Abstract
SRm160 is an SR-like protein implicated in multiple steps of RNA processing and nucleocytoplasmic export. Although its biochemical functions have been extensively described, its genetic interactions and potential participation in signaling pathways remain largely unknown, despite the fact that it is highly phosphorylated in both mammalian cells and Drosophila. To begin elucidating the functions of the protein in signaling and its potential role in developmental processes, we characterized mutant and overexpression SRm160 phenotypes in Drosophila and their interactions with the locus encoding the LAMMER protein kinase, Doa. SRm160 mutations are recessive lethal, while its overexpression generates phenotypes including roughened eyes and highly disorganized internal eye structure, which are due at least in part to aberrantly high levels of apoptosis. SRm160 is required for normal somatic sex determination, since its alleles strongly enhance a subtle sex transformation phenotype induced by Doa kinase alleles. Moreover, modification of SRm160 by DOA kinase appears to be necessary for its activity, since Doa alleles suppress phenotypes induced by SRm160 overexpression in the eye and enhance those in genital discs. Modification of SRm160 may occur through direct interaction because DOA kinase phosphorylates it in vitro. Remarkably, SRm160 protein was concentrated in the nuclei of precellular embryos but was very rapidly excluded from nuclei or degraded coincident with cellularization. Also of interest, transcripts are restricted almost exclusively to the developing nervous system in mature embryos.
Keywords: SR proteins, sex determination, apoptosis, phosphorylation, splicing
ALTERNATIVE splicing is a tightly regulated process through which multiple mRNAs can be generated from a single gene, contributing substantially to proteome complexity (Pan et al. 2008; Wang et al. 2008) as well as to diverse cellular processes, including development, differentiation (Xu et al. 2005; Gabut et al. 2011; Grabowski 2011; Li et al. 2013), and apoptosis (Schwerk and Schulze-Osthoff 2005; Moore et al. 2010). Its misregulation contributes to a large number of diseases, notably cancer (Srebow and Kornblihtt 2006; Venables et al. 2008; Yoshida et al. 2011; Kaida et al. 2012), but many others as well (Cooper et al. 2009; Fan and Tang 2013; Fu et al. 2013). Splicing requires the precise assembly and function of the large spliceosome complex, which is composed of small nuclear ribonucleoproteins (snRNP) U1, U2, U4/6, U5, and >100 additional proteins (Herold et al. 2009). The composition and structure of the spliceosome is largely conserved, at least between Drosophila and humans.
Among those proteins required for proper splicing are SR and SR-related proteins; for reviews see Long and Caceres (2009) and Zhong et al. (2009), for which a new gene nomenclature was proposed (Manley and Krainer 2010). SR proteins contain one or two N-terminal RNA recognition motifs (RRMs) and a C-terminal “RS” domain rich in serine and arginine repeats. SRm160 (SRRM1) is an SR-related protein that contains several RS domains. Although lacking RRM motifs (Blencowe et al. 1998), it binds nucleic acids directly through a conserved “PWI” motif (Blencowe and Ouzounis 1999; Szymczyna et al. 2003). Initially referred to as B1C8, SRm160 was first identified in a screen for proteins associated with the nuclear matrix (Wan et al. 1994), and the domains responsible for this association were mapped (Wagner et al. 2003). Proteins connected with the nuclear matrix are often distributed in perichromatin fibrils and or interchromatin granule clusters (IGC), also referred to as “nuclear speckles” due to their punctate appearance. SRm160 was also isolated as an IGC component under the name “plenty of prolines” (Mintz et al. 1999). IGCs or speckles serve as concentration or storage sites for snRNPs, SR proteins, and the hyperphosphorylated form of the large subunit of RNA polymerase II. Although IGCs are not sites of active splicing, splicing factors fail to associate with pre-mRNA and spliced mRNAs are almost undetectable if IGCs are disrupted (Sacco-Bubulya and Spector 2002). Intriguingly, SRm160 nuclear localization is dependent upon the availability of ATP, suggesting regulation of its mobility (Wagner et al. 2004).
SRm160 activates splicing in vitro, and interacts with a number of RNA-binding proteins. Among them is the mammalian ortholog of the Drosophila protein Transformer 2 (Blencowe et al. 1998; Eldridge et al. 1999), another SR-related protein which influences alternative-splice site selection. In Drosophila, TRA2 directly binds and influences splicing of the dsx transcript as part of a SR-protein complex essential to somatic sex determination (Forch and Valcarcel 2003; Rabinow and Samson 2010). SRm160 and SR proteins function together in the first step of spliceosome formation to facilitate the interaction of the U1 subunit of the spliceosome with its target pre-mRNA (Blencowe et al. 1998).
Among those proteins in addition to SR proteins associating with SRm160 is Sam68, a KH-domain RNA-binding protein (Cheng and Sharp 2006). The activity of Sam68 and SRm160 affects the alternative splicing of mammalian CD44, which is required for tumor invasiveness, suggesting a possible connection with cancer progression. SRm160 also associates with TLS/FUS (Meissner et al. 2003), a proto-oncoprotein associated with familial amyotrophic lateral sclerosis (Kwiatkowski et al. 2009). A recent study demonstrated that SRm160 interacts with the long noncoding RNA MALAT-1, which it helps localize to nuclear speckles (Miyagawa et al. 2012). Mammalian SRm160 also complexes with cohesin throughout the cell cycle, suggestive of a role in chromatin organization, segregation, or transcriptional regulation (McCracken et al. 2005). The protein is localized to the mitotic spindle during M phase (Blencowe et al. 1998), although its function there remains unknown.
Known SRm160 functions are not restricted to splicing. As pre-mRNA is spliced, a cluster of proteins, including SRm160, is deposited 20–24 nucleotides upstream of the exon–exon junction (Tange et al. 2004; Andersen and Le Hir 2008). Proteins in this exon junction complex (EJC) are important for transport of the mRNA into the cytoplasm and for nonsense-mediated decay (NMD) (Chamieh et al. 2008; Ivanov et al. 2008). SRm160 also stimulates 3′-end cleavage in cultured cells independently of its action in the EJC (McCracken et al. 2002, 2003).
Splicing of transcripts encoding MAP kinase in Drosophila cells is affected by RNAi-induced depletion of EJC components, including SRm160 (Ashton-Beaucage et al. 2010; Roignant and Treisman 2010). The locus-encoding Drosophila MAP kinase is spread over a long distance and is embedded in heterochromatin, and it was speculated that the splicing of other heterochromatic genes may also be affected by EJC components such as SRm160. For the moment, however, no further examples have been reported.
SRm160 is among the most phosphorylated proteins in HeLa cell and 293T embryonic kidney cell nuclei (Beausoleil et al. 2004; Molina et al. 2007), as well as in Drosophila embryos (Bodenmiller et al. 2008; Zhai et al. 2008). It possesses over 25 and 45 different phosphorylation sites in mammalian cells and Drosophila embryos respectively, for which not all the kinases responsible have been identified. In particular, it has been suggested to be a substrate for MAP kinase in HeLa and 203T cells (Cheng and Sharp 2006), influencing splice-site selection for CD44. Both mammalian and Drosophila SRm160 orthologs contain predicted phosphorylation sites for the LAMMER protein kinases as well (Y.-J. F. and L. R., unpublished results; Nikolakaki et al. 2002), several of which are phosphorylated in vivo (Zhai et al. 2008). LAMMER (also known as CLK) kinases participate in the regulation of alternative splicing through the phosphorylation of SR and SR-like proteins, such as SRm160 (Colwill et al. 1996; Duncan et al. 1997; Nayler et al. 1997; Du et al. 1998; Nikolakaki et al. 2002; Prasad and Manley 2003; Savaldi-Goldstein et al. 2003).
Analyses in cultured Drosophila cells somewhat surprisingly demonstrated that several EJC components, including SRm160, are dispensable for general RNA export and cellular viability (Gatfield and Izaurralde 2002), suggesting the possibility that the functions of these proteins may be important only during specific developmental processes, in particular cell types or in combination with developmentally specific cofactors (Venables et al. 2012). Other studies demonstrate that specific combinations of SR protein and SRm160 activity are required for oocyte viability in Caenorhabditis elegans, although elimination of SRm160 activity alone had no discernable phenotypes (Longman et al. 2000; Longman et al. 2001).
To provide further understanding of SRm160 function and its potential role in the development of a multicellular organism, we characterized mutant and overexpressing alleles of the Drosophila SRm160 ortholog, CG11274. Unlike C. elegans, SRm160 activity is required for viability in Drosophila. SRm160 also plays a role in the cascade of Drosophila somatic sex determination, since heterozygosity for mutant SRm160 alleles strongly enhances cryptic sex transformations produced by mild heteroallelic mutant combinations in the locus encoding the Drosophila LAMMER protein kinase Doa. SRm160 overexpression in the developing eye imaginal disc induces apoptosis and severe disorganization of the adult retina. These and other SRm160 overexpression phenotypes are suppressed by Doa alleles, suggesting that phosphorylation by this kinase is required for SRm160 activity, an inference supported by in vitro phosphorylation results. Intriguingly, SRm160 protein is eliminated from blastoderm embryonic nuclei coincident with cellularization.
