Abstract
Directional export of messenger RNA (mRNA) protein particles (mRNPs) through nuclear pore complexes (NPCs) requires multiple factors. In Saccharomyces cerevisiae, the NPC proteins Nup159 and Nup42 are asymmetrically localized to the cytoplasmic face and have distinct functional domains: a phenylalanine-glycine (FG) repeat domain that docks mRNP transport receptors and domains that bind the DEAD-box ATPase Dbp5 and its activating cofactor Gle1, respectively. We speculated that the Nup42 and Nup159 FG domains play a role in positioning mRNPs for the terminal mRNP-remodeling steps carried out by Dbp5. Here we find that deletion (Δ) of both the Nup42 and Nup159 FG domains results in a cold-sensitive poly(A)+ mRNA export defect. The nup42ΔFG nup159ΔFG mutant also has synthetic lethal genetic interactions with dbp5 and gle1 mutants. RNA cross-linking experiments further indicate that the nup42ΔFG nup159ΔFG mutant has a reduced capacity for mRNP remodeling during export. To further analyze the role of these FG domains, we replaced the Nup159 or Nup42 FG domains with FG domains from other Nups. These FG “swaps” demonstrate that only certain FG domains are functional at the NPC cytoplasmic face. Strikingly, fusing the Nup42 FG domain to the carboxy-terminus of Gle1 bypasses the need for the endogenous Nup42 FG domain, highlighting the importance of proximal positioning for these factors. We conclude that the Nup42 and Nup159 FG domains target the mRNP to Gle1 and Dbp5 for mRNP remodeling at the NPC. Moreover, these results provide key evidence that character and context play a direct role in FG domain function and mRNA export.
Keywords: nuclear pore complex, nuclear transport, DEAD-box protein, nucleoporin, mRNA export
IN eukaryotes, messenger RNA (mRNA) export from the nucleus is an essential process that is highly regulated, with events sequentially coordinated to ensure proper RNA processing and cytoplasmic fate (reviewed in Moore 2005; Muller-McNicoll and Neugebauer 2013). In general, transcripts exit the nucleus through nuclear pore complexes (NPCs), large (60 MDa in yeast, 100 MDa in vertebrates) proteinaceous structures that provide an aqueous channel for ions, metabolites, proteins, and ribonuclear protein complexes (RNPs) to cross the nuclear envelope (reviewed in Wente and Rout 2010; Bilokapic and Schwartz 2012). For molecules >40 kDa, transport through the NPC is facilitated via transport receptors (TRs) that bind both the cargo and NPC proteins (nucleoporins, or Nups). For mRNA, binding to TRs is carefully coordinated to allow export only after the message is fully processed in the nucleus (reviewed in Muller-McNicoll and Neugebauer 2013).
A key aspect of NPC function involves different NPC structures performing specific roles to ensure efficient transport. During NPC translocation, TRs interact with a class of Nups known as phenylalanine-glycine (FG) Nups (reviewed in Wente and Rout 2010). Each FG Nup has an unstructured domain enriched with FG, glycine-leucine-phenylalanine-glycine (GLFG), or phenylalanine-any amino acid-phenylalanine-glycine (FxFG) repeats, which are flanked by characteristic polar spacer sequences (Denning et al. 2003; Terry and Wente 2009). Each FG Nup is anchored in a specific NPC substructure via non-FG domains that interact with scaffold Nups. In Saccharomyces cerevisiae, the 11 FG Nups are located either symmetrically or asymmetrically (on the nuclear or cytoplasmic face) in the NPC (Rout et al. 2000; Alber et al. 2007). FG Nups are required for the NPC permeability barrier (Hulsmann et al. 2012), and TRs overcome this barrier by interacting with phenylalanine residues of the FG domains (reviewed in Wente and Rout 2010). Using a large-scale genetic approach in S. cerevisiae, we previously generated a collection of FG deletion mutants in higher-order combinations among the 11 different FG Nups. This work defined the minimal, functionally important FG domains and revealed functionally distinct FG pathways used by different TRs (Strawn et al. 2004; Terry and Wente 2007; Fiserova et al. 2010).
Importantly, NPC structures involved in the terminal steps of the protein import and export pathways are critical for the action of TRs in the Ran-dependent karyopherin (Kap) family. For example, in addition to their FG repeat domains that bind Kaps, Nups localized asymmetrically at the nuclear NPC face contain key binding sites for coordinated Kap-import cargo disassembly (Gilchrist et al. 2002; Gilchrist and Rexach 2003; Matsuura et al. 2003; Matsuura and Stewart 2005; Sun et al. 2013). Additionally, at the cytoplasmic NPC face, the mammalian FG Nup358 (RanBP2) functions as a multi-subunit SUMO E3 complex that coordinates SUMO ligase activity, export cargo disassembly through binding the RanGAP, and Kap binding (Werner et al. 2012). Also at the cytoplasmic face, the conserved FG Nups, Nup42 (yeast)/hCG1 (vertebrate), and Nup159 (yeast)/Nup214 (vertebrate) provide important binding sites for the Kap protein export receptor Crm1 (Floer and Blobel 1999; Sabri et al. 2007; Roloff et al. 2013). Thus, within individual Nups, juxtaposed binding sites for TRs and TR-cargo release factors function to integrate terminal steps of transport. It has not been directly investigated whether proximal positioning of binding sites for TRs and cargo release factors is required for mRNP cargo disassembly. We speculate that an mRNP might require combinatorial interactions between Nups, TRs, and mRNP-remodeling factors at the NPC cytoplasmic face.
The mRNA export cargo is defined as an mRNA protein (mRNP) complex that contains the TR heterodimer Mex67-Mtr2 (Tap-p15 in vertebrates), which is bound through adaptor proteins, such as the poly(A)+ RNA-binding protein Nab2 (reviewed in Rodriguez-Navarro and Hurt 2011; Natalizio and Wente 2013). When the mRNP reaches the NPC cytoplasmic face, it undergoes mRNP remodeling where a subset of RNP proteins, including Mex67 and Nab2, are selectively removed (Lund and Guthrie 2005; Tran et al. 2007). The conserved DEAD-box ATPase Dbp5 (DDX19B in vertebrates) catalyzes this mRNP remodeling in a manner that is dependent upon Gle1 and the small molecule inositol hexakisphosphate (IP6) (Alcazar-Roman et al. 2006; Weirich et al. 2006; Tran et al. 2007). Dbp5/DDX19B is localized to the NPC cytoplasmic face through interaction with Nup159/Nup214 (Hodge et al. 1999; Schmitt et al. 1999; Weirich et al. 2004), and Gle1 interacts with Nup42/hCG1 (Murphy and Wente 1996; Hodge et al. 1999; Schmitt et al. 1999; Strahm et al. 1999; Weirich et al. 2004; Kendirgi et al. 2005). The cycling of interactions between Dbp5, Gle1-IP6, Nup159, and RNA has been investigated in depth (Folkmann et al. 2011; Hodge et al. 2011; Montpetit et al. 2011; Noble et al. 2011). Additionally, we recently discovered that Gle1 self-associates via a conserved coiled-coil domain in its amino-terminal region and that this self-association is required during mRNA export (Folkmann et al. 2013). However, it is unknown how mRNP-remodeling selectivity—wherein Mex67-Mtr2, Nab2, and potentially other mRNP components are specifically targeted for remodeling—is dictated.
In addition to having binding sites for the mRNP-remodeling factors Dbp5 and Gle1-IP6, Nup159 and Nup42 also contain FG domains (Gorsch et al. 1995; Kraemer et al. 1995; Stutz et al. 1995). These FG domains bind Mex67 in vitro (Strasser et al. 2000). Therefore, we hypothesized that the Nup159 and Nup42 FG domains serve to position the mRNP for remodeling by Dbp5 and potentially target the TR Mex67-Mtr2 for release. Using a combination of genetic and biochemical approaches, we have uncovered a role during mRNP remodeling for these FG domains at the cytoplasmic NPC face.
Materials and Methods
Yeasts and growth
In the Supporting Information, Table S1 lists the yeast strains used in this study. Yeast genetic methods including mating, sporulation, dissection, and transformations were conducted according to standard procedures (Sherman et al. 1986). Yeast strains were grown at indicated temperatures in either YPD (2% peptone, 2% dextrose, 1% yeast extract) or selective minimal media lacking appropriate amino acids and supplemented with 2% dextrose and 5-fluoroorotic acid (5-FOA; United States Biological) as needed at 1.0 mg/ml.
Vector construction
Table S2 lists the vectors used in this study. Vector cloning was performed according to standard molecular biology strategies. FG swap vectors were generated by amplifying a wild-type NUP vector to replace the FG domain with a unique restriction site. FG domains were then amplified with compatible cohesive restriction sites and cloned into the nupΔFG vector, with DNA sequencing to confirm the construct.
