Abstract
One strategy being evaluated for HIV-1 vaccine development is focusing immune responses towards neutralizing epitopes on the gp120 outer domain (OD) by removing the immunodominant, but non-neutralizing, inner domain. Previous OD constructs have not elicited strong neutralizing antibodies (nAbs). We constructed two immunogens, a monomeric gp120-OD and a trimeric gp120-OD×3, based on an M group consensus sequence (MCON6). Their biochemical and immunological properties were compared with intact gp120. Results indicated better preservation of critical neutralizing epitopes on gp120-OD×3. In contrast to previous studies, our immunogens induced potent, cross-reactive nAbs in rabbits. Although nAbs primarily targeted Tier 1 viruses, they exhibited significant breadth. Epitope mapping analyses indicated that nAbs primarily targeted conserved V3 loop elements. Although the potency and breadth of nAbs were similar for all three immunogens, nAb induction kinetics indicated that gp120-OD×3 was superior to gp120-OD, suggesting that gp120-OD×3 is a promising prototype for further gp120 OD-based immunogen development.
Introduction
According to the 2013 UNAIDS report on the global Acquired Immunodeficiency Syndrome (AIDS) epidemic, an estimated 35.3 million people were infected with human immunodeficiency virus type 1 (HIV-1) in 2012. Despite the existence of highly effective anti-viral drugs, there were still 2.3 million new HIV-1 infections in 2012. A vaccine that could elicit broadly neutralizing antibodies (bnAbs) is hypothesized to be the most effective means to stop the AIDS pandemic (Haynes and Montefiori, 2006; Hoxie, 2010; Mascola and Montefiori, 2010). Unfortunately, eliciting bnAbs against HIV-1 has been a major scientific challenge (Ross et al., 2010). Several factors contribute to this difficulty, including the high mutation rate, extensive glycosylation (Wyatt et al., 1998), and significant conformational flexibility of the envelope glycoprotein (Karlsson Hedestam et al., 2008; Kwong et al., 2011), as well as low envelope spike density on the virion surface (J. S. Klein and Bjorkman, 2010; J. S. Klein et al., 2009).
The HIV-1 envelope glycoprotein is the sole known target of nAbs against the virus. It is comprised of two subunits; a surface glycoprotein, gp120, that contains the receptor (CD4) and co-receptor (viz. CCR5 or CXCR4) binding sites; and a transmembrane glycoprotein, gp41, that mediates fusion between the viral and the host cellular membranes (reviewed in LaBranche et al., 2001; Pantophlet and Burton, 2006; Wyatt and Sodroski, 1998). These two subunits form a trimeric heterodimer (Center et al., 2002; 2001; Zhu et al., 2003). Structurally, gp120 is divided into three domains: an inner domain (ID), an outer domain (OD), and a bridging sheet that connects the two (Kwong et al., 1998). In the native trimeric envelope spike, the ID is buried internally while the OD is exposed on the surface (Kwong et al., 2000). The envelope glycoprotein has four conserved regions that are targeted by potent bnAbs isolated from HIV-1 infected patients: (1) the CD4 binding site (e.g. b12, VRC01, NIH45-46, 3BNC117 (Burton et al., 1991; Diskin et al., 2011; Scheid et al., 2011; X. Wu et al., 2010)), (2) glycans around N160 along with conserved elements on V1/V2 (e.g. PG9, PG16 (Walker et al., 2009)), (3) glycans and the base of V3 loop around N332 (e.g. 2G12 and PGT antibodies, including PGT128 (Buchacher et al., 1994; Walker et al., 2011)), and (4) the membrane-proximal external region (MPER) of gp41 (e.g. 2F5, Z13e1, 4E10 and 10E8 (Huang et al., 2012; Muster et al., 1994; Purtscher et al., 1994; Stiegler et al., 2001; Zwick et al., 2001; also reviewed in Kwong and Mascola, 2012; Kwong et al., 2013). A number of bnAbs that target other novel epitopes have also been isolated more recently: 3BC176 and 3BC315 that target a region near the V3 loop and the CD4i site (F. Klein et al., 2012), 8ANC195 that recognizes portions of gp41 and N-linked glycans adjacent to the CD4BS (Scharf et al., 2014), and PGT151 that targets a glycan-dependent epitope at the interface of gp120 and gp41 (Blattner et al., 2014; Falkowska et al., 2014).