Materials and Methods
Drosophila culture, mutagenesis, and crosses
Drosophila stocks and crosses were maintained on standard cornmeal agar medium. Crosses were performed at 25°, except as noted. Doa alleles and crosses to generate heteroallelic individuals were previously described (Kpebe and Rabinow 2008b; Rabinow et al. 1993). An EP P-element insertion at nucleotide 13030111 of chromosome 3L, GE25979, was purchased from Genexel, Daejeon, South Korea, and the site was verified by sequencing. This site lies within the 5′ end of the SRm160-transcribed region. Two additional alleles were recovered from imprecise excisions of the GE25979 insertion: DrMio/TMS, P{ry{+t7.2} ∆2-3} males were crossed with yw67c23; P{w+ SRm160} GE25979/TM3 females. F1 ∆2-3/ P{w+ SRm160} GE25979 males were crossed with yw67c23; TM2 /TM3 females. F2 white-eyed males were mated to w; TM3/TM6 females, stocks were established, and PCR was performed to test for imprecise excisions. A total of 178 white-eyed males were analyzed by PCR, 2 of which contained imprecise excisions of the P element (B103, C52), both of which carried deletions internal to SRm160 of at least 1.5 kb. To remove any accompanying recessive lethal loci, the B103 allele was outcrossed to a wild-type w1118 stock and balanced w− males were selected in the next generation. Individuals carrying the B103 chromosome were selected for viability over the SRm160GE25979 chromosome of origin in the next generation to eliminate any flanking lethals. A w− /TM6B stock was then established and confirmed as carrying the B103 deficiency by PCR. G18603 is a second P[EP]-element insertion in SRm160 at nucleotide 13030112 of chromosome 3L, obtained from the Bloomington Drosophila stock center. RNAi alleles 11274R-1 and R-3 were obtained from the National Institute of Genetics, Genetic Strains Research Center (NIG-Fly), Mishima, Japan.
Molecular biology
Northern transfers were performed as previously described (Yun et al. 1994; Kpebe and Rabinow 2008a). SRm160 probes were synthesized by in vitro transcription of BDGP EST clone LD17438, sequencing of which confirmed that it contains the full-length open-reading frame and terminates in a poly(A) tail. RNA was prepared by Trizol extraction (Invitrogen, Carlsbad, CA). Controls for RNA loading were performed by rehybridizing the blot with a probe for rp49 (O’Connell and Rosbash 1984). Quantification was performed with a Molecular Dynamics “Storm” phosphorimager and ImageQuant software. Levels were normalized to loadings by dividing the SRm160 signal by the corresponding rp49 signal.
Histology, scanning electron microscopy, immunocytochemistry, and in situ hybridization
Semithin retinal sections, scanning electron microscopy and in situ hybridization were performed as described (Tautz and Pfeifle 1989; Kpebe and Rabinow 2008b). Staging of embryos was performed per Campos-Ortega and Hartenstein (1997). Immunohistochemical staining of embryos was performed as described (Ashburner 1989), using the Vectastain ABC kit (Vector Laboratories). Primary antibody incubations were performed overnight at 4° in 1:1000 anti-human SRm160, a generous gift of Jeffrey Nickerson (Blencowe et al. 1998). The anti-human SRm160 serum recognizes the same protein as anti-Drosophila SRm160 on immunoblots (see Results), although the latter is not effective for immunocytochemistry (Gatfield and Izaurralde 2002). Anti-cleaved caspase 3 was obtained from Cell Signaling Technology, Danvers, MA USA.
Recombinant protein expression and in vitro phosphorylation
SRm160 fragments coding amino acid residues 1–137 (16 kDa) and 1–200 (23 kDa) were amplified from Drosophila cDNA by PCR, subcloned into pET-28a and verified by DNA sequencing. The N-terminal 6HIS-tagged proteins were expressed in Escherichea coli BL21(DE3) and purified by affinity binding to Ni–NTA agarose (Qiagen, Courtaboeuf, France). The expression and purification of recombinant DOA kinase catalytic domain and in vitro phosphorylation reactions were previously described (Lee et al. 1996; Nikolakaki et al. 2002).
Drosophila protein preparations, immunoblots, and antibody production
Adults, 0–24 hr old, were collected, washed in PBS and homogenized in TBS (0.1 M NaCl, 20 mM Tris pH 7.4), containing 0.2 mM PMSF, 1 μg/ml leupeptin, and 0.2 μg/ml pepstatin A. Homogenates were cleared by centrifugation at 13,000 rpm at 4° for 20 min in a microcentrifuge. Proteins were heated but not boiled prior to gel loading to avoid degradation, as suggested by David Gatfield (personal communication) and resolved by electrophoresis on 8% polyacrylamide SDS gels prior to blotting. Antibody to Drosophila SRm160 was produced in rabbits against the peptide DTRFSDKEKKLMKQMC (amino-acid residues 11–25) and affinity-purified on a peptide-agarose column. Other anti-Drosophila and anti-human SRm160 antibodies, generous gifts of Elisa Izaurralde (Gatfield and Izaurralde 2002) and Jeffrey Nickerson (Blencowe et al. 1998), were used at dilutions of 1:500 and 1:1000 in 5% skimmed milk–TBST. Transfers were stripped prior to probing with second or third antibodies.
Results
Variation in SRm160 transcript accumulation during development
The locus encoding the Drosophila ortholog of SRm160 (CG11274) is located at cytological position 69F6 on chromosome 3L. The four exons of the gene are widely expressed throughout development (http://flybase.org/).
A single SRm160 transcript of ∼3.7 kb was observed on Northern transfers at all developmental stages (Figure 1A), consistent with gene model data from Flybase predicting a transcript of 3862 nt. RNA-Seq data from modENCODE available on Flybase show an alternative 3′ splice acceptor site for the first intron, which would yield an mRNA ∼15 bp shorter than the major transcript encoding a protein lacking 5 amino-acid residues. We would not have detected this minor transcript on Northern transfers given the small size difference. Only 13 sequence reads were detected for this isoform throughout development compared with >5000 of the major isoform according to Flybase, and thus it is at most a minor contribution to the total population of SRm160 transcripts.
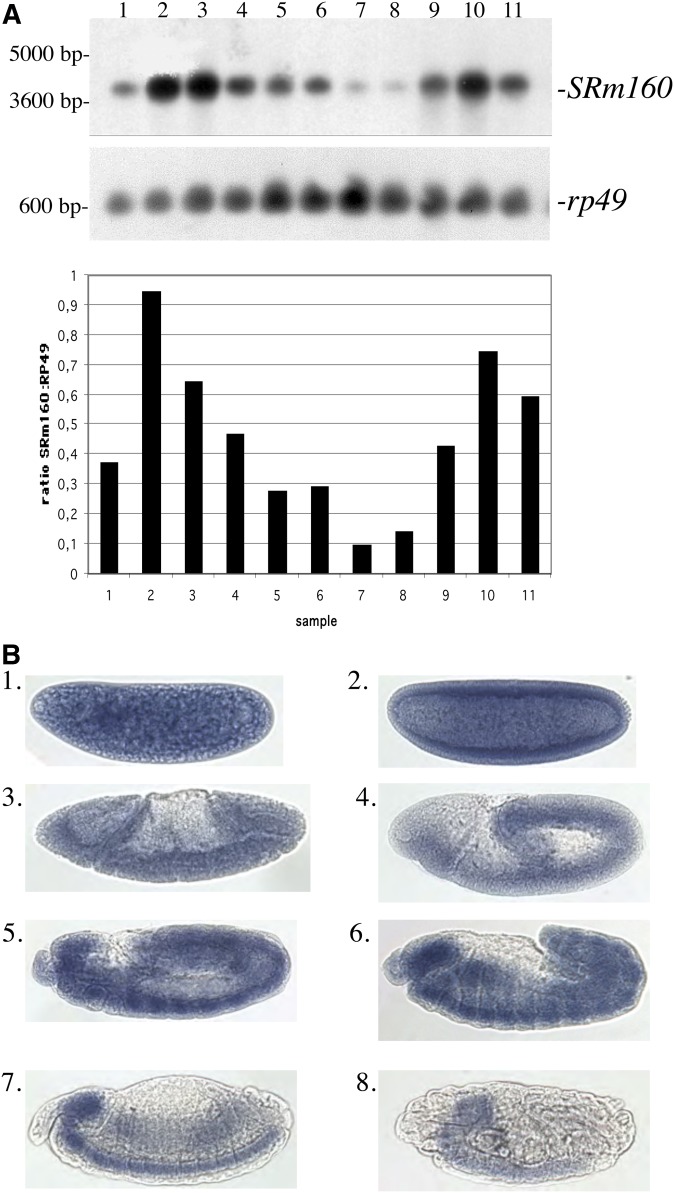
Figure 1.