In situ hybridization, immunofluorescence, and live-cell microscopy
Yeast strains were grown to mid-log phase (OD600 ∼0.5) in YPD media at the indicated temperatures. For live-cell imaging, cultures were collected, resuspended in synthetic complete media, and imaged. For in situ hybridization, yeast were processed as described (Wente and Blobel 1993), hybridized with 1 ng/μl Cy3-conjugated oligo(dT), and stained with 0.1 mg/ml DAPI to visualize the nucleus as described in Folkmann et al. (2013). For indirect immunofluorescence, yeast were processed as above and labeled as in Ho et al. (2000). Briefly, samples were incubated with anti-Nup159 mouse mAbs (mAb 165C10; Kraemer et al. 1995) for 16 hr at 4° followed by incubation with anti-Nup116-C rabbit antibodies (WU600; Iovine et al. 1995) for 1 hr at room temperature. Bound primary antibodies were detected with Alexa Flour 594-conjugated goat anti-rabbit and 488-conjugated goat anti-mouse IgG (1:200; Molecular Probes), and samples were stained with 0.1 mg/ml DAPI. Wide-field images were acquired using a microscope (BX50; Olympus) equipped with a motorized stage (model 999000, Ludl), Olympus 100× numerical aperture 1.3 UPlanF1 objective, and digital charge-coupled device camera (Orca-R2; Hamamatsu). Images were processed with ImageJ (National Institutes of Health) or Adobe Photoshop CS6.
UV cross-linking and immunoblotting
UV cross-linking was performed essentially as described (Anderson et al. 1993; Tran et al. 2007). Cultures were grown to late log phase (OD600 ∼0.8) at the indicated temperatures, cross-linked using a UV Stratalinker (Stratagene) with 254-nm UV light for 2 × 2.5 min, and lysed by bead beating; poly(A)+ RNA was purified by antisense chromatography using oligo(dT) resin (Applied Biosystems, Foster City, CA) and the manufacturer’s protocol. After RNase treatment, samples were separated by SDS-PAGE, and immunoblotting was conducted with affinity-purified rabbit anti-Nab2 (ASW 44; Tran et al. 2007) and affinity-purified rabbit anti-Cbp80 (Gorlich et al. 1996) antibodies. Enriched protein was quantitated by densitometry and reported as the amount of protein bound to RNA vs. the total in lysate. This ratio was normalized between strains with wild type set to 1.0.
For whole cell lysate immunoblotting, cells were grown to mid-log phase, and equal OD600 units of cell numbers were collected by centrifugation. Cells were resuspended in SDS loading buffer (20% glycerol, 4% SDS, 50 mM Tris–HCl, pH 6.8, 100 mM DTT, 0.01% bromophenol blue), lysed by vortexing with glass beads, and boiled 5 min. Lysates were then separated by SDS-PAGE and blotted using affinity-purified guinea pig anti-Gle1 (ASW 43 Alcazar-Roman et al. 2010), affinity-purified chicken anti-Nup159-NTD (ASW 55), or anti-yPgk1 (Invitrogen). The anti-Nup159-NTD antibody was generated against purified His6-Nup159-NTD (Noble et al. 2011) and affinity-purified using the same antigen. Alexa Flour 650- or 700-conjugated secondary antibodies (1:5000; Molecular Probes) were visualized with the Li-Cor Odyssey scanner (Lincoln, NE).
Quantitative PCR
Quantitative PCR was performed as described in Burns and Wente (2014). Indicated strains were grown to OD600 0.5–1.0 at 30° or 16°, and total RNA was extracted using the hot acidic phenol method. RNA was DNase-treated and reverse-transcribed using MultiScribe Reverse Transcriptase (Invitrogen). Complementary DNA levels were quantified with real-time PCR using iQ SYBR green PCR master mix and a CFX96 Quantitative PCR Thermocycler (Bio-Rad). Levels of transcripts from the polyadenylated transcripts PGK1 and ACT1 were normalized to the non-polyadenylated RNA SCR1, and this ratio was normalized between strains with wild type set to 1.0. Primers used were the following: ACT1 Fo—CTCCACCACTGCTGAAAGAGAA; ACT1 Re—CGAAGTCCAAGGCGACGTAA; PGK1 Fo—TTCTCTGCTGATGCCAACAC; PGK1 Re—ATTCGAAAACACCTGGTGGA; SCR1 Fo: AACCGTCTTTCCTCCGTCGTAA; and SCR1 Re—CTACCTTGCCGCACCAGACA.
Assay for heat-shock protein production
The [35S]methionine incorporation assay was performed as described (Carmody et al. 2010). Briefly, strains were grown to early log phase (OD600 ∼0.2) in synthetic complete medium lacking methionine and leucine (SC −Met −Leu) at 25°, collected, and resuspended in 500 μl SC −Met −Leu. The sample was split in half, and 250 μl SC −Met −Leu at 59° was added to one tube that was then incubated for 15 min at 42° (heat-shocked sample), and 250 μl SC −Met −Leu at 25° was added to the other tube and incubated at room temperature for 15 min (control sample). The samples were then radiolabeled by addition of 50 μCi of [35S]methionine, and incubated an additional 15 min at the indicated temperature before harvesting by centrifugation at 4°. The cells were washed with 4° SC −Met −Leu and lysed in 25 μl SDS loading buffer at 100° for 5 min. Samples were separated by SDS-PAGE, and the resulting gel was dried and exposed to autoradiography film.
Results
FG domains at the NPC cytoplasmic face function in mRNP remodeling
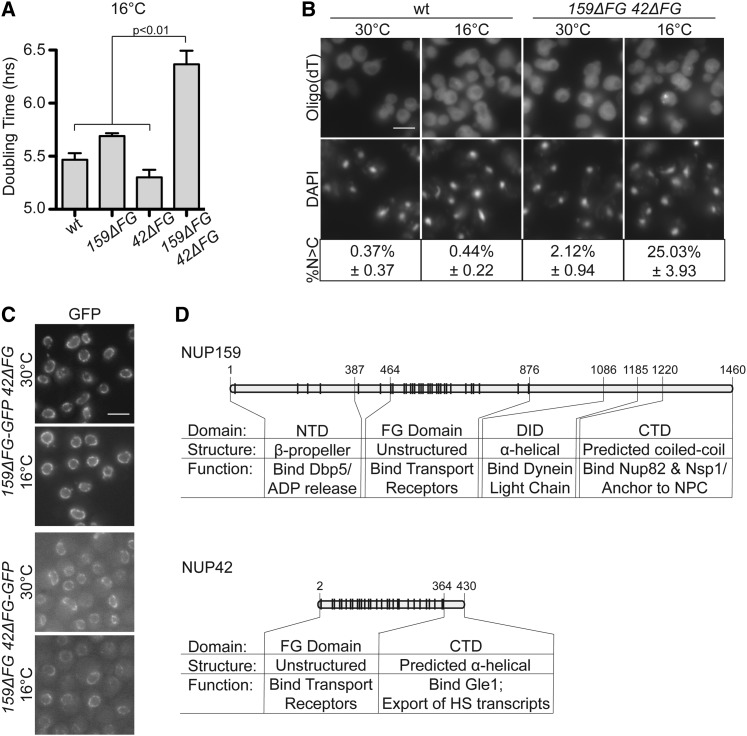
Previous studies reported that a nup42ΔFG nup159ΔFG double mutant lacking the FG domains of both Nup159 and Nup42 is viable (Strawn et al. 2004; Zeitler and Weis 2004). We carefully analyzed the growth of this mutant and observed cold sensitivity but no growth defects at higher temperatures (Figure S1A). In liquid culture at 16°, the nup42ΔFG nup159ΔFG strain displayed a significantly longer doubling time compared to either single mutant or a wild-type strain (Figure 1A). To analyze whether this growth defect corresponds to an mRNA export defect, we performed in situ analysis for poly(A)+ localization after growth at 30° or shifting to 16° for 12 hr. At 16°, we observed a reproducible mRNA export defect in nup42ΔFG nup159ΔFG compared to wild type (Figure 1B). Specifically, poly(A)+ RNA localized to puncta within the nucleus (as judged by localization relative to DAPI signal) in ∼25% of the cells. This phenotype correlates with other reported mRNA export defects in nup mutants, e.g., nup60Δ (Powrie et al. 2011). Importantly, this increase in nuclear signal of poly(A)+ RNA was not due to increased transcription as quantitative PCR showed that transcript levels in nup42ΔFG nup159ΔFG compared to wild-type cells were actually lower (Figure S1D). This decrease in transcript levels is likely due to feedback between the inhibition of mRNA export with transcription and nuclear mRNA decay (reviewed in Rodriguez-Navarro and Hurt 2011). These results suggested that the Nup159 and Nup42 FG domains perform a function, although not an essential one, during mRNA export.