Considering that many of the epitopes targeted by bnAbs are on the gp120 OD and that the highly immunodominant ID elicits non-neutralizing antibodies, it has been hypothesized that immunogens comprised of only the OD are better able to induce bnAbs than the intact gp120 (Kwong et al., 2011; Nabel et al., 2011; Zhou et al., 2007). Unfortunately, previous immunization studies using OD-based immunogens have largely been unsuccessful. Yang et al. (Yang et al., 2004) expressed a clade B YU2 OD construct, termed OD1. The OD1 protein could be recognized by 2G12 and various anti-V3 human mAbs, but not by b12. In a later study, however, it was shown by surface plasmon resonance (SPR) that b12 could bind OD1, but with a high dissociation rate (Zhou et al., 2007). The protein also failed to bind CD4. Not surprisingly, rabbits immunized with OD1 failed to mount detectable levels of nAbs against HIV-1KB9 or HIV-1YU2.
In another study, Chen et al. (Chen et al., 2007; 2008) immunized mice with a clade C virus OD (CN54) fused to the human IgG1 Fc domain as an immune enhancer. However, there were no neutralizing activities in the mice sera. Bhattacharyya et al. (Bhattacharyya et al., 2010) designed an OD-based immunogen (ODEC) from HIV-1 HxBc2. The ODEC protein was expressed in E. coli and, therefore, it was not glycosylated. It also lacked the variable loops V1/V2 and V3, and 11 mutations were introduced to increase binding affinity to CD4 and IgG1 b12. However, the sera from immunized rabbits showed only marginal neutralizing activity against four clade B viruses and one clade C virus. Wu et al. (L. Wu et al., 2009) generated a membrane-anchored OD containing residues 252–482 from the HIV-1 clade B virus TAI-R3A. This OD variant had a truncated β20–β21 hairpin and a shortened V3 loop, fused to the transmembrane region of CD4. This optimized, membrane-anchored OD could be recognized by b12 and b13, but was not recognized by other CD4 binding site (CD4BS) antibodies. Joyce et al. (Joyce et al., 2013) recently reported an antigenically optimized OD4.2.2 from the clade A KER2018 strain, which could bind to VRC01 with nanomolar affinity. However, its immunogenic properties were not reported.
Despite these unsuccessful attempts to induce bnAbs using immunogens based on gp120 OD, it is our hypothesis that focusing immune responses to the OD of gp120 is still worth pursuing based on one or more of the following reasons: (1) the immunogens tested thus far have not been tested rigorously for their proper antigenic conformation, (2) the immunogens were based on a single HIV-1 isolate, and (3) the adjuvants used in these studies may not have been ideal to maintain correct antigenic structures. In this report, we describe our attempt to design and evaluate gp120 OD-based immunogens to induce bnAbs in rabbits. The results suggest that our immunogens and/or the antigen delivery system we used are better than others reported thus far in inducing nAbs against HIV-1.
Results
Construction, expression and purification of monomeric and trimeric forms of gp120 OD, and gp120
In past studies of generating immunogens based on gp120 OD, envelope protein sequences from a single HIV-1 isolate from a given clade were used. Considering that centralized HIV-1 antigens based on ancestral, consensus or a phylogenetic tree center sequences have been suggested to have the potential to increase the breadth of immune responses (Gao et al., 2004; Gaschen et al., 2002; Nickle, 2003), we have chosen to use an M group consensus sequence for generating our immunogens (MCON6, (Gao et al., 2005)). Antisera from guinea pigs immunized with recombinant MCON6 gp120 have previously been shown to neutralize several HIV-1 primary isolates, albeit weakly and somewhat sporadically (Gao et al., 2005).