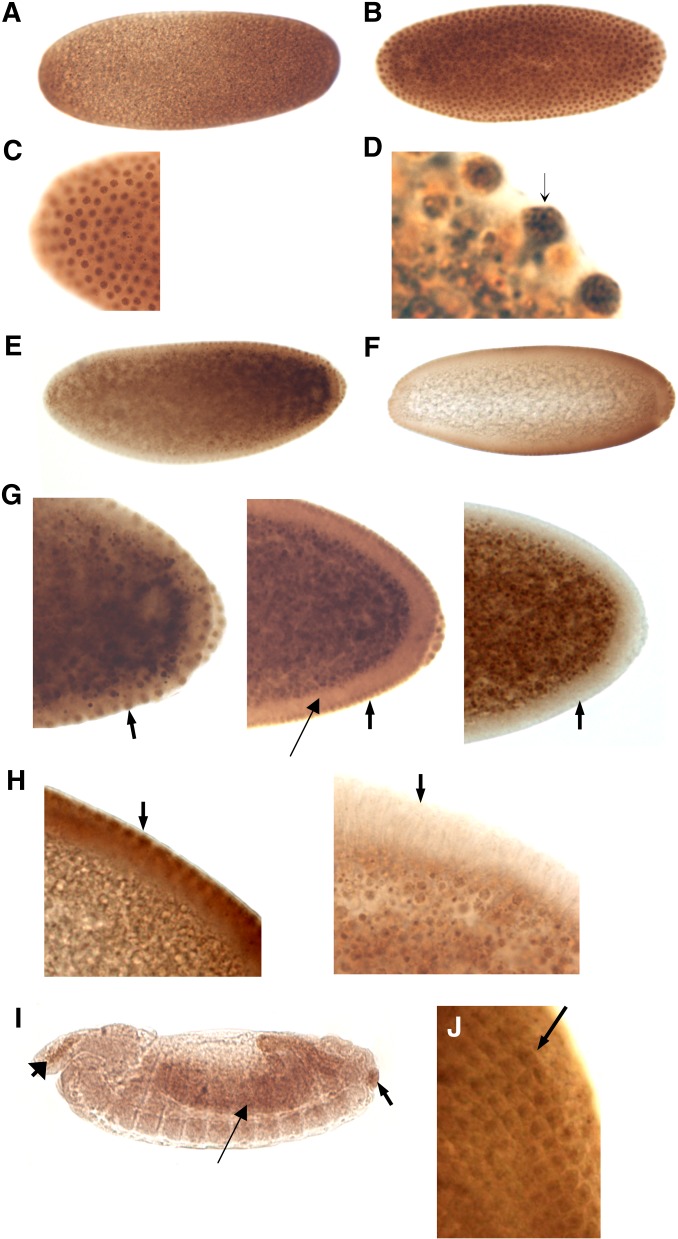
Analysis of SRm160 transcript structure and expression (A) Northern transfer. Lane 1: 0- to 24-hr embryos. Lane 2: 0- to 4-hr embryos. Lane 3: 4- to 8-hr embryos. Lane 4: 8- to 12-hr embryos. Lane 5: 12- to 16-hr embryos. Lane 6: 16- to 20-hr embryos. Lane 7: 20- to 24-hr embryos. Lane 8: first-instar larvae. Lane 9: third-instar larvae. Lane 10: 0- to 48-hr pupae. Lane 11: adults, mixed sex. Top: SRm160 probe. Bottom: rp49 probe of the same blot as a loading control. Decreasing levels of SRm160 signal are observed throughout embryonic development and increase in third-instar and pupal samples. Quantification of SRm160 signal throughout development is displayed as a ratio to rp49 signal at each developmental stage, as quantified in a phosphorimager. (B) Whole-mount in situ hybridization of SRm160 during embryonic development. Embryos were hybridized with a digoxigenin-labeled antisense SRm160 RNA probe. (1) 0- to 1-hr embryo (precellular blastoderm, stages 1–2). (2) 2- to 3-hr embryo (cellular blastoderm, stage 5). (3) 3- to 4-hr embryo (early to mid-gastrulation, stage 6). (4) 3- to 4-hr embryo (germ-band elongation, stages 7–8). (5) 4- to 5-hr embryo, germ-band segmentation, stages 9–10). (6) 7- to 9-hr embryo (germ-band retracting, segmentation appearing, stages 11–12). (7) 9- to 11-hr embryo (germ band retracted, stage 13–14). (8) ∼16-hr embryo (ventral nerve cord condensed, stage ±15). Hybridization with the opposite strand “sense” probe for SRm160 produced no signal (Figure S1).
SRm160 transcript accumulation peaks in 0- to 4-hr embryos, declines throughout subsequent embryonic stages, remains low until beginning to rise again in third-instar larvae, and reaches a second peak in 0- to 48-hr pupae. Adults show intermediate levels of transcript accumulation. The decline in transcript levels throughout the progression of embryonic development was linear (y = −0.1568x + 1.0021, r2 = 0.9294) and is reduced by 90% at late stages. By the second peak in the pupal stage, transcript levels climb back to 79% of their initial value. These data are consistent with RNA-Seq data available on Flybase.
SRm160 transcripts undergo progressive spatial restriction during embryogenesis
Whole-mount in situ hybridization with an antisense probe against SRm160 reveals widespread transcript distribution during early embryonic development (Figure 1B). Embryos hybridized with an SRm160 sense probe as a control show no staining (Supporting Information, Figure S1). Consistent with data from Northern transfers, SRm160 transcripts are ubiquitous and at high levels in 0- to 1-hr embryos (Figure 1B, 1). In 2- to 3-hr embryos, transcripts accumulate around the embryonic periphery, consistent with early zygotic gene transcription (Figure 1B, 2). At 4–5 hr of development, SRm160 transcripts are distributed in the germ band in a segmental pattern (Figure 1B, 5). Transcripts accumulate in the developing ventral nerve cord and brain regions, as well as the retracting germ band (Figure 1B6) as the nervous system becomes more defined and the first neurons appear, ∼7–8 hr after fertilization. SRm160 transcripts are primarily restricted to the developing central nervous system (CNS) at 10–11 hr of development, with some expression in the elongating midgut (Figure 1B, 7), and by 16 hr, label is found exclusively in the condensed CNS (Figure 1B, 8). The potential role of SRm160 in the nervous system is unknown, although alternative splicing in neurons is particularly complex and gives rise to numerous nervous system-specific RNA and protein isoforms (Loya et al. 2010; Calarco et al. 2011; Grabowski 2011).
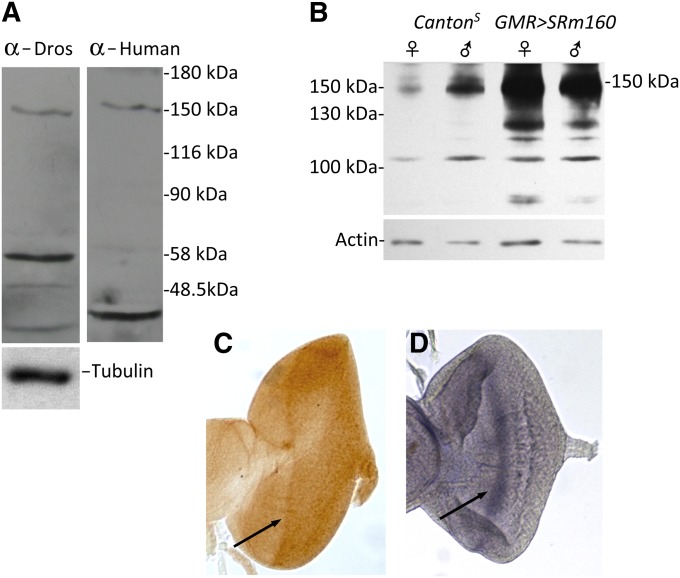
Figure 2.
Anti-human SRm160 recognizes Drosophila SRm160. (A) The same protein of 150 kDa is recognized by both anti-human anti-SRm160 antibody and anti-Drosophila SRm160. Protein samples are from adult flies. The transfer was stripped in between probing and no signal was detected following stripping, prior to redetection with the second antiserum. Lower-molecular-weight bands detected by the two sera may be SRm160 degradation products, because the protein is very labile and is undetectable if samples are boiled prior to loading (D. Gatfield, personal communication, confirmed by our results). Importantly, while a protein of identical molecular weight is observed at 150 kDa in both samples, the secondary bands are different, demonstrating no carryover of signal between probings of the blot with different sera. (B) GMR > SRm160 expression induces SRm160 protein overaccumulation (150 kDa) in head protein extracts compared with wild type (CS). The immunoblot was probed with anti-Drosophila SRm160. (C) An anti-SRm160 antibody-stained third-instar eye antennal disc shows higher-protein-level accumulation at the level of the morphogenetic furrow (arrow). (D) In situ hybridization of a third-instar SRm160 anti-sense probe hybridized to an eye antennal disc, revealing higher transcript accumulation in the morphogenetic furrow (arrow), confirming the specificity of the anti-human SRm160 for the Drosophila protein and also demonstrating increased concentration of the protein as well as the RNA in cells initially differentiating to the neuronal cell fate.
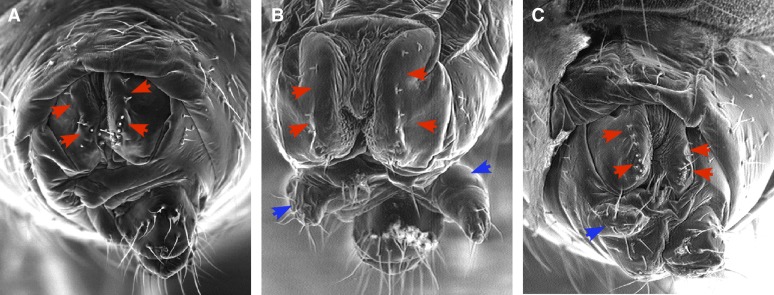
Figure 5.
SRm160 is required for normal somatic sex determination. (A) Essentially normal female genital morphology of DoaHD/DoaDEM. Very slight asymmetry and a few missing vaginal bristles may be seen (red arrowheads), typical of this genotype. SRm160B103 strongly enhances the sex transformation phenotype of Doa alleles. (B) SRm160B103 DoaDEM/+ DoaHD; (C) Doa105/ SRm160B103 DoaDEM. Note the appearance of male claspers (blue arrowheads) in the double-mutant XX individuals.
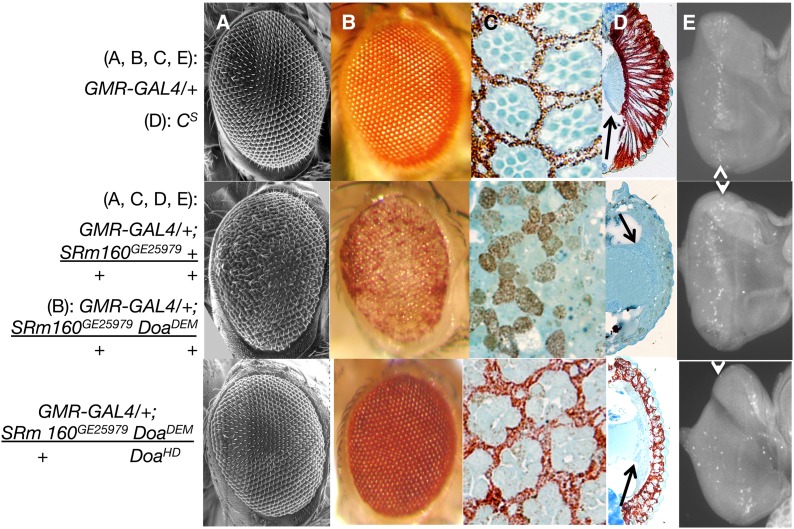
Figure 7.