Figure 1.
Deletion of FG domains on the cytoplasmic face of the NPC results in a cold-sensitive mRNA export defect. (A) nup42ΔFG nup159ΔFG has a cold-sensitive growth defect. The indicated strains were grown to early log phase (OD600 ∼0.2) at 16°, with OD600 measurements taken every 3 hr and normalized to time = 0, and doubling times were determined. Graph displays average of three independent experiments, and error bars indicate SEM. (B) nup42ΔFG nup159ΔFG has an mRNA export defect at 16°. In situ hybridization with an oligo(dT) probe for poly(A)+ RNA localization was conducted on indicated mutants after growth at 30° and shifting to 16° or 30° overnight. DAPI staining marks the nucleus. Bar, 5 μm. Quantification of three independent experiments of >100 cells for each strain is shown below images. “%N>C” indicates the percentage of cells with increased nuclear poly(A)+ signal (as puncta or diffuse nuclear signal). Uncertainty (±) indicates SEM. (C) nup42ΔFG-GFP and nup159ΔFG-GFP are localized at the NPC. Cells were grown at 30°, shifted to 16° or 30° for 12 hr, and imaged by wide-field live-cell direct fluorescence microscopy. Bar, 5 μm. (D) Diagram depicting functional and structural domains of Nup159 and Nup42 as determined by prior studies (Del Priore et al. 1997; Saavedra et al. 1997; Stutz et al. 1997; Belgareh et al. 1998; Hurwitz et al. 1998; Schmitt et al. 1999; Strahm et al. 1999; Bailer et al. 2000; Vainberg et al. 2000; Denning et al. 2003; Strawn et al. 2004; Weirich et al. 2004; Stelter et al. 2007; Noble et al. 2011; Yoshida et al. 2011) to scale according to primary structure. NTD, amino-terminal domain; DID, dynein-interacting domain; CTD, carboxy-terminal domain.
Others have shown that the temperature-sensitive mRNA export defect in a nup159 mutant (rat7-1) is due to loss of Nup159 association with the NPC and its subsequent degradation (Del Priore et al. 1996). Therefore, as controls, the sequence encoding GFP was fused to either nup42ΔFG or nup159ΔFG to determine whether Nup localization was altered. At either the permissive or nonpermissive growth temperature (30° or 16°), nup42ΔFG-GFP and nup159ΔFG-GFP localized to the nuclear envelope rim (Figure 1C). This observation is concordant with the fact that domains outside of the FG region are sufficient for NPC localization (Figure 1D) (Del Priore et al. 1997; Stutz et al. 1997). Furthermore, addition of the GFP tags did not alter the function of these proteins, as minimal enhanced growth defects were observed with tagged strains (Figure S1B). As determined by immunofluorescence microscopy (Figure S1C), GFP-tagged and untagged nup159ΔFG proteins also localized similarly to wild-type Nup159 (Figure S1C). Therefore, we concluded that the nup42ΔFG nup159ΔFG growth defect is due to loss of function of the FG domains.
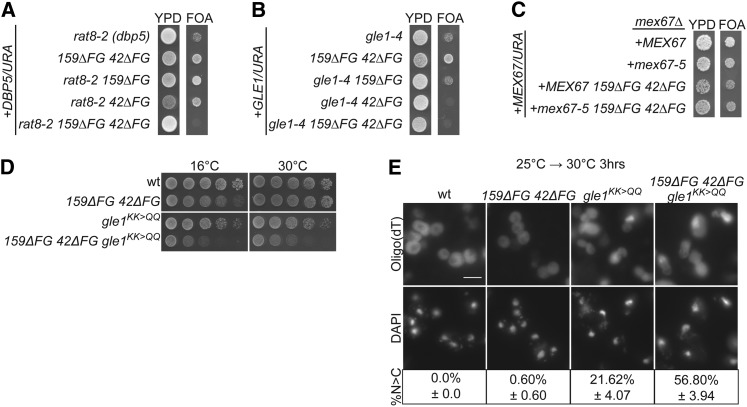
Roles for Nup159 and Nup42 in mRNA export were previously attributed to their respective binding of Dbp5 and Gle1 at the NPC (reviewed in Folkmann et al. 2011). The regions that interact with Dbp5 and Gle1 are adjacent to the FG domains in Nup159 and Nup42 (Figure 1D). Therefore, we tested for genetic interactions between the nup42ΔFG nup159ΔFG mutant and mutants of factors involved in mRNA export. Synthetic lethality was observed when nup42ΔFG nup159ΔFG was combined with a temperature-sensitive dbp5 mutant, rat8-2 (Figure 2A) (Snay-Hodge et al. 1998), and when nup42ΔFG alone was combined with the gle1-4 temperature-sensitive mutant (Figure 2B) (Murphy and Wente 1996). Notably, synthetic lethality was not observed when nup42ΔFG nup159ΔFG was combined with mex67-5, where mRNA export is perturbed prior to NPC docking (Figure 2C) (Segref et al. 1997; Santos-Rosa et al. 1998).
Figure 2.
nup42ΔFG nup159ΔFG exhibits genetic interactions with mRNP-remodeling mutants. (A) nup42ΔFG nup159ΔFG is synthetically lethal with rat8-2 (dbp5). Strains bearing the indicated alleles in addition to a DBP5/URA vector were spotted onto YPD or 5-FOA at 25°. Failure to grow on 5-FOA indicates synthetic lethality. (B) nup42ΔFG is synthetically lethal with gle1-4. Strains bearing the indicated alleles, in addition to a GLE1/URA vector, were spotted onto YPD or 5-FOA at 25°. (C) nup42ΔFG nup159ΔFG is not synthetically lethal with mex67-5. Strains bearing the indicated alleles in addition to a MEX67/URA vector were spotted onto YPD or 5-FOA at 25°. (D) nup42ΔFG nup159ΔFG gle1KK>QQ has an enhanced growth defect. Yeast strains were grown at 25° and fivefold serially diluted on YPD plates for growth at the indicated temperature. (E) nup42ΔFG nup159ΔFG gle1KK>QQ has an enhanced mRNA export defect. In situ hybridization with an oligo(dT) probe for poly(A)+ RNA localization was conducted on indicated mutants after growth at 25° and shifting to 30° for 3 hr. DAPI staining marks the nucleus. Bar, 5 μm. Quantification of three independent experiments of >100 cells for each strain is shown below images. “%N>C” indicates the percentage of cells with increased nuclear poly(A)+ signal. Uncertainty (±) indicates SEM.
We also observed an enhanced growth defect when we combined the nup42ΔFG nup159ΔFG mutant with the gle1KK>QQ mutant in which the IP6-binding sites on Gle1 are altered (Figure 2D) (Alcazar-Roman et al. 2010). We next tested whether this growth defect correlated with an increased mRNA export defect at 30°. After shifting to 30° for 3 hr, there was an increased mRNA export defect in nup42ΔFG nup159ΔFG gle1KK>QQ (57%) relative to gle1KK>QQ alone (22%) (Figure 2E). Notably, triple-mutant cells appeared larger and differently shaped, likely due to sickness caused by the mRNA export defect. Therefore, deletion of the Nup159 and Nup42 FG domains contributed to an mRNA export defect and resulted in lethality when combined with mutations in genes encoding factors involved in mRNP remodeling at the NPC cytoplasmic face.