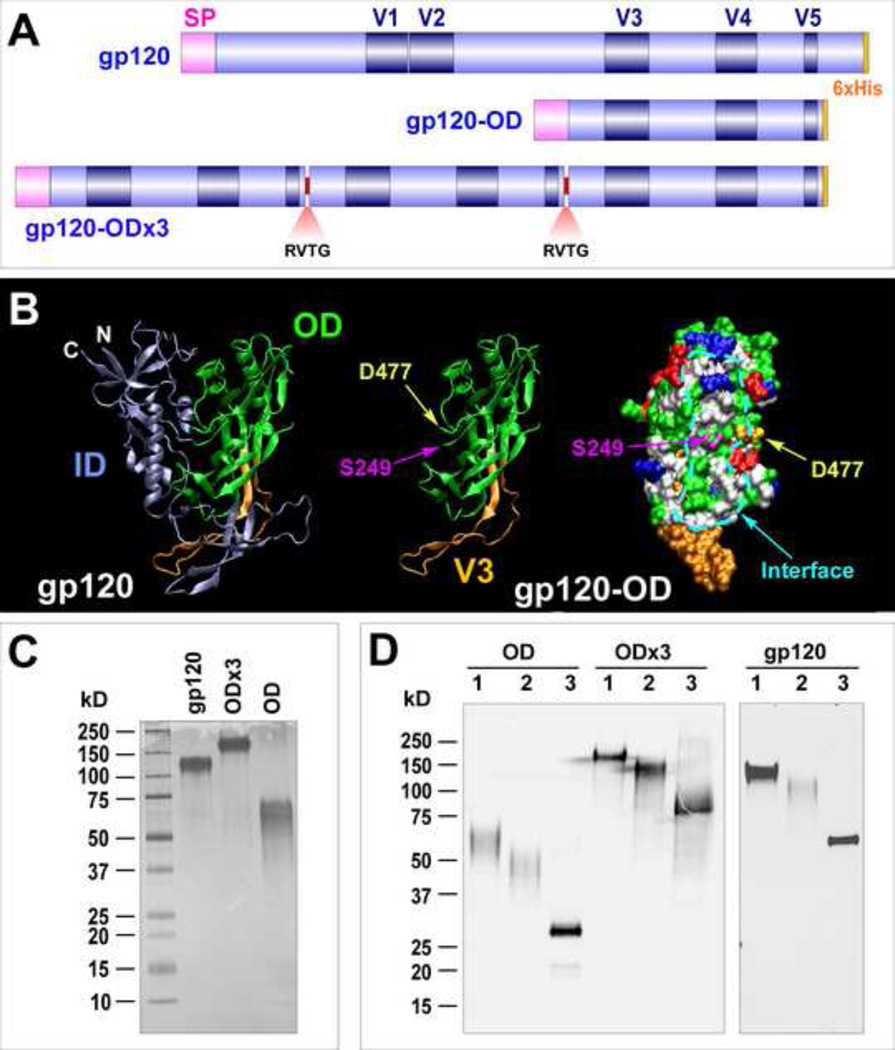
First, we generated gp120-OD by cloning 249STQLLL…RPGGGD477 (numbering based on MCON6) fused to the signal peptide (Fig. 1A). In addition to monomeric gp120-OD, we also constructed a trimeric form (gp120-OD×3), which contains three tandem copies of OD covalently linked by a four amino acid linker (RVTG). The rationale for generating OD×3 was as follows: When the ID is removed from gp120, the interface between the two domains would be exposed (Fig. 1B). The newly exposed surface could potentially be immunogenic, which might decrease immunogenicity of critical neutralizing epitopes on the OD. Because the N-terminal and the C-terminal ends of gp120-OD (S249 and D477, respectively) are on the same face of the protein and in close proximity, we hypothesized that it might be possible to “mask” the newly exposed surface from the immune system by bringing the three tandem copies close together by covalently linking them with a short, four amino acid linker. One potential benefit of trimerizing gp120-OD was enhancement of immunogenicity by increasing the valency of epitopes. In addition to gp120-OD and gp120-OD×3, we constructed the intact gp120 for comparing immunogenicity.
Fig. 1.
Schematic diagram, expression and purification of gp120, gp120-OD and -OD×3. (A) A schematic diagram of three immunogens. All proteins contain N-terminal signal peptide (SP) and C-terminal 6xHis tag. For gp120-OD×3, the OD segments are connected by a four amino acid linker (RVTG). (B) Structural representations of gp120 and gp120-OD. The OD and ID are shown in green and ice blue, respectively. The V3 loop is shown in orange. The N- and C-terminal ends of gp120-OD (S249 and D477, respectively), as well as the interface region between the ID and OD are indicated. The crystal structure of trimeric gp140 SOSIP (4NCO) was used (Khayat et al., 2013). (C) Silver stain of purified proteins expressed in HEK293 cells. (D) Western Blot analyses of deglycosylated proteins. Lane 1: untreated; lane 2: EndoH; lane 3: PNGaseF.