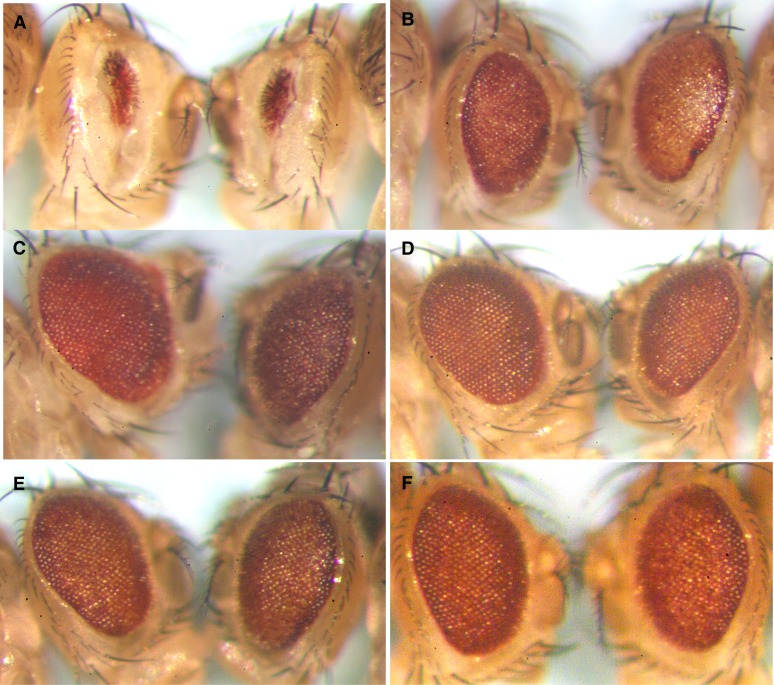
Overexpression of SRm160 in the eye induces loss of differentiation, disorganization, and apoptosis. (A) Scanning electron microscopy; (B) light microscopy; (C) semithin saggital sections, (D) Transverse sections; (E) eye imaginal discs stained with anti-activated caspase-3. Genotypes are as indicated. GMR > SRm160 DoaDEM/+ + and GMR > SRm160 DoaDEM/+ DoaHD samples were from female siblings, raised at 25°. GMR > SRm160 eyes (middle row) show significant eye aberrations, including disorganized facets in A and B, disrupted ommatidial pigment cell lattice and photoreceptor organization (C), loss of the elongation of photoreceptors and pigment cells, as well as the lamina (D), and higher levels of activated caspase (E), all as compared with GMR–GAL4 or CS as indicated (top row). The arrows in D indicate the lamina, present in Cs (top), but eliminated in GMR > SRm160 (second from top). Higher relative fluorescent signal (E, arrowheads) of activated caspase 3 in GMR > SRm160 (second from top) relative to GMR–GAL4/+ alone (top) (arrow). Simultaneous mutation of Doa (GMR > SRm160; DoaDEM/DoaHD, bottom row), almost completely suppresses the smooth glassy eye of GMR > SRm160 (A and B), restoring organization and pigment cell differentiation to the retina (C and D), and even to the lamina (D, arrow) and also strongly reduces caspase 3 activation (arrow, bottom). GMR–GAL4 as a homozygote is capable of autonomously inducing eye roughness and apoptosis, but has neither effect as a heterozygote.
Figure 8.
Phenotypes of modifiers of apoptosis alter or are altered by SRm160 genotypes. Flies are female siblings segregating from a single cross. (A) Ectopic apoptosis induced by GMR-directed expression of hid is reduced slightly by the B103 null allele of SRm160 (left), compared with its TM6 balancer sibling (right). (B) Deficiency (3L)H99 removes three pro-apoptotic loci reaper, hid and grim. Df(3L)H99 (left) partially suppresses the disorganized and glassy-eye phenotype induced by GMR > SRm160GE25979, compared with a TM6 sibling (right). (C) Expression of p35 protein in GMR > p35 (left), a potent negative regulator of apoptosis, strongly suppresses the phenotype induced by GMR > SRm160GE25979, yielding a more organized and larger eye compared with a TM6 sibling (right). (D) Expression of the endogenous inhibitor of apoptosis dIAP1 (left) also suppresses the GMR > SRm160GE25979 phenotype compared with its TM6 balancer sibling. (E) Heterozygosity for mutation of the gene encoding the apical caspase DRONC (left; droncI24) partially suppresses the unmodified GMR > SRm160GE25979 phenotype (right). A second allele, droncI29, produces a similar effect (not shown). (F) Mutation of the effector caspase-encoding locus Dcp1 (CG5370) (Dcp1k5606, left) partially suppresses GMR > SRm160GE25979 in the CyO sibling (right).
Anti-human SRm160 antibodies recognize Drosophila SRm160
An antibody to Drosophila SRm160 raised against amino acids 1–192 is not effective for immunocytochemistry (Gatfield and Izaurralde 2002; E. Izaurralde and D. Gatfield, personal communication). We raised another anti-peptide antibody to Drosophila SRm160 (AA residues 11–25), but it also failed in immunocytochemical detection of the protein. However, sequences at the N terminus of the Drosophila SRm160 protein corresponding to the 153 amino acids (AA 7–160) at the N terminus of human SRm160 are 55% identical and 78% similar in amino-acid sequence to the human protein. Anti-human SRm160 antibody directed against these initial 153 residues (Blencowe et al. 1998) was very generously provided by Jeffrey Nickerson. The specificity of this heterospecific antibody was verified on immunoblots (Figure 2). Anti-human and anti-Drosophila antibodies reveal a protein of identical mobility at ∼150 kDa (Figure 2A), instead of the 108 kDa predicted by the SRm160 amino-acid sequence of 954 AA residues. This observation is similar to that made with human SRm160, where the predicted protein molecular weight is ∼90 kDa but observed at 160 kDa. The identical molecular weight of the proteins recognized by anti-Drosophila and anti-human SRm160 provides evidence that the anti-human antibody specifically recognizes the Drosophila protein. Moreover, the discrepancy between the predicted and observed molecular weights for the Drosophila protein reinforces previous findings that SRm160 is highly phosphorylated in both Drosophila and humans, consistent with phosphoproteomic studies (Beausoleil et al. 2004; Molina et al. 2007; Bodenmiller et al. 2008; Zhai et al. 2008).
We tested the specificity of the Drosophila antibody for SRm160 on immunoblots by expressing the gene in fly heads using GMR–GAL4 and GE25979, a P-element inserted at the 5′ end of the locus permitting its expression (see below). The results demonstrated strongly increased expression of SRm160 protein under these conditions (Figure 2B). Finally, using the anti-human SRm160 for immunocytochemical detection, we noted recognition of distinctly higher protein accumulation in or near the morphogenetic furrow of developing eye imaginal discs (Figure 2C) coincident with higher SRm160 mRNA accumulation revealed by in situ hybridization (Figure 2D). We were therefore confident in using the anti-human SRm160 to examine distribution of the protein in developing Drosophila embryos.
Dynamic distribution of SRm160 protein in early embryos
In early precellular blastoderm embryos, immunostaining was ubiquitous (Figure 3A). Strong nuclear staining was observed during the precellular blastoderm stage, where the nuclei of an embryo in about the eighth nuclear division are clearly defined (Figures 3, B and C). Inspection of these nuclei under higher magnification revealed that SRm160 was distributed in the nucleus in a punctate pattern, consistent with its association with the nuclear matrix and nuclear speckles (Figure 3D, arrow), typical of SR proteins in general and SRm160 in particular.
Figure 3.
Immunocytochemical localization of SRm160 protein during embryonic development. (A) Stage 1–2: 0–30 min, pre-cellular blastoderm. (B) Stage 3: 1- to 2-hr precellular blastoderm. (C) SRm160 is localized to nuclei of 0- to 1-hr embryos undergoing synchronous nuclear divisions. (D) Higher magnification of the same embryo. The punctate nuclear staining is consistent with previous characterizations of SRm160 intranuclear localization in mammalian cells. An example is marked with an arrow. (E) Stage 4: 1- to 2-hr old embryo. Note the staining of the nuclei around the entire embryonic cortex and in the pole cells. Also a slight anterior to posterior gradient of SRm160 accumulation is observed. (F) Stage 4: 1- to 2-hr embryo. A slightly lighter staining than E, emphasizing SRm160 localization to peripheral nuclei, pole cells, and the anterior to posterior gradient. (G) SRm160 nuclear localization is quickly eliminated during cellularization. Left: early stage 4 embryo. SRm160 is seen in the nuclei of cells in the cortex (arrow), as well as in those of the pole cells. Center: Late stage 4 embryo. As cellularization begins, SRm160 appears to already be lessening in the cortex and pole-cell nuclei (arrow). The thin arrow indicates the cytoplasm of the newly forming cells. Right: Stage 5 embryo. SRm160 is no longer observed in the nuclei of the newly formed cells or the pole cells at the completion of cellularization (arrow). (H) A closer view of the cortex of the embryo immediately before (left) and immediately following cellularization (right). SRm160 localization to the nuclei (left, arrow) or indeed within the newly formed cells is no longer seen following cellularization (right). (I) Stage 13–14: 9- to 11-hr embryo. Arrowhead indicates area of hemocyte generation. Later staged embryos show essentially uniform and ubiquitous SRm160 expression (not shown), except here, where noticeably higher protein concentrations are observed in the area of hemocyte generation (anterior, left arrow), the developing midgut (arrow), and the anus (posterior, right arrow). (J) A third-instar eye-antennal disc showing higher SRm160 protein accumulation in differentiating neuronal prephotoreceptor cell clusters (arrow).
During nuclear migration to the embryonic cortex and pole cell formation at the posterior end of the embryo, SRm160 protein remained concentrated in nuclei and formed punctae of more intense nucleoplasmic staining (Figure 3D). An anterior-to-posterior concentration gradient of SRm160 was also observed (Figure 3, E and F), suggesting a potential role for it in formation of the embryonic anterior/posterior axis. Of particular note was very rapid clearing of SRm160 protein from the nuclei coinciding with cellularization (Figure 3, G and H). Little or no immunostaining was subsequently seen in the newly formed cells in either the cytoplasm or the nucleus. At ∼11 hr of development, widely distributed protein was observed at low levels with heavier concentrations in the elongating midgut (arrow, Figure 3I) and in the anus (posterior arrowhead). A small area of more intense staining was seen in the head area and small structures on the dorsal side of the embryo (anterior arrowhead), apparently corresponding to the area of hemocyte development.
We also noted relatively uniform distribution of SRm160 protein in imaginal discs (not shown) with the exception of the eye disc (Figure 2C, above). Higher magnification of the eye imaginal disc revealed that SRm160 protein accumulates to a greater level in differentiating photoreceptor precursors (Figure 2J), suggestive of a role for the protein in neuronal differentiation, as suggested by the accumulation of its mRNA in the CNS in terminal stage embryos (Figure 1B8).
Characterization of SRm160 mutant and RNAi phenotypes
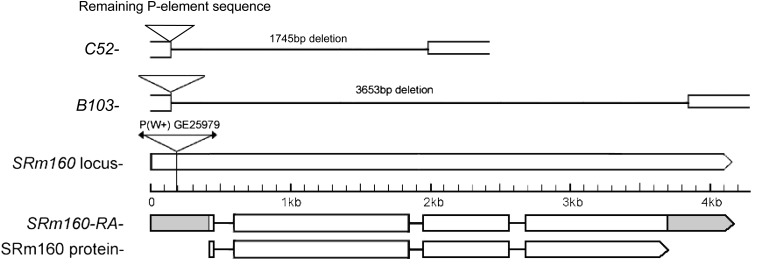
The line GE25979 carries an EP P-element insertion parallel to CG11274, SRm160, which potentially permitted its overexpression but also provided a reagent for the generation of additional alleles. The chromosome carried at least one recessive lethal allele. Mobilization of the inserted w+ P-element in GE25979 yielded two imprecise excisions generating deficiencies of 1.6 kb or more and retaining part of the initial P-element. The first of these alleles, B103, is a deletion of 3653 bp. completely eliminating SRm160 coding sequences (Figure 4; accession no. HM572038.1). It is thus a null allele and is recessive lethal. The second allele, C52, deletes 1.7 kb of the ORF (accession no. KF447873) and is thus also a null. We also examined G18603, another SRm160 allele carrying a P-element insertion at or immediately adjacent to the element inserted in GE25979 (Bellen et al. 2011; Y.-J. Fan and L. Rabinow, unpublished results). G18603 is also recessive lethal, although rare homozygous pupae can be seen on vial walls. It partially complements GE25979 (Table S1), suggesting that both alleles are hypomorphs. Surviving GE25979/G18603 heteroallelic flies are normal in all cuticular phenotypes and fertile.
Figure 4.
Structure of the SRm160 locus (CG11274), its products, the insertion site of the P-element in GE29510, and the deletion alleles B103 and C52. A single mRNA and protein are predicted by FlyBase, consistent with our transcript and immunoblot analysis. The insertion site and orientation of the EP P-element in GE29510 permit GAL4-directed expression of the gene. The B103 allele removes the entire coding and parts of the noncoding region of the gene, whereas C52 removes only the two 5′ exons.
To eliminate possible secondary recessive lethals on the B103 deletion chromosome, B103 was outcrossed to w1118 as described in Materials and Methods. Complementation testing showed that the cleansed B103 chromosome remained recessive lethal, with embryos dying prior to hatching. B103 was also lethal with the C52 deletion allele, but partially complemented both GE25979 and G18603, in each case with a less drastic effect on male survival rates relative to females (Table S1).
Finally, two lines expressing interfering RNAs for SRm160 from NIG-FLY (http://www.shigen.nig.ac.jp/fly/nigfly/) display identical phenotypes, including lethality at 25° and 29° when expressed ubiquitously with da–GAL4 and slightly roughened eyes when expressed posterior to the morphogenetic furrow of the developing eye disc with GMR–GAL4 (Table S2). Somewhat surprisingly, no lethality or other phenotypes were observed when either of these RNAi lines was expressed in neurons using three different elav–GAL4 driver elements (C155, G1-9, and G1-10). When expression of the RNAi constructs was driven in the dorsal developing wing with MS1096–GAL4, wings were severely blistered and failed to expand completely (Table S2; not shown).
SRm160 functions in somatic sex determination
Somatic sex determination in Drosophila is regulated by a cascade of alternative splicing, under the control of the SR-like TRA and TRA2 proteins, among others (Forch and Valcarcel 2003; Rabinow and Samson 2010). Mammalian SRm160 interacts directly with the mammalian TRA2 ortholog (Eldridge et al. 1999), suggesting that the Drosophila protein might participate in somatic sex determination. Alleles of Doa, the locus encoding the unique LAMMER protein kinase of Drosophila, are almost always lethal and so homozygotes are usually not recoverable, but combinations of specific hypomorphic alleles survive to adulthood (Kpebe and Rabinow 2008b; Rabinow and Birchler 1989; Rabinow et al. 1993). Among other phenotypes, heteroallelism for Doa induces female-to-male sexual transformations of varying degrees due to the hypophosphorylation of TRA and TRA2 and aberrant splicing of dsx transcripts (Du et al. 1998; Kpebe and Rabinow 2008b). Moreover, epistasis experiments demonstrated that Doa function intervenes at or after the level of tra in sex determination. Combined with biochemical data, the results of the epistasis experiments argue strongly that DOA kinase phosphorylates TRA directly to effect female-specific splicing of dsx transcripts. Sex transformations observed in Doa heteroallic females range from undetectable to barely visible reductions in the number of vaginal “teeth,” to a complete phenocopy of dsx (Du et al. 1998; Kpebe and Rabinow 2008b). We used the variable expressivity of these female-to-male transformations as a sensitized system to test whether SRm160 mutations affect somatic sex determination. SRm160B103 was crossed with two different Doa heteroallelic combinations that produce very mild or undetectable sex transformations, respectively, HD/DEM (Figure 5A) and DEM/105 (not shown). Strong enhancement of somatic sex transformation was observed in both double mutant SRm160B103 DoaDEM/+ DoaHD (Figure 5B), SRm160B103 DoaDEM/+ Doa105 (Figure 5C), and DoaDEM SRm160GE25979/+ DoaHD (not shown), with the appearance of the male genital arch and “claspers” in XX individuals. These results demonstrate a role for SRm160 in Drosophila somatic sex determination, most likely through the regulation of splicing and its interaction with the splicing-enhancer complex on the dsx transcript.
Severe phenotypes provoked by SRm160 overexpression
We used the P-element insertion in SRm160GE25979 to overexpress the gene via the GAL4–UAS system (Brand and Perrimon 1993) (Table S2). Ubiquitous overexpression using da–GAL led to lethality at 25°. Of the few survivors from a mass cross at room temperature (>1000 F1 of the control genotype da > GAL4; TM6B/+ were scored), no males and only 25 female da > SRm160 escaped lethality, but these showed no visible mutant phenotypes. Lethality in these animals occurs early in development for the most part, since only 59% of the F1 embryos hatch from the cross da-GAL4 × SRm160GE25979/+, 75% of the first-instar larvae survive to pupation and 86% of pupae eclose as adults.
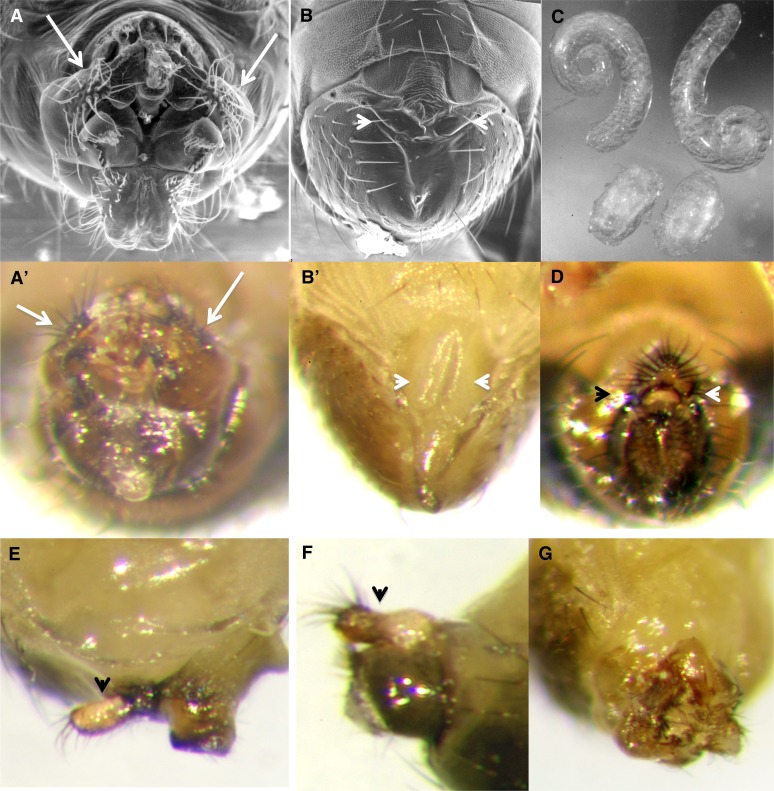
Driving SRm160 expression in the genital disc using esg–GAL4 yielded mostly normal progeny at 25°, of which both sexes were fertile. However, 10% of the male F1 showed abnormal genital structure, including rotated genitals or extra appendages or completely lacking external genitalia and analia, although having normal sex combs and pigmentation (N = 157; Figure 6). Animals with aberrant genital morphology had testes that were poorly developed and had failed to elongate (Figure 6C), but these contained sperm of normal morphology when squashed and examined under a microscope. Only a single esg > SRm160 female showed loss of external analia (N > 100).
Figure 6.
Overexpression of SRm160 interferes with normal male genital development: In wild-type males, the external genitals of the males are easily seen (A, A′), including the “claspers” (arrows). In ∼10% of male esg > SRm160GE25979 animals, external genitals and or analia are defective or even completely missing (B, B′, arrowheads). Pigmentation and sex combs were not affected, however. Testes dissected from esg > SRm160GE25979 animals without external phenotypes (C, top) are near normal, whereas those from their brothers lacking external genitalia fail to elongate (C, bottom). Both, however, have sperm of normal morphology and movement. Other defects observed include animals with normal analia but missing genitalia (D, arrowheads), extra appendages of unknown origin (E and F, arrowheads), and rotated genitals (G).
SRm160 overexpression in the eye imaginal disc induces apoptosis
GMR > SRm160 expression produced rough and shiny eyes, observable at 25° (Figure 7, A and B, middle), with males more strongly affected. At 29° these phenotypes were significantly enhanced: very few GMR > SRm160 flies survived and these were all females possessing glassy eyes with little pigment (not shown). Male lethality occurred during pupation, given the large number of dead and blackened pupae observed on the vial walls. Semithin transverse and tangential sections of fly retinas from GMR > SRm160 flies grown at 25° reveal loss of differentiation and complete disorganization of the ommatidial array compared with wild-type controls (GMR–GAL4 (Figure 7, C and D, middle vs. top) or Cs (not shown). No sign of the highly organized pigment cell lattice is visible (compare Figure 7C, top, GMR-GAL4, and middle, GMR > SRm160). However, at least partially functional cone cells must be present to secrete the lens, which is visible in scanning electron micrographs, photomicrographs (A, B), and transverse sections (D) of these genotypes. This observation suggests that differentiation may begin normally in the eye disc, since formation of cone cells depends upon induction of their progenitors by the photoreceptors, but either pigment cell differentiation is blocked or those cells degenerate, if formed. We also note that formation of the lamina, the subretinal cell layer, is abnormal in GMR > SRm160 (arrow, Figure 7D, middle), compared with wild type (top).
Immunocytochemical staining of eye imaginal discs with anti-activated caspase 3 from animals overexpressing SRm160 revealed elevated and more dispersed signal compared with GMR–GAL4; + (Figure 7E, compare top, GMR–GAL4 and GMR > SRm160, middle). Although GMR–GAL4 is capable of inducing apoptosis on its own when homozygous or even heterozygous at 29° (Kramer and Staveley 2003), neither of those conditions were fulfilled in these experiments. That GMR > SRm160 induces gross disorganization of the eye, loss of differentiation, and elevated levels of activated caspase well beyond those observed with the GMR driver alone demonstrates that the defects observed in GMR > SRm160 animals are due to abnormally high levels of apoptosis induced by SRm160 overexpression.
We further tested whether SRm160 promotes apoptosis by crossing mutant and overexpressing alleles with loci encoding various modifiers and effectors of apoptosis (Table 1 and Figure 8). Balanced males carrying a modifier or construct affecting apoptosis were crossed with females either mutant for or overexpressing SRm160 in the eye (GMR > SRm160GE25979). In the first case tested, slight reduction of ectopic apoptosis induced by GMR > hid is observed by reduction of SRm160 dose in the null allele B103, compared with siblings carrying the TM6 balancer (Figure 8A). In five subsequent tests, slight to strong suppression of GMR > SRm160GE259789-induced rough eyes with fused facets is observed by either expression of apoptotic inhibitor proteins (P35, dIAP1, Figure 8, C and D, respectively) or by mutation/deletion of apoptotic inducers/effectors. These include Df(3L)H99, which deletes three loci encoding the apoptotic inducers hid, rpr, and grm (Figure 8B), two alleles of the apical caspase dronc (Figure 8E and not shown), and the effector caspase Dcp1 (Figure 8F). Based upon these results and those of the immunocytochemical staining described above, we conclude that overexpression of SRm160 induces authentic apoptosis.
Table 1. Interactions between SRm160 alleles and loci encoding apoptotic modifiers and effectors.
| Apoptotic modifier locus/construct (male) X | SRm160 allele/balancer (female) | Observation/figure |
|---|---|---|
| GMR > hid/CyO | SRm160B103/TM6 | Slight suppression of ectopic apoptosis induced by GMR > hid by SRm160B103 (Figure 8A) |
| Df(3L)H99/TM6 | GMR–GAL4/+; SRm160GE25979/TM6 | Suppression of GMR > SRm160-induced eye roughness by Df(3L)H99 (Figure 8B) |
| UAS–p35/TM6 | GMR–GAL4/+; SRm160GE25979/TM6 | Strong suppression of GMR > SRm160-induced eye roughness by GMR > p35 (Figure 8C) |
| UAS-dIAP1/TM6 | GMR–GAL4/+; SRm160GE25979/TM6 | Strong suppression of GMR > SRm160-induced eye roughness by GMR > dIAP1 (Figure 8D) |
| droncI24/TM3 | GMR–GAL4/+; SRm160GE25979/TM6 | Slight suppression of GMR > SRm160-induced eye roughness by droncI24/+ (Figure 8E) |
| droncI29/TM3 | GMR–GAL4/+; SRm160GE25979/TM6 | Slight suppression of GMR > SRm160-induced eye roughness by droncI29/+ (not shown) |
| Dcp1k5606/CyO | GMR–GAL4/+; SRm160GE25979/TM6 | Slight suppression of GMR > SRm160-induced eye roughness by Dcp1k5606 (Figure 8F) |
All effects are described in female F1 of the cross listed comparing those carrying the modifier with their balancer siblings.
In contrast to these results, SRm160B103 does not apparently suppress apoptosis induced by GMR > p53 (not shown), nor does heterozygosity for either of the alleles p535A-1-4 or p5311-1B-1 suppress the phenotype of GMR > SRm160GE25979 (not shown), suggesting that apoptosis induced by SRm160 overexpression is independent of the strong apoptotic-inducer p53.
Genetic interactions between Doa and SRm160 alleles suggest a kinase–substrate relationship
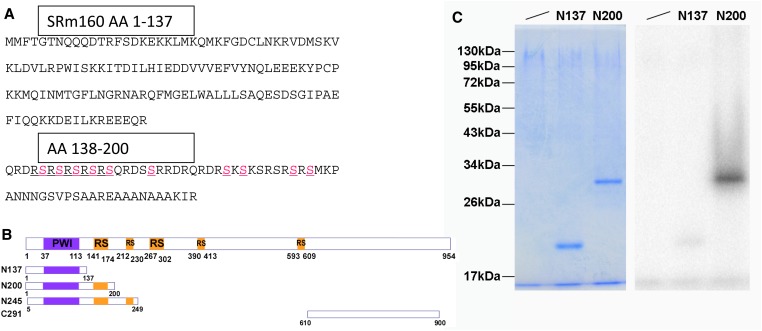
As described above, Doa alleles induce female-to-male sex transformations of varying degrees due to the hypophosphorylation of SR and SR-like proteins (Du et al. 1998; Kpebe and Rabinow 2008b), which are strongly enhanced by SRm160 alleles. Also as described, homozygosity for Doa is usually lethal but combinations of specific alleles can produce adults displaying variable phenotypes depending on those used. Given that SRm160 is among the most highly phosphorylated proteins in both mammals and Drosophila and that its sequence contains several possible DOA phosphorylation sites (e.g., those highlighted in Figure 9A, between residues 138 and 200, as well as others), we further asked whether additional SRm160 phenotypes would modify or be modified by mutations in Doa.
Figure 9.
In vitro phosphorylation of SRm160 by DOA kinase. (A) The sequence of the N-terminal 200 amino-acid residues of Drosophila SRm160 protein. No DOA-consensus phosphorylation sites are observed within the first 138 AA. Between AA 138 and 200, 10 possible phosphorylation sites for DOA are found, indicated in pink. (B) A schematic of SRm160 and constructs expressing peptides for in vitro phosphorylation studies. The first of three RS-domains lies between AA141–306, a second between AA391–424, and the third at 593–609. (C) In vitro phosphorylation of an RS-domain containing peptide derived from SRm160. Peptides were expressed in bacteria, purified, and phosphorylated in vitro by DOA kinase, prior to being resolved by electrophoresis and stained for total protein (left). Phosphorylation signal was observed only with the N200 peptide (right) and N245 peptide (not shown), despite equal amounts of protein having been loaded on the gel (left).
Genetic interactions between Doa and SRm160 were observed in crosses with GAL4-directed expression of SRm160. As described above, flies of genotype da–GAL4 >SRm160GE25979 were early lethals, which was partially rescued by heterozygosity for the DoaHD allele (Table S3). Flies escaping lethality displayed no cuticular phenotypes. Curiously, further reductions in the dose of active Doa are lethal, as da–GAL4/+; SRm160GE25979 DoaDEM/+ DoaHD heteroallelic flies failed to survive. This observation suggests that the cumulative detrimental effects of Doa mutations combined with those due to SRm160 overexpression lead to lethality. Alternatively, it is formally possible that partial dephosphorylation of SRm160 in Doa mutants produces a protein with toxic effects.
As shown above, GMR > SRm160 expression leads to roughened eyes, complete ablation of the pigment cell lattice, and disorganization of the retina, caused by elevated levels of apoptosis. These phenotypes are strongly suppressed by heteroallelism at Doa (Figure 7, middle compared with bottom). Tangential sections demonstrate substantial recovery of the underlying ommatidial organization and cellular differentiation in GMR–GAL4/+; SRm160GE25979 DoaDEM/+ DoaHD flies, lost in GMR–GAL4/+; SRm160GE25979 DoaDEM/+ + (Figure 7, middle vs. bottom). The various cell types and layers (e.g., photoreceptors, pigment cells, even some of the lamina structure; Figure 7D, arrow) and at least some of their internal organelles (i.e., rhabdomeres, Figure 7C) are all identifiable, albeit somewhat perturbed in organization in the GMR > SRm160; Doa/Doa heteroallelic flies. Transverse sections further confirm that suppression of GMR > SRm160-induced disorganization is remarkable albeit incomplete. Although differentiated pigment cells are observed and cone cells secrete a cornea of normal appearance in the double mutants, the pigment and photoreceptor cells fail to elongate to form a normal retina (Figure 7D bottom vs. top). Importantly, the suppression of GMR > SRm160-induced eye phenotypes is accompanied by a reduction in caspase 3 activation (Figure 7E, arrowheads, compare middle and bottom rows). Together, these observations demonstrate that DOA kinase potentiates the activity of SRm160, suggesting that the protein requires phosphorylation by DOA to achieve full activity.
Also described above, esg > SRm160 induced loss of external genitalia in 10% of the males (N = 157). In contrast to the suppression of GMR > SRm160 GE25979 phenotypes, the loss of external genitalia was substantially enhanced by heterozygosity for either DoaDEM or DoaHD, since 30% of these F1 showed the phenotype (N = 185). Very few esg > SRm160 GE25979 DoaDEM /+ DoaHD animals survived. Of these, 8 of 13 males (61%) escaping lethality were completely lacking genitalia and analia and died shortly following eclosion. Additionally, unlike esg > SRm160GE25979 alone, 2.7% of esg > SRm160GE25979; DoaDEM females lacked external genitals and anal plates (N = 293).
DOA kinase phosphorylates SRm160 in vitro
To investigate further the basis for interactions between SRm160 and Doa, we cloned and expressed various domains of SRm160 and tested them as in vitro substrates for the kinase. Constructs encoding the first 137, the first 200, and the amino acids 5–249 of SRm160 were expressed in E. coli, and the recombinant peptides were purified and tested for in vitro phosphorylation by recombinant DOA as previously described (Lee et al. 1996; Nikolakaki et al. 2002; Fan et al. 2010). Of those tested, peptides including sequences from the first of three RS-domains (AA 141–424) present in SRm160 (constructs N200 and N245) were the only ones phosphorylated by DOA, while peptide fragments lacking these sequences were not (Figure 9, A–C and not shown). This observation supports the hypothesis that SRm160 is a substrate in vivo for DOA kinase. It further suggests that the suppression of SRm160 overexpression phenotypes by Doa alleles may be due to hypophosphorylation of SRm160, yielding a reduction in its activity.
Discussion
Our results demonstrate that correct dosage of SRm160 function is vital to the survival of the animal, since its overexpression or depletion via mutation or RNA interference is lethal. The lethality observed when SRm160 is mutated or depleted is in contrast to the situation in C. elegans, where RNAi depletion of SRm160 produced no recognizable phenotypes (Longman et al. 2000; Longman et al. 2001). This finding suggests that Drosophila, which possesses more widespread alternative splicing than the nematode, requires functions that may already exist in evolutionarily simpler organisms but that are nonvital for them. It might be relevant that SRm160 of C. elegans possesses highly reduced RS-domains compared with either the human or Drosophila orthologs (not shown).
During embryogenesis just prior to cellularization when the nuclei have migrated to the embryonic cortex, an anterior-to-posterior gradient of SRm160 protein was observed. SRm160 and Y14 proteins are both part of the exon junction complex, which is deposited at splice junctions on mature mRNAs (Tange et al. 2004; Andersen and Le Hir 2008). The Y14 ortholog of Drosophila, Tsunagi, is essential for the localization of maternally derived oskar RNA, which is crucial for proper anterior–posterior axis formation of the oocyte and formation of the germline in the Drosophila embryo (Micklem et al. 1997; Hachet and Ephrussi 2001; Mohr et al. 2001; Hachet and Ephrussi 2004; Lewandowski et al. 2010). Our observation of an anterior–posterior gradient of SRm160 in the early embryo makes it tempting to speculate that SRm160 may also play a role in these processes.
As embryogenesis comes to a close, SRm160 transcripts are found exclusively in the central nervous system, although accumulation of the protein is more widespread, in particular in regions of the head likely corresponding to sites of hemocyte production and the hemocytes themselves. The role of SRm160 protein in these locations remains to be explored. The restriction of the SRm160 transcript accumulation in the central nervous system is intriguing in light of extensive neural-specific post-transcriptional regulation (e.g., Loya et al. 2010).
Characterization of SRm160 protein localization in the embryo also revealed dramatic clearing from blastoderm nuclei coincident with cellularization. Cellularization occurs about 3 hr after the onset of development, and the onset of zygotic gene expression occurs slightly afterward. It is possible that the rapid clearing of SRm160 from the nuclei results from the production and export of mRNA molecules at the onset of zygotic gene expression. Either nuclear SRm160 is exported to the cytoplasm, which has not been previously reported, or the protein is degraded simultaneously with cellularization. The latter possibility is particularly intriguing, given that data from the nematode Ascaris lumbricoides demonstrate the existence of a “switch” in SR protein activity and localization during embryonic development, coincident with a transition in their phosphorylation state (Sanford and Bruzik 1999; Sanford and Bruzik 2001). We speculate that the ATP-dependent regulation of SRm160 intranuclear localization previously noted in mammalian cells (Wagner et al. 2004) may be related to our observation.
Our data show that DOA kinase is required for full SRm160 activity. This requirement may be due to direct or indirect effects of DOA kinase on SRm160, since although direct phosphorylation is likely, intermediary factors are not ruled out by the present data. Moreover, given the extensive in vivo phosphorylation of SRm160 in both mammalian cells and Drosophila embryos, multiple kinases with this or similar effects are likely.
As might have been anticipated given previously described protein–protein interactions between mammalian SRm160 and TRA2 (Eldridge et al. 1999), SRm160 alleles strongly enhanced female-to-male somatic sex transformations in a sensitized genetic background. SRm160 must thus be involved in in vivo regulation of the dsx splicing enhancer, as suggested by the in vitro studies. This observation supports a conserved role for SRm160 in the regulation of alternative splicing and is consistent with the hypothesis that the protein is required for exonic splicing enhancer-dependent alternative splicing (Eldridge et al. 1999; Longman et al. 2001).
The observation that SRm160 overexpression induces apoptosis serves as an example of the role of splicing factors in the regulation of programmed cell death. Although it has been recognized for some time that alternative splicing is an important regulator of apoptosis (Schwerk and Schulze-Osthoff 2005), the identity of many splicing regulators affecting apoptosis and their apoptosis-related target transcripts are only more recently being defined (e.g., Moore et al. 2010).
The genetic and immunocytochemical evidence supporting the induction of apoptosis by SRm160 overexpression is supported by the observation that among the plethora of other proteins interacting with it is the structurally related protein RED120 (Fortes et al. 2007). Overexpression of RED120, also known as RBM25, increases apoptosis, at least in part through affecting the splicing pattern of Bxl–Xs (Zhou et al. 2008).
Finally, it was recently found that components of the EJC, of which SRm160 is a part, regulate the splicing of apoptotic regulators (Michelle et al. 2012). However, in contrast to our findings, where SRm160 overexpression leads to ectopic apoptosis, depletion of some of the core EJC components leads to increased apoptosis. This apparent discrepancy is likely to be dependent upon the functions of the individual proteins and their precise roles in the processing of target transcripts.
Our results open many new lines of inquiry. Further investigation of the biological functions of SRm160 in genetically tractable systems such as C. elegans and Drosophila will open important new insights into its regulation of biological processes, as well as help further identify its partners and the signaling pathways regulating its activity.
Supplementary Material
Acknowledgments
We thank Elisa Izaurralde and especially Jeffrey Nickerson for generous gifts of anti-SRm160 antibodies, and David Gatfield for suggestions on their use. We thank Jim Birchler, Bill Mattox, Andreas Bergmann, the Indiana University Drosophila stock center, the Genetic Strains Research Center, National Institute of Genetics, Mishima, Japan, and the Drosophila Genetic Resource Center, Kyoto, Japan for Drosophila strains, and Severine Domenichini (Plateforme Microscopie Electronique, Imagerie-Biocellulaire IFR87, Orsay-Gif, Université Paris 11), for the scanning electron micrography. Funding was provided by the Université Paris Sud 11, the Centre National de la Recherche Scientifique (CNRS) and IFCPAR award 4903-A to L.R., and the National Natural Science Foundation of China grant (31000340), and the Knowledge Innovation Program of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (2010KIP307) to Y.-J. Fan. We also gratefully acknowledge the Division de la Recherche, Université Paris Sud for supplemental support. A.G. was supported by a fellowship from the Fulbright Commission for Educational Exchange. Y.-J. Fan was supported by graduate fellowships from the Université Paris Sud 11 and the China Scholarship Council during early stages of this work.