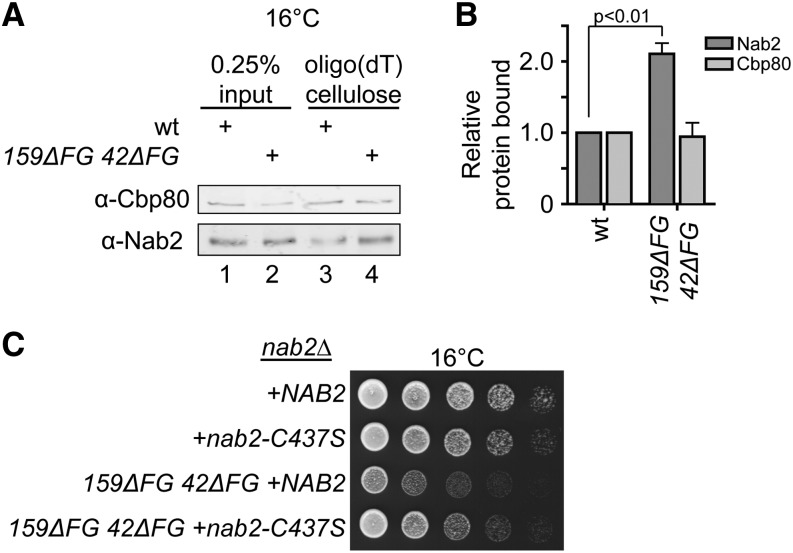
Given that the Nup42 and Nup159 FG domains are adjacent to binding sites for mRNP-remodeling factors (Gle1-IP6 and Dbp5, respectively), and that genetic interactions are observed with these same mRNP-remodeling factors, we hypothesized that the FG domains at the cytoplasmic NPC face function in mRNP remodeling. If so, since Nab2 is a target of Dbp5 remodeling in vivo, these mutants should show increased levels of Nab2 association with mRNA. A UV cross-linking and poly(A)+ mRNP isolation procedure was used to analyze the level of Nab2 bound to mRNA in nup42ΔFG nup159ΔFG cells after growth at 16° for 12 hr. Cbp80 was used as a control for a protein that is removed after the Dbp5 mRNP-remodeling process [with displacement occurring during the pioneer round of translation (Ishigaki et al. 2001)]. An approximately twofold increased association of Nab2 with poly(A)+ RNA was observed in nup42ΔFG nup159ΔFG relative to wild type (Figure 3, A and B). Importantly, the levels of Cbp80 associated with poly(A)+ RNA did not change in the nup42ΔFG nup159ΔFG mutant compared to wild type. If the increased levels of Nab2 associated with poly(A)+ RNA reflected a defect in mRNP remodeling, we speculated that the nup42ΔFG nup159ΔFG cold sensitivity should be rescued by decreasing the stability of the mRNP. The nab2-C437S mutant alters one of the Nab2 zinc-finger motifs and results in decreased affinity for RNA and a less stable mRNP, which allows partial rescue of the rat8-2 (dbp5) phenotype (Tran et al. 2007; Brockmann et al. 2012). We found that expression of nab2-C437S also partially rescued the cold sensitivity of nup42ΔFG nup159ΔFG, indicating that the growth defect was due at least in part to defective mRNP remodeling (Figure 3C).
Figure 3.
Deletion of the FG domains on the cytoplasmic face of the NPC results in an mRNP remodeling defect in vivo. (A and B) nup42ΔFG nup159ΔFG has increased Nab2 association with poly(A)+ RNA. The association of Nab2 and Cbp80 proteins with poly(A)+ RNA was assessed by shifting strains to 16° overnight, cross-linking with 254-nm UV light, isolating RNA by antisense chromatography, and immunoblotting after treatment with RNase. (A) Representative immunoblot. (B) The level of Nab2 and Cbp80 bound is indicated as a ratio of protein bound to poly(A)+ RNA relative to total cellular protein in each strain, with the wild-type ratio normalized to 1. Graph indicates the average of three independent experiments, and error bars indicate SEM. (C) nab2-C437S partially rescues the cold sensitivity of nup42ΔFG nup159ΔFG. Yeast strains were grown at 30° and fivefold serially diluted on YPD plates for growth at 16°.
FG domains have intrinsically distinct functions
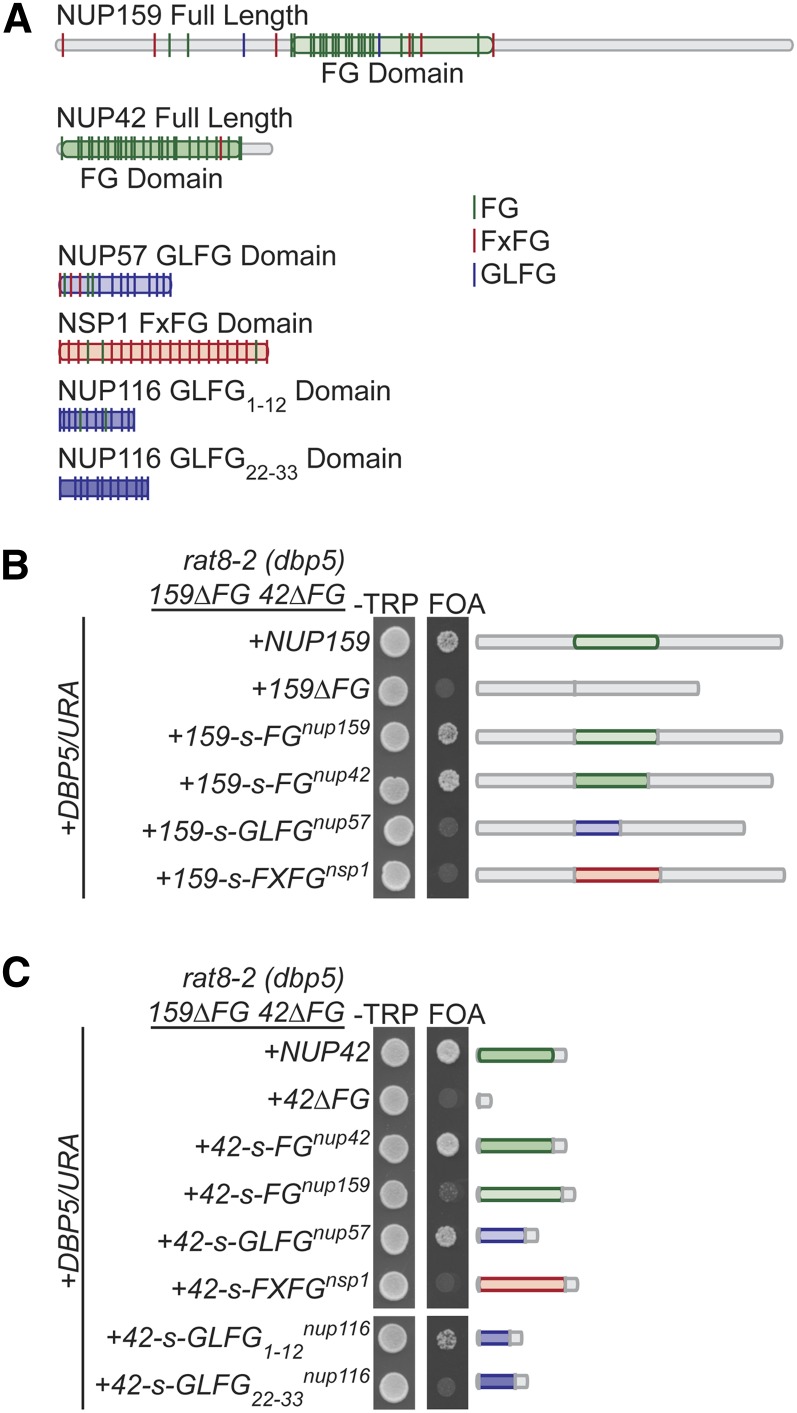
Because the Nup159 and Nup42 FG domains were essential in combination with the rat8-2 (dbp5) mutant, we used the rat8-2 (dbp5) nup42ΔFG nup159ΔFG triple mutant to analyze whether other FG domains can functionally compensate when anchored to the same location in the NPC. A series of “FG swap” expression vectors were generated in which the endogenous FG domain was replaced with an FG domain from another Nup (Figure 4). We tested the normally symmetrically localized Nup57-GLFG and Nsp1-FXFG domains as well as the Nup42-FG and Nup159-FG domains. Expression of all of the FG swap constructs was validated genetically and/or by immunoblotting (Figure S2). As expected, vectors expressing only nup42ΔFG or nup159ΔFG did not rescue the synthetic lethality of rat8-2 (dbp5) nup42ΔFG nup159ΔFG. This result indicates that the defect in this strain was not due to the HA/myc and LoxP sites that remain from generation of the chromosomal FG deletions in the nup42ΔFG nup159ΔFG mutant, as we had observed with one FG deletion strain (nup49ΔGLFG; Terry and Wente 2007). Moreover, when the endogenous FG domain was swapped in, lethality was rescued (Figure 4, B and C). Interestingly, when the Nup42 FG domain was swapped into Nup159, the lethality was rescued. However, when the Nup159 FG domain was swapped into Nup42, the lethality was not rescued. Additionally, the Nup57 GLFG domain rescued only when swapped into Nup42, and the FXFG domain of Nsp1 did not rescue when swapped into either Nup. These results reinforced our previous conclusion that FG domains are functionally distinct (Terry and Wente 2007). Furthermore, these results demonstrated that, although the FG domains at the cytoplasmic NPC face perform partially redundant functions, they also have distinct features as evidenced by the differential rescue.
Figure 4.
FG swaps reveal specificity of FG domain function. (A) Diagram depicting type and location of FG repeats for indicated FG domains. The FG domain is shown as delineated by Strawn et al. (2001) (for nup116GLFG1-12 and nup116GLFG22-33, see Strawn et al. 2004). Diagram is to scale according to primary structure. (B) The Nup42 FG domain can functionally compensate for the Nup159 FG domain. rat8-2 (dbp5) nup42ΔFG nup159ΔFG mutants containing nup159-s-FG/TRP FG swap vectors in addition to a DBP5/URA vector were spotted onto −Trp synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation. (C) The Nup57 GLFG domain and a Nup116 GLFG subdomain that bind Mex67 can functionally compensate for the Nup42 FG domain. rat8-2 (dbp5) nup42ΔFG nup159ΔFG mutants containing nup42-s-FG/TRP FG swap vectors in addition to a DBP5/URA vector were spotted onto −Trp synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation.