All three proteins were expressed efficiently in HEK293 cells, and all of them could be purified to near homogeneity using a combination of affinity chromatography (Con A and Ni-NTA) and ionexchange chromatography using Q-sepharose (Fig. 1C). Approximate molecular weights of gp120, gp120-OD, and gp120-OD×3 were 120kD, 60kD and 180kD, respectively. Gp120-OD migrated as a broad smear, suggesting significant heterogeneity in glycosylation. Deglycosylation analyses with EndoH and PNGaseF showed that proteins are glycosylated with both high-mannose and complex carbohydrates (Fig. 1D). Fully deglycosylated gp120-OD, gp120-OD×3 and gp120 migrated as expected theoretical molecular weights of 26 kD, 75 kD and 55 kD, respectively.
Antigenic properties of gp120, gp120-OD and gp120-OD×3
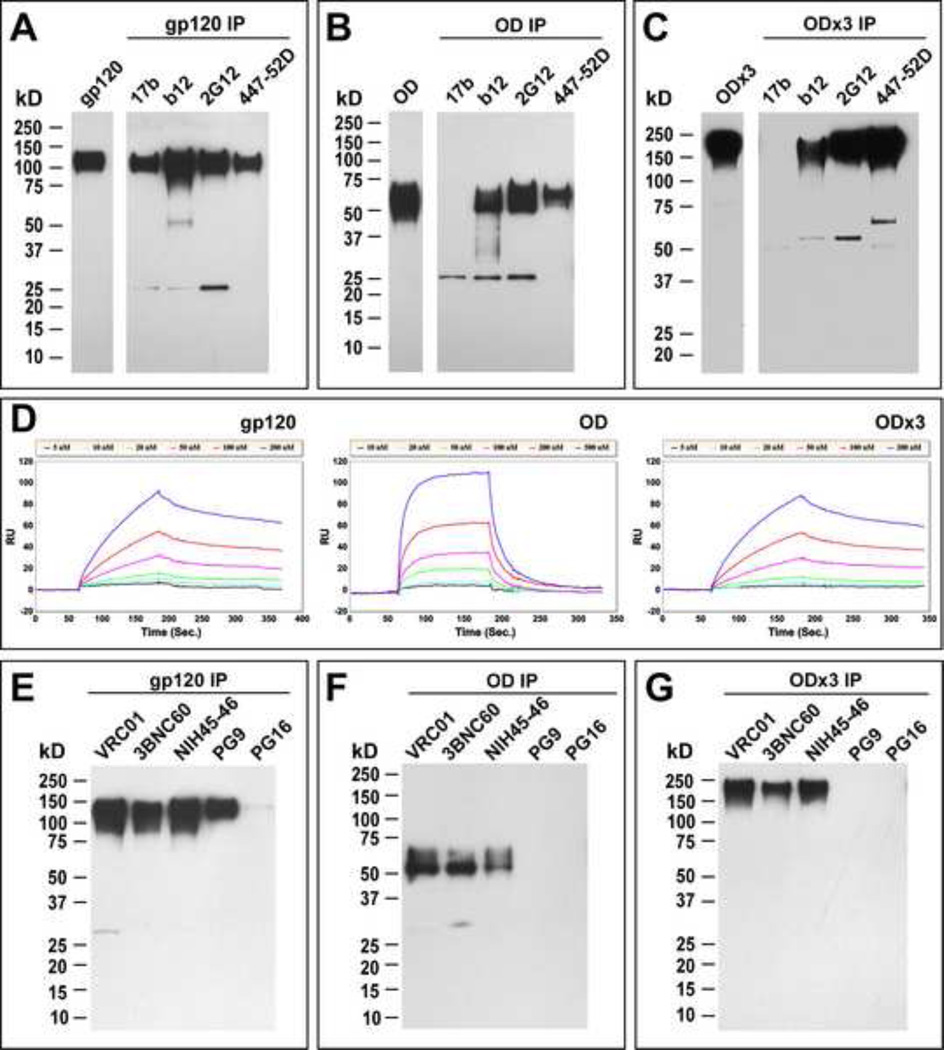
To evaluate whether OD-based immunogens were conformationally intact, their antigenic properties were examