol. Biol. Cell 23: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregio A. P., Cano V. P., Avaca J. S., Valentini S. R., Zanelli C. F., 2009. eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 380: 785–790. [DOI] [PubMed] [Google Scholar]
- Gutierrez E., Shin B. S., Woolstenhulme C. J., Kim J. R., Saini P., et al. , 2013. eIF5A promotes translation of polyproline motifs. Mol. Cell 51: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauske-Abel H. M., Park M. H., Hanauske A. R., Popowicz A. M., Lalande M., et al. , 1994. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim. Biophys. Acta 1221: 115–124. [DOI] [PubMed] [Google Scholar]
- Hanazawa M., Kawasaki I., Kunitomo H., Gengyo-Ando K., Bennett K. L., et al. , 2004. The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech. Dev. 121: 213–224. [DOI] [PubMed] [Google Scholar]
- Imamura H., Tanaka K., Hihara T., Umikawa M., Kamei T., et al. , 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16: 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao D. L., Chen K. Y., 2006. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 97: 583–598. [DOI] [PubMed] [Google Scholar]
- Kang H. A., Hershey J. W., 1994. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J. Biol. Chem. 269: 3934–3940. [PubMed] [Google Scholar]
- Kitzing T. M., Wang Y., Pertz O., Copeland J. W., Grosse R., 2010. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene 29: 2441–2448. [DOI] [PubMed] [Google Scholar]
- Kono K., Saeki Y., Yoshida S., Tanaka K., Pellman D., 2012. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell 150: 151–164. [DOI] [PubMed] [Google Scholar]
- Landau G., Bercovich Z., Park M. H., Kahana C., 2010. The role of polyamines in supporting growth of mammalian cells is mediated through their requirement for translation initiation and elongation. J. Biol. Chem. 285: 12474–12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P. L., Palin M. F., Murphy B. D., 2011. Polyamines on the reproductive landscape. Endocr. Rev. 32: 694–712. [DOI] [PubMed] [Google Scholar]
- Li C. H., Ohn T., Ivanov P., Tisdale S., Anderson P., 2010. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS ONE 5: e9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., et al. , 2011. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Santiago-Tirado F. H., Bretscher A., 2012. Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation. Mol. Biol. Cell 23: 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga F., Poincloux R., Romao M., Montagnac G., Le Dez G., et al. , 2009. Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res. 69: 2792–2800. [DOI] [PubMed] [Google Scholar]
- Magdolen V., Klier H., Wohl T., Klink F., Hirt H., et al. , 1994. The function of the hypusine-containing proteins of yeast and other eukaryotes is well conserved. Mol. Gen. Genet. 244: 646–652. [DOI] [PubMed] [Google Scholar]
- Matheos D., Metodiev M., Muller E., Stone D., Rose M. D., 2004. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J. Cell Biol. 165: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Dudin O., Martin S. G., 2013. Mate and fuse: how yeast cells do it. Open Biol. 3: 130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel A., Monton F., Li T., Gomez-Herreros F., Chavez S., et al. , 2013. External conditions inversely change the RNA polymerase II elongation rate and density in yeast. Biochim. Biophys. Acta 1829: 1248–1255. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Lee S. B., Park J. H., Park M. H., 2012. Essential role of eIF5A–1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 42: 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. H., Nishimura K., Zanelli C. F., Valentini S. R., 2010. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., et al. , 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297: 612–615. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A., 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15: 4971–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Elion E. A., 2005. Formin-induced actin cables are required for polarized recruitment of the Ste5 scaffold and high level activation of MAPK Fus3. J. Cell Sci. 118: 2837–2848. [DOI] [PubMed] [Google Scholar]
- Sagot I., Klee S. K., Pellman D., 2002a Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4: 42–50. [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D., 2002b An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4: 626–631. [DOI] [PubMed] [Google Scholar]
- Saini P., Eyler D. E., Green R., Dever T. E., 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier J., Schwelberger H. G., Smit-McBride Z., Kang H. A., Hershey J. W., 1991. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader R., Young C., Kozian D., Hoffmann R., Lottspeich F., 2006. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J. Biol. Chem. 281: 35336–35346. [DOI] [PubMed] [Google Scholar]
- Slaughter B. D., Smith S. E., Li R., 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb. Perspect. Biol. 1: a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J., Fink G. R., 1989. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc. Natl. Acad. Sci. USA 86: 9916–9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S., Lassak J., Starosta A. L., Kraxenberger T., Wilson D. N., et al. , 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85. [DOI] [PubMed] [Google Scholar]
- Valentini S. R., Casolari J. M., Oliveira C. C., Silver P. A., McBride A. E., 2002. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics 160: 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. W., Guan X. Y., Xie D., 2013. Roles of eukaryotic initiation factor 5A2 in human cancer. Int. J. Biol. Sci. 9: 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohl T., Klier H., Ammer H., Lottspeich F., Magdolen V., 1993. The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. Mol. Gen. Genet. 241: 305–311. [DOI] [PubMed] [Google Scholar]
- Zanelli C. F., Valentini S. R., 2005. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics 171: 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C. F., Valentini S. R., 2007. Is there a role for eIF5A in translation? Amino Acids 33: 351–358. [DOI] [PubMed] [Google Scholar]
- Zanelli C. F., Maragno A. L., Gregio A. P., Komili S., Pandolfi J. R., et al. , 2006. eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 348: 1358–1366. [DOI] [PubMed] [Google Scholar]
- Zhu X. L., Zeng Y. F., Guan J., Li Y. F., Deng Y. J., et al. , 2011. FMNL2 is a positive regulator of cell motility and metastasis in colorectal carcinoma. J. Pathol. 224: 377–388. [DOI] [PubMed] [Google Scholar]
- Zuk D., Jacobson A., 1998. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 17: 2914–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.