Based on the distinct results with the Nup57 GLFG and Nsp1 FXFG swaps, we further tested for rescue with the Nup116 GLFG domain that shows preferential binding in different regions for specific TRs. Yeast two-hybrid analysis has shown that the amino-terminal GLFG repeat region of Nup116, GLFG1-12, interacts with Mex67 and that the carboxy-terminal repeat region GLFG22-33 interacts with Kap95 (Strawn et al. 2001). We swapped these subdomains into Nup42 to analyze whether each domain could rescue the synthetic lethality of the rat8-2 (dbp5) nup42ΔFG nup159ΔFG mutant. Importantly, nup42-s-GLFG1-12nup116 rescued lethality, but nup42-s-GLFG22-33nup116 did not (Figure 4C), suggesting that FG binding to Mex67 was potentially critical in this mutant.
A gle1-FG fusion bypasses the requirement for the endogenous Nup42 FG domain
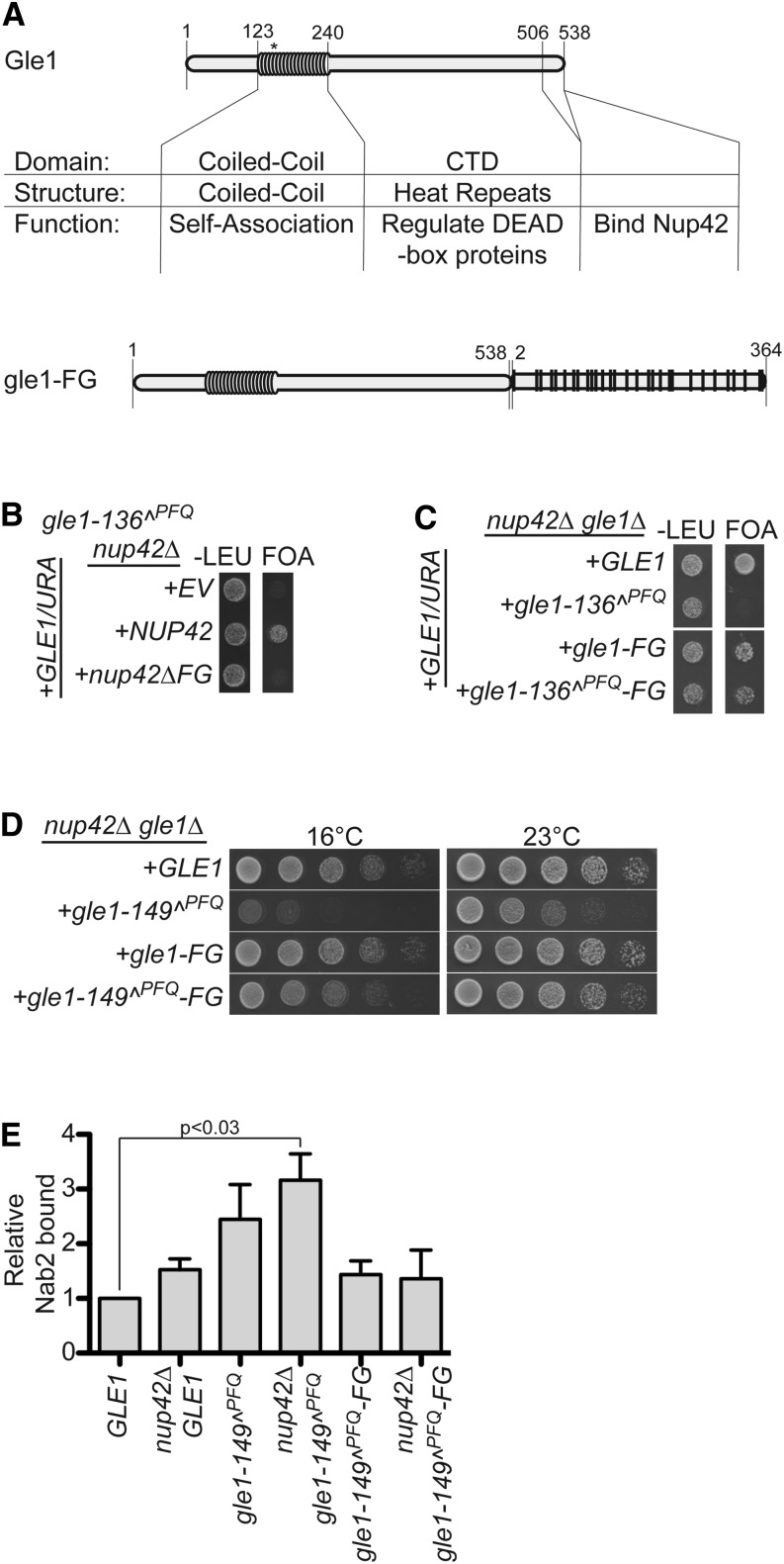
The carboxy-terminal, non-FG domain (CTD) of Nup42 is required for heat-shock mRNA export and in part for localization of Gle1 to the NPC (Stutz et al. 1995, 1997; Saavedra et al. 1997). We recently uncovered an essential function for Nup42 when Gle1 self-association is perturbed by insertion of PFQ residues in the Gle1 coiled-coil region after residues 136 or 149 (designated gle1^PFQ mutants) (Folkmann et al. 2013). As we reported, the nup42Δ gle1-136^PFQ mutant is synthetically lethal, the nup42Δ gle1-149^PFQ has fitness defects, and the nup42Δ gle1-157^PFQ mutant serves as a control with no enhanced defects. To analyze which domain of Nup42 is important in the nup42Δ gle1-136^PFQ mutant, vector complementation studies were conducted. Although expression of full-length NUP42 rescued the synthetic lethality of nup42Δ gle1-136^PFQ, nup42ΔFG did not (Figure 5B). This indicated that the FG domain of Nup42 is essential when Gle1 self-association is perturbed.
Figure 5.
A gle1-FG chimera complements nup42Δ gle1 mutants. (A) Diagram depicting functional and structural domains of Gle1, to scale according to primary structure, and a diagram of the gle1-FG fusion. Asterisk indicates location of PFQ insertions. (B) The Nup42 FG domain is essential when Gle1 self-association is perturbed. nup42Δ gle1-136^PFQ mutants containing a GLE1/URA vector in addition to an empty vector (EV) or nup42/TRP vectors were spotted onto −Trp synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation. (C) gle1-136^PFQ-FG rescues synthetic lethality of nup42Δ gle1-136^PFQ. nup42Δ gle1Δ mutants containing a GLE1/URA vector in addition to gle1-136^PFQ/LEU vectors without or with carboxy-terminal FG fusions were spotted onto −Leu synthetic media or 5-FOA at 25°. (D) gle1-149^PFQ-FG partially rescues the synthetic growth defect of nup42Δ gle1-149^PFQ. nup42Δ gle1Δ mutants containing gle1-149^PFQ/LEU vectors without or with carboxy-terminal FG fusions were spotted onto YPD plates to grow at the indicated temperatures. (E) gle1-149^PFQ-FG rescues the mRNP-remodeling defect of nup42Δ gle1-149^PFQ. The association of Nab2 protein with poly(A)+ RNA was assessed by shifting strains to 16° for 3 hr, UV cross-linking, isolating RNA by antisense chromatography, and immunoblotting after treatment with RNase. The level of Nab2 bound is indicated as a ratio of Nab2 bound to poly(A)+ RNA relative to total cellular protein in each strain, with the GLE1 ratio being normalized to 1. Graph indicates the average of three independent experiments, and error bars indicate SEM.
Taken together, we hypothesized that Nup159 and Nup42 serve as a scaffold to allow coincident, adjacent binding of the exporting mRNP with remodeling factors. To investigate this further, we generated a construct expressing a fusion between Gle1 and the FG domain of Nup42, gle1-FG (Figure 5A). We first tested whether this fusion rescued the synthetic lethality of nup42Δ gle1-136^PFQ, because this lethality is due to the loss of the Nup42 FG domain. To do this, we inserted PFQ residues after amino acid residues 136, 149, or 157 in the gle1-FG, generating gle1-136^PFQ-FG, gle1-149^PFQ-FG, and gle1-157^PFQ-FG constructs. All gle1-FG constructs were expressed and rescued a gle1Δ mutant with minimal growth defects (Figure S3). Importantly, nup42Δ gle1-136^PFQ-FG was not synthetically lethal, whereas nup42Δ gle1-136^PFQ was (Figure 5C). We also observed that, although nup42Δ gle1-149^PFQ was not synthetically lethal, it did result in an enhanced growth defect, particularly at cold temperatures (Figure 5D). Fusing the Nup42 FG domain to gle1-149^PFQ rescued the nup42Δ gle1-149^PFQ enhanced growth defect (Figure 5D). Using the UV cross-linking assay, the level of Nab2 bound to poly(A)+ RNA was measured in the gle1-149^PFQ mutant strains. At 16°, nup42Δ gle1-149^PFQ had approximately threefold more Nab2 bound to transcripts than the GLE1 wild type strain (Figure 5E, fourth and first columns, respectively; Figure S4). However, nup42Δ gle1-149^PFQ-FG had a decreased level of Nab2 bound that was not statistically different from wild-type levels (Figure 5E, sixth column). We concluded that the interaction between Gle1 and the Nup42-CTD, at least in part, serves to scaffold the Nup42 FG domain adjacent to Gle1 and that this positioning is important for mRNP remodeling.