Footnotes
Communicating editor: M. Wolfner
Literature Cited
- Andersen G. R., Le Hir H., 2008. Structural Insights into the exon junction complex. Curr. Opin. Struct. Biol. 18: 112–119. [DOI] [PubMed] [Google Scholar]
- Ashburner M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ashton-Beaucage D., Udell C. M., Lavoie H., Baril C., Lefrancois M., et al. , 2010. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143: 251–262. [DOI] [PubMed] [Google Scholar]
- Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Ville J., et al. , 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101: 12130–12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J., Ouzounis C. A., 1999. The PWI motif: a new protein domain in splicing factors. Trends Biochem Sci 24: 179–180. [DOI] [PubMed] [Google Scholar]
- Blencowe B. J., Issner R., Nickerson J. A., Sharp P. A., 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., et al. , 2008. PhosphoPep: a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 26: 1339–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Calarco J. A., Zhen M., Blencowe B. J., 2011. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA 17: 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega J. A., Hartenstein V., 1997. The Embryonic Development of Drosophila melanogaster. Springer, Berlin. [Google Scholar]
- Chamieh H., Ballut L., Bonneau F., Le Hir H., 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 15: 85–93. [DOI] [PubMed] [Google Scholar]
- Cheng C., Sharp P. A., 2006. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell. Biol. 26: 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J. L., et al. , 1996. The Clk/STY protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15: 265–275. [PMC free article] [PubMed] [Google Scholar]
- Cooper T. A., Wan L., Dreyfuss G., 2009. RNA and disease. Cell 136: 777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., McGuffin M. E., Dauwalder B., Rabinow L., Mattox W., 1998. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol. Cell 2: 741–750. [DOI] [PubMed] [Google Scholar]
- Duncan P. I., Stojdl D. F., Marius R. M., Bell J. C., 1997. In vivo regulation of alternative pre-mRNA splicing by the Clk1 protein kinase. Mol. Cell. Biol. 17: 5996–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge A. G., Li Y., Sharp P. A., Blencowe B. J., 1999. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl. Acad. Sci. USA 96: 6125–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Tang L., 2013. Aberrant and alternative splicing in skeletal system disease. Gene 528: 21–26. [DOI] [PubMed] [Google Scholar]
- Fan Y., Schlierf M., Cuervo-Gaspar A., Dreux C., Kpebe A., et al. , 2010. Drosophila translational elongation factor-1{gamma} is modified in response to DOA kinase activity and is essential for cellular viability. Genetics 184: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch P., Valcarcel J., 2003. Splicing regulation in Drosophila sex determination. Prog. Mol. Subcell. Biol. 31: 127–151. [DOI] [PubMed] [Google Scholar]
- Fortes P., Longman D., McCracken S., Ip J. Y., Poot R., et al. , 2007. Identification and characterization of RED120: a conserved PWI domain protein with links to splicing and 3′-end formation. FEBS Lett. 581: 3087–3097. [DOI] [PubMed] [Google Scholar]
- Fu R. H., Liu S. P., Huang S. J., Chen H. J., Chen P. R., et al. , 2013. Aberrant alternative splicing events in Parkinson’s disease. Cell Transplant. 22: 653–661. [DOI] [PubMed] [Google Scholar]
- Gabut M., Samavarchi-Tehrani P., Wang X., Slobodeniuc V., O’Hanlon D., et al. , 2011. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147: 132–146. [DOI] [PubMed] [Google Scholar]
- Gatfield D., Izaurralde E., 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski P., 2011. Alternative splicing takes shape during neuronal development. Curr. Opin. Genet. Dev. 21: 388–394. [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A., 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666–1674. [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A., 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963. [DOI] [PubMed] [Google Scholar]
- Herold N., Will C. L., Wolf E., Kastner B., Urlaub H., et al. , 2009. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 29: 281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P. V., Gehring N. H., Kunz J. B., Hentze M. W., Kulozik A. E., 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D., Schneider-Poetsch T., Yoshida M., 2012. Splicing in oncogenesis and tumor suppression. Cancer Sci. 103: 1611–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kpebe A., Rabinow L., 2008a Alternative promoter usage generates multiple evolutionarily conserved isoforms of Drosophila DOA kinase. Genesis 46: 132–143. [DOI] [PubMed] [Google Scholar]
- Kpebe A., Rabinow L., 2008b Dissection of DOA kinase isoform functions in Drosophila. Genetics 179: 1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., Staveley B. E., 2003. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet. Mol. Res. 2: 43–47. [PubMed] [Google Scholar]
- Kwiatkowski T. J., Jr, Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., et al. , 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208. [DOI] [PubMed] [Google Scholar]
- Lee K., Du C., Horn M., Rabinow L., 1996. Activity and autophosphorylation of LAMMER protein kinases. J. Biol. Chem. 271: 27299–27303. [DOI] [PubMed] [Google Scholar]
- Lewandowski J. P., Sheehan K. B., Bennett P. E., Jr, Boswell R. E., 2010. Mago Nashi, Tsunagi/Y14, and Ranshi form a complex that influences oocyte differentiation in Drosophila melanogaster. Dev. Biol. 339: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Carter G., Romero J., Gower K. M., Watson J., et al. , 2013. Clk/STY (cdc2-like kinase 1) and Akt regulate alternative splicing and adipogenesis in 3T3–L1 pre-adipocytes. PLoS ONE 8: e53268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. C., Caceres J. F., 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417: 15–27. [DOI] [PubMed] [Google Scholar]
- Longman D., Johnstone I. L., Caceres J. F., 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19: 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., McGarvey T., McCracken S., Johnstone I. L., Blencowe B. J., et al. , 2001. Multiple interactions between SRm160 and SR family proteins in enhancer-dependent splicing and development of C. elegans. Curr. Biol. 11: 1923–1933. [DOI] [PubMed] [Google Scholar]
- Loya C. M., Van Vactor D., Fulga T. A., 2010. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 24: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Krainer A. R., 2010. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24: 1073–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Lambermon M., Blencowe B. J., 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22: 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Longman D., Johnstone I. L., Caceres J. F., Blencowe B. J., 2003. An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J. Biol. Chem. 278: 44153–44160. [DOI] [PubMed] [Google Scholar]
- McCracken S., Longman D., Marcon E., Moens P., Downey M., et al. , 2005. Proteomic analysis of SRm160-containing complexes reveals a conserved association with cohesin. J. Biol. Chem. 280: 42227–42236. [DOI] [PubMed] [Google Scholar]
- Meissner M., Lopato S., Gotzmann J., Sauermann G. and A. Barta, 2003. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell Res. 283: 184–195. [DOI] [PubMed] [Google Scholar]
- Michelle L., Cloutier A., Toutant J., Shkreta L., Thibault P., et al. , 2012. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol. Cell. Biol. 32: 954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem D. R., Dasgupta R., Elliott H., Gergely F., Davidson C., et al. , 1997. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 7: 468–478. [DOI] [PubMed] [Google Scholar]
- Mintz P. J., Patterson S. D., Neuwald A. F., Spahr C. S., Spector D. L., 1999. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 18: 4308–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa R., Tano K., Mizuno R., Nakamura Y., Ijiri K., et al. , 2012. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18: 738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S. E., Dillon S. T., Boswell R. E., 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15: 2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H., Horn D. M., Tang N., Mathivanan S., Pandey S., 2007. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 104: 2199–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J., Wang Q., Kennedy C. J., Silver P. A., 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayler O., Stamm S., Ullrich A., 1997. Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem. J. 326: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E., Du C., Lai J., Cantley L., Giannakouros T., et al. , 2002. Phosphorylation by LAMMER protein kinases: determination of a consensus site, identification of in vitro substrates and implications for substrate preferences. Biochemistry 41: 2055–2066. [DOI] [PubMed] [Google Scholar]
- O’Connell P. O., Rosbash M., 1984. Sequence, structure and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 12: 5495–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J., 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Prasad J., Manley J. L., 2003. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol. Cell. Biol. 23: 4139–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow L., Birchler J. A., 1989. A dosage-sensitive modifier of retrotransposon induced alleles of the Drosophila white locus. EMBO J. 8: 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow L., Samson M. L., 2010. The role of the Drosophila LAMMER protein kinase DOA in somatic sex determination. J. Genet. 89: 271–277. [DOI] [PubMed] [Google Scholar]
- Rabinow L., Chiang S. L., Birchler J. A., 1993. Mutations at the Darkener of apricot locus modulate transcript levels of copia and copia-induced mutations in Drosophila melanogaster. Genetics 134: 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J. Y., Treisman J. E., 2010. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143: 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco-Bubulya P., Spector D. L., 2002. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J. R., Bruzik J. P., 1999. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 13: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J. R., Bruzik J. P., 2001. Regulation of SR protein localization during development. Proc. Natl. Acad. Sci. USA 98: 10184–10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Aviv D., Davydov O., Fluhr R., 2003. Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell 15: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C., Schulze-Osthoff K., 2005. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell 19: 1–13. [DOI] [PubMed] [Google Scholar]
- Srebow A., Kornblihtt A. R., 2006. The connection between splicing and cancer. J. Cell Sci. 119: 2635–2641. [DOI] [PubMed] [Google Scholar]
- Szymczyna B. R., Bowman J., McCracken S., Pineda-Lucena A., Lu Y., et al. , 2003. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 17: 461–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange T. O., Nott A., Moore M. J., 2004. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16: 279–284. [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C., 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81–85. [DOI] [PubMed] [Google Scholar]
- Venables J. P., Klinck R., Bramard A., Inkel L., Dufresne-Martin G., et al. , 2008. Identification of alternative splicing markers for breast cancer. Cancer Res. 68: 9525–9531. [DOI] [PubMed] [Google Scholar]
- Venables J. P., Tazi J., Juge F., 2012. Regulated functional alternative splicing in Drosophila. Nucleic Acids Res. 40: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Chiosea S., Nickerson J. A., 2003. The spatial targeting and nuclear matrix binding domains of SRm160. Proc. Natl. Acad. Sci. USA 100: 3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Chiosea S., Ivshina M., Nickerson J. A., 2004. In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. J. Cell Biol. 164: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan K. M., Nickerson J. A., Krockmalnic G., Penman S., 1994. The B1C8 protein is in the dense assemblies of the nuclear matrix and relocates to the spindle and pericentriolar filaments at mitosis. Proc. Natl. Acad. Sci. USA 91: 594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., et al. , 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yang D., Ding J. H., Wang W., Chu P. H., et al. , 2005. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120: 59–72. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., et al. , 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. [DOI] [PubMed] [Google Scholar]
- Yun B., Farkas R., Lee K., Rabinow L., 1994. The Doa locus encodes a member of a new protein kinase family, and is essential for eye and embryonic development in Drosophila melanogaster. Genes Dev. 8: 1160–1173. [DOI] [PubMed] [Google Scholar]
- Zhai B., Villen J., Beausoleil S. A., Mintseris J., Gygi S. P., 2008. Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X. Y., Wang P., Han J., Rosenfeld M. G., Fu X. D., 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 35: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A., Ou A. C., Cho A., Benz E. J., Jr, Huang S. C., 2008. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell. Biol. 28: 5924–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.