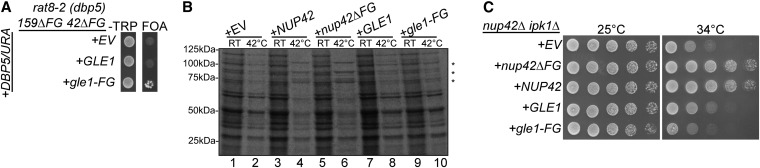
To test whether the gle1-FG fusion rescued other defects associated with Nup42, we transformed the gle1-FG construct into a variety of mutants with distinct requirements for these domains. Clearly, the FG domain of Nup42 was essential when combined with rat8-2 (dbp5) and nup159ΔFG mutants (Figure 2). Expression of gle1-FG rescued this synthetic lethality (Figure 6A). Thus, the essential function of the Nup42 FG domain was linked with its adjacent positioning to Gle1.
Figure 6.
The Nup42 FG domain and CTD have distinct functions. (A) gle1-FG rescues synthetic lethality due to loss of the Nup42 FG domain. rat8-2 (dbp5) nup42ΔFG nup159ΔFG containing empty vector (EV) or gle1/LEU vectors were spotted onto −Leu synthetic media or 5-FOA at 25°. Growth on 5-FOA indicates functional complementation. (B) gle1-FG does not rescue the heat-shock mRNA export defect of nup42Δ mutants. nup42Δ strains containing EV, nup42/LEU, or gle1/LEU vectors were grown at 25° to early log phase, kept at 25° or shifted to 42° for 15 min, labeled with [35S]methionine for an additional 15 min, and lysed. Lysates were separated by SDS-PAGE, and proteins were visualized by autoradiography. The positions of Hsp proteins, induced upon heat shock, are indicated by asterisks. (C) gle1-FG does not rescue temperature sensitivity of nup42Δ ipk1Δ. nup42Δ ipk1Δ containing EV, nup42/LEU, or gle1/LEU vectors were spotted onto −Leu synthetic media at the indicated temperatures.
Others have published functions for the Nup42 CTD that are independent from the FG domain. Specifically, Nup42 CTD is required for export of heat-shock protein transcripts (hsp) after heat shock and for function in a ipk1Δ mutant (lacking IP6 production) (Stutz et al. 1995, 1997; Miller et al. 2004). We tested whether gle1-FG rescued this defect by monitoring for new protein synthesis using a [35S]methionine incorporation assay. Whereas expression of NUP42 and nup42ΔFG resulted in heat-shock protein production and rescued the nup42Δ phenotype (Figure 6B, lanes 4 and 6), expression of gle1-FG did not (Figure 6B, lane 10). This indicated that nuclear export of hsp transcripts is inhibited in the gle1-FG nup42Δ cells. Expression of gle1-FG also did not rescue the nup42Δ ipk1Δ growth defect (Figure 6C). Therefore, we concluded that the Nup42 CTD has a separate function in addition to positioning the FG domain in proximity to Gle1.
Discussion
From this work, we conclude that the Nup159 and Nup42 FG domains at the NPC cytoplasmic face function in mRNP remodeling. Furthermore, we have identified a requirement for the proximal positioning of remodeling factors and FG domains in this process. Given the conservation of the general domain structures of the vertebrate orthologs of Nup159 and Nup42 (Nup214 and hCG1, respectively) (Kraemer et al. 1994; Strahm et al. 1999), we predict that FG domains at the NPC cytoplasmic face play roles in mRNP remodeling across eukaryotes. Others have previously used a FG swap approach to investigate the cytoplasmic and nuclear asymmetric FG domains (i.e., exchanging the cytoplasmic Nup159 and nuclear Nup1 FG domains) and have concluded that changing the location of these FG domains in the NPC does not dominantly affect bulk transport (Zeitler and Weis 2004). In contrast, here we analyzed whether FG swaps could rescue a defect due to loss of the FG domains. The differential rescue observed in this study indicates an inherent specificity to a given FG domain. Moreover, given the function of a gle1-FG fusion protein, these studies reveal a previously undefined step in the mRNA export mechanism, wherein mRNPs are recruited to the FG domains on the NPC cytoplasmic face for remodeling.
The differential rescue by FG swaps uncovers an inherent specialization of FG domain function. This specialization might be due to the different type of FG repeats (FG, GLFG, or FxFG) enriched in the respective domains, or it might be due to the distinct biochemical and biophysical properties that are governed by the spacer sequences between the FG repeats. For example, due to its high content of charged residues, the Nup159 FG domain forms a noncohesive extended coil, whereas the Nup42 FG domain is a cohesive collapsed coil (Yamada et al. 2010). The Nup57 GLFG domain is also a collapsed coil, and this characteristic may partially explain why it can functionally replace the Nup42 FG domain and not the Nup159 FG domain. The Nsp1 FxFG domain is an extended coil, and it did not compensate for either FG domain, suggesting that this property alone is not sufficient for function. Indeed, using a directed approach by swapping in FG domains with distinct binding capabilities, we determined that binding to Mex67 is likely the important function of the Nup42 FG domain. Therefore, we speculate that the combination of biophysical properties (determined by spacer sequences) and preferences for TR binding (determined by type of FG repeat and adjacent residues) underlie the specificity of FG domain function.
In addition to mRNA export roles for FG domains on the cytoplasmic NPC face, these FG domains also function in Kap (Crm1)-mediated export (Floer and Blobel 1999; Sabri et al. 2007; Roloff et al. 2013), and it is postulated that SAFGxPSFG repeats on Nup159 and Nup42 provide specialized binding sites for Crm1 (Denning and Rexach 2007). It is established that structured regions on the FxFG Nups located at the nuclear NPC face contribute to import cargo disassembly (Gilchrist et al. 2002; Gilchrist and Rexach 2003; Matsuura et al. 2003; Matsuura and Stewart 2005; Sun et al. 2013). These structured domains are located adjacent to the FxFG domains that bind importing Kaps (Pyhtila and Rexach 2003). Thus, although the asymmetric FG domains are not strictly required for NPC transport (Strawn et al. 2004; Zeitler and Weis 2004), these domains likely play a general function in placing TRs at the optimal locale for cargo disassembly. Based on these roles at both the nuclear and cytoplasmic NPC faces, FG Nups do not simply function as a cargo permeability barrier but also are actively involved in providing spatial context to the transport mechanism through providing specialized binding sites.
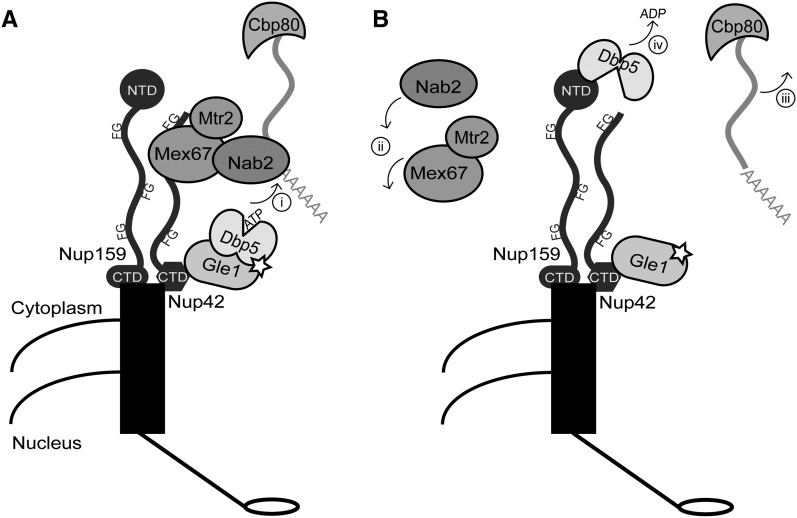
There are several potential models for the molecular function of the FG domains during mRNP remodeling. The schematic in Figure 7 summarizes several of these possible steps. First, the FG domains potentially act in docking the mRNA at the cytoplasmic face of the NPC until Mex67-Mtr2 has been remodeled off (Figure 7A, i). Real-time single-molecule experiments show that steps at the nuclear and cytoplasmic face of the NPC are rate limiting during the mRNA export process (Grunwald and Singer 2010). These FG domains potentially provide important binding sites during this process and possibly facilitate multiple rounds of mRNP remodeling. DEAD-box proteins are not processive enzymes, and there is evidence that flexible extensions outside of the helicase core on other DEAD box proteins aid in allowing additional rounds of activity of the enzyme toward a substrate (Mallam et al. 2011). We speculate that the FG domains at the NPC cytoplasmic face might have a similar role during mRNA export wherein, if multiple Mex67-Mtr2 dimers associate with an mRNP, the FG repeats could interact with multiple Mex67 molecules to direct rounds of mRNP remodeling by Dbp5.
Figure 7.
Schematic diagram for model by which FG domains on the cytoplasmic face of the NPC recruit exporting mRNPs for remodeling by Gle1-activated Dbp5. (A) Mex67-Mtr2 mediate export of mature mRNPs, and the dimer is bound to the transcript via adaptors such as the poly(A)+ binding protein Nab2. At the cytoplasmic face of the NPC, the Nup42 and Nup159 FG domains bind Mex67-Mtr2 to bring the mRNP in close proximity to Dbp5. Gle1 stimulates Dbp5 ATP loading, and (i) Dbp5 binds the RNA to trigger remodeling. Dbp5 might also be a constituent of the exporting mRNP. (B) Coincident with ATP hydrolysis, Dbp5 remodels specific proteins such as Nab2 and Mex67 off the mRNP. (ii) These proteins are then recycled into the nucleus for additional rounds of mRNA export. (iii) The mRNP, still bound to other mRNA-binding proteins such as Cbp80, is free for cytoplasmic functions. (iv) The Nup159 NTD facilitates ADP release from Dbp5, allowing additional rounds of mRNP remodeling. It is unknown where the Nup159 NTD is localized relative to its CTD in vivo.
Another possible function for FG domains during mRNP remodeling is to specify which RNA binding proteins are removed during the remodeling process (Figure 7B). Mex67 and Nab2 are bona fide targets for Dbp5 remodeling in vivo (Lund and Guthrie 2005; Tran et al. 2007), and other proteins, such as the Cap-binding protein (Cbp20/80 in yeast) and presumably the poly(A)+ binding protein Pab1 are retained on the mRNA (reviewed in Schoenberg and Maquat 2012) (Figure 7B, iii). DEAD-box proteins bind RNA in a sequence-independent manner via the phosphate backbone of the polynucleotide chain (reviewed in Russell et al. 2013). While other DEAD-box proteins contain extensions that direct their activity toward specific substrates, Dbp5 does not (Tieg and Krebber 2013). Indeed, Dbp5 can remodel nonphysiological targets such as Pab1 from a model poly(A)+ substrate in vitro (Tran et al. 2007). Therefore, it is tempting to speculate that the FG domains provide a mechanism for dictating the in vivo removal of specific mRNP proteins during mRNA export. In this way, the specificity for remodeling is not conferred by RNA sequence or direct protein recognition of the RNA-binding protein to be removed, but is achieved by spatial proximity between the FG domain (binding the TR) and remodeling factors, and thus is dictated by the NPC structure (Figure 7A, i). As such, the FG domains at the NPC cytoplasmic face would function to confer specificity to the mRNP-remodeling mechanism.
During mRNA export cycles, an alternate function for FG domains at the NPC cytoplasmic face might be to bind Mex67 after it is removed from the RNP and facilitate efficient transport back into the nucleus for additional export events (Figure 7B, ii). This “recycling” of Mex67 would be an indirect function for FG domains in mRNP remodeling. However, our data indicate a direct role in mRNP remodeling due to an accumulation of Nab2 on poly(A)+ RNA when the FG domains are deleted. Moreover, the nab2-C437S mutant rescues the cold sensitivity of the nup42ΔFG nup159ΔFG mutant. Additionally, we did not observe increased cytoplasmic Mex67-GFP localization in nup42ΔFG nup159ΔFG cells (data not shown).
Our studies uncover an unexpected role for the FG domain of Nup42 in mRNP remodeling. However, it is clear that there is an additional function for the CTD of Nup42: expression of the CTD alone rescues the hsp mRNA export in nup42Δ cells (Stutz et al. 1995, 1997; Saavedra et al. 1997), and the gle1-FG fusion did not rescue defects associated with Nup42 CTD (Figure 6). It has been proposed that the Nup42 CTD is required for anchoring Gle1 to the NPC both during normal growth conditions and during heat shock (Strahm et al. 1999; Rollenhagen et al. 2004), and it has been determined that hCG1 is required for steady-state human Gle1 localization to the nuclear envelope (Kendirgi et al. 2005). It is also possible that the Nup42 CTD modulates Gle1 stimulation of Dbp5 at the NPC. Overall, it is clear that there are still unresolved steps in this mechanism.
The Dbp5–Gle1 interaction has been compared to the interaction between eIF4A and eIF4G, as the structural rearrangements of the DEAD box protein (Dbp5 or eIF4A) are similar when the binding partner (Gle1 or eIF4G) interacts (Oberer et al. 2005; Montpetit et al. 2011). eIF4G is considered the major scaffold of the eIF4F translation initiation complex: through multiple interaction partners, eIF4G functions to recruit the mRNP to the 40S ribosomal subunit, circularize the transcript, and directly stimulate the ATPase activity of eIF4A to allow structural rearrangements of the mRNP that are necessary for translation (reviewed in Andreou and Klostermeier 2013). Thus, eIF4G plays an important role in coordinating mRNP recruitment to functional interaction partners during translation initiation. We propose that Gle1 is playing a similar role as a scaffold during mRNA export: Nup42 interacts with the mRNP through Mex67 binding to its FG repeats, the Nup42 CTD interacts with Gle1, and Gle1 binds to Dbp5 to regulate its ATPase activity. Thus, recruitment of these factors through distinct interactions effectively increases the likelihood of Dbp5–RNA interaction and thus directs the mRNP-remodeling capacity of Dbp5. Furthermore, Gle1 self-association is likely also required for this scaffolding activity (Folkmann et al. 2013). Together, this work highlights key mechanisms coupling mRNP export and mRNP remodeling at the cytoplasmic face of the NPC that allow efficient gene expression.
Supplementary Material
Acknowledgments
The authors thank D. Görlich for the rabbit anti-Cbp80 antibody and M. Rout for the mouse anti-Nup159 antibody; members of the Wente laboratory, C. Cole, E. Bowman, and E. Tran for discussions and critical reading of the manuscript; K. Noble for generating the chicken anti-Nup159-NTD antibody; and R. Dawson and Y. Zhou for technical assistance. This work was supported by National Institutes of Health grant R37GM051219 (to S.R.W.) and training position grant T32HD007502 (to R.L.A.).
Footnotes
Communicating editor: O. Cohen-Fix
Literature Cited
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701. [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman A. R., Tran E. J., Guo S., Wente S. R., 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8: 711–716. [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman A. R., Bolger T. A., Wente S. R., 2010. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J. Biol. Chem. 285: 16683–16692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. T., Wilson S. M., Datar K. V., Swanson M. S., 1993. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 13: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou A. Z., Klostermeier D., 2013. The DEAD-box helicase eIF4A: Paradigm or the odd one out? RNA Biol. 10: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer S. M., Balduf C., Katahira J., Podtelejnikov A., Rollenhagen C., et al. , 2000. Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. J. Biol. Chem. 275: 23540–23548. [DOI] [PubMed] [Google Scholar]
- Belgareh N., Snay-Hodge C., Pasteau F., Dagher S., Cole C. N., et al. , 1998. Functional characterization of a Nup159p-containing nuclear pore subcomplex. Mol. Biol. Cell 9: 3475–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S., Schwartz T. U., 2012. 3D ultrastructure of the nuclear pore complex. Curr. Opin. Cell Biol. 24: 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann C., Soucek S., Kuhlmann S. I., Mills-Lujan K., Kelly S. M., et al. , 2012. Structural basis for polyadenosine-RNA binding by Nab2 Zn fingers and its function in mRNA nuclear export. Structure 20: 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns L. T., Wente S. R., 2014. Casein kinase II regulation of the Hot1 transcription factor promotes stochastic gene expression. J. Biol. Chem. 289: 17668–17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody S. R., Tran E. J., Apponi L. H., Corbett A. H., Wente S. R., 2010. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol. Cell. Biol. 30: 5168–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore V., Snay C. A., Bahr A., Cole C. N., 1996. The product of the Saccharomyces cerevisiae RSS1 gene, identified as a high-copy suppressor of the rat7–1 temperature-sensitive allele of the RAT7/NUP159 nucleoporin, is required for efficient mRNA export. Mol. Biol. Cell 7: 1601–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore V., Heath C., Snay C., MacMillan A., Gorsch L., et al. , 1997. A structure/function analysis of Rat7p/Nup159p, an essential nucleoporin of Saccharomyces cerevisiae. J. Cell Sci. 110(Pt 23): 2987–2999. [DOI] [PubMed] [Google Scholar]
- Denning D. P., Rexach M. F., 2007. Rapid evolution exposes the boundaries of domain structure and function in natively unfolded FG nucleoporins. Mol. Cell. Proteomics 6: 272–282. [DOI] [PubMed] [Google Scholar]
- Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M., 2003. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 100: 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J., Richards S. A., Wente S. R., Goldberg M. W., 2010. Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J. Cell Sci. 123: 2773–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M., Blobel G., 1999. Putative reaction intermediates in Crm1-mediated nuclear protein export. J. Biol. Chem. 274: 16279–16286. [DOI] [PubMed] [Google Scholar]
- Folkmann A. W., Noble K. N., Cole C. N., Wente S. R., 2011. Dbp5, Gle1–IP6 and Nup159: a working model for mRNP export. Nucleus 2: 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann A. W., Collier S. E., Zhan X., Aditi M. D. Ohi et al, 2013. Gle1 functions during mRNA export in an oligomeric complex that is altered in human disease. Cell 155: 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist D., Rexach M., 2003. Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin alpha in the nucleoplasm. J. Biol. Chem. 278: 51937–51949. [DOI] [PubMed] [Google Scholar]
- Gilchrist D., Mykytka B., Rexach M., 2002. Accelerating the rate of disassembly of karyopherin.cargo complexes. J. Biol. Chem. 277: 18161–18172. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Kraft R., Kostka S., Vogel F., Hartmann E., et al. , 1996. Importin provides a link between nuclear protein import and U snRNA export. Cell 87: 21–32. [DOI] [PubMed] [Google Scholar]
- Gorsch L. C., Dockendorff T. C., Cole C. N., 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129: 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D., Singer R. H., 2010. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature 467: 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A. K., Shen T. X., Ryan K. J., Kiseleva E., Levy M. A., et al. , 2000. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol. Cell. Biol. 20: 5736–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C. A., Colot H. V., Stafford P., Cole C. N., 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J. 18: 5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge C. A., Tran E. J., Noble K. N., Alcazar-Roman A. R., Ben-Yishay R., et al. , 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 25: 1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmann B. B., Labokha A. A., Gorlich D., 2012. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150: 738–751. [DOI] [PubMed] [Google Scholar]
- Hurwitz M. E., Strambio-de-Castillia C., Blobel G., 1998. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc. Natl. Acad. Sci. USA 95: 11241–11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine M. K., Watkins J. L., Wente S. R., 1995. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 131: 1699–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L. E., 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Kendirgi F., Rexer D. J., Alcazar-Roman A. R., Onishko H. M., Wente S. R., 2005. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol. Biol. Cell 16: 4304–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D., Wozniak R. W., Blobel G., Radu A., 1994. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc. Natl. Acad. Sci. USA 91: 1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D. M., Strambio-de-Castillia C., Blobel G., Rout M. P., 1995. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J. Biol. Chem. 270: 19017–19021. [DOI] [PubMed] [Google Scholar]
- Lund M. K., Guthrie C., 2005. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell 20: 645–651. [DOI] [PubMed] [Google Scholar]
- Mallam A. L., Jarmoskaite I., Tijerina P., Del Campo M., Seifert S., et al. , 2011. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc. Natl. Acad. Sci. USA 108: 12254–12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Stewart M., 2005. Nup50/Npap60 function in nuclear protein import complex disassembly and importin recycling. EMBO J. 24: 3681–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Lange A., Harreman M. T., Corbett A. H., Stewart M., 2003. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 22: 5358–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. L., Suntharalingam M., Johnson S. L., Audhya A., Emr S. D., et al. , 2004. Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J. Biol. Chem. 279: 51022–51032. [DOI] [PubMed] [Google Scholar]
- Montpetit B., Thomsen N. D., Helmke K. J., Seeliger M. A., Berger J. M., et al. , 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472: 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J., 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518. [DOI] [PubMed] [Google Scholar]
- Muller-McNicoll M., Neugebauer K. M., 2013. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 14: 275–287. [DOI] [PubMed] [Google Scholar]
- Murphy R., Wente S. R., 1996. An RNA-export mediator with an essential nuclear export signal. Nature 383: 357–360. [DOI] [PubMed] [Google Scholar]
- Natalizio B. J., Wente S. R., 2013. Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol. 23: 365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble K. N., Tran E. J., Alcazar-Roman A. R., Hodge C. A., Cole C. N., et al. , 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 25: 1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberer M., Marintchev A., Wagner G., 2005. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 19: 2212–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie E. A., Zenklusen D., Singer R. H., 2011. A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae. RNA 17: 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhtila B., Rexach M., 2003. A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J. Biol. Chem. 278: 42699–42709. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Hurt E., 2011. Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 23: 302–309. [DOI] [PubMed] [Google Scholar]
- Rollenhagen C., Hodge C. A., Cole C. N., 2004. The nuclear pore complex and the DEAD box protein Rat8p/Dbp5p have nonessential features which appear to facilitate mRNA export following heat shock. Mol. Cell. Biol. 24: 4869–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roloff S., Spillner C., Kehlenbach R. H., 2013. Several phenylalanine-glycine motives in the nucleoporin Nup214 are essential for binding of the nuclear export receptor CRM1. J. Biol. Chem. 288: 3952–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., et al. , 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148: 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R., Jarmoskaite I., Lambowitz A. M., 2013. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol. 10: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra C. A., Hammell C. M., Heath C. V., Cole C. N., 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 11: 2845–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri N., Roth P., Xylourgidis N., Sadeghifar F., Adler J., et al. , 2007. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J. Cell Biol. 178: 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., et al. , 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18: 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C., von Kobbe C., Bachi A., Pante N., Rodrigues J. P., et al. , 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 18: 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg D. R., Maquat L. E., 2012. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 13: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., et al. , 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B., Cold Spring Harbor Laboratory , 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory, New York, NY. [Google Scholar]
- Snay-Hodge C. A., Colot H. V., Goldstein A. L., Cole C. N., 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P., Kunze R., Flemming D., Hopfner D., Diepholz M., et al. , 2007. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat. Cell Biol. 9: 788–796. [DOI] [PubMed] [Google Scholar]
- Strahm Y., Fahrenkrog B., Zenklusen D., Rychner E., Kantor J., et al. , 1999. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 18: 5761–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K., Bassler J., Hurt E., 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150: 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Wente S. R., 2001. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J. Biol. Chem. 276: 6445–6452. [DOI] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R., 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6: 197–206. [DOI] [PubMed] [Google Scholar]
- Stutz F., Neville M., Rosbash M., 1995. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell 82: 495–506. [DOI] [PubMed] [Google Scholar]
- Stutz F., Kantor J., Zhang D., McCarthy T., Neville M., et al. , 1997. The yeast nucleoporin rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 11: 2857–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Fu G., Ciziene D., Stewart M., Musser S. M., 2013. Choreography of importin-alpha/CAS complex assembly and disassembly at nuclear pores. Proc. Natl. Acad. Sci. USA 110: E1584–E1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R., 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178: 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R., 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 8: 1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieg B., Krebber H., 2013. Dbp5: from nuclear export to translation. Biochim. Biophys. Acta 1829: 791–798. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Zhou Y., Corbett A. H., Wente S. R., 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28: 850–859. [DOI] [PubMed] [Google Scholar]
- Vainberg I. E., Dower K., Rosbash M., 2000. Nuclear export of heat shock and non-heat-shock mRNA occurs via similar pathways. Mol. Cell Biol. 20: 3996–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Berger J. M., Weis K., 2004. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16: 749–760. [DOI] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., et al. , 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8: 668–676. [DOI] [PubMed] [Google Scholar]
- Wente S. R., Blobel G., 1993. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 123: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Rout M. P., 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2: a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A., Flotho A., Melchior F., 2012. The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol. Cell 46: 287–298. [DOI] [PubMed] [Google Scholar]
- Yamada J., Phillips J. L., Patel S., Goldfien G., Calestagne-Morelli A., et al. , 2010. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9: 2205–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Seo H. S., Debler E. W., Blobel G., Hoelz A., 2011. Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc. Natl. Acad. Sci. U S A 108: 16571–16576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler B., Weis K., 2004. The FG-repeat asymmetry of the nuclear pore complex is dispensable for bulk nucleocytoplasmic transport in vivo. J. Cell Biol. 167: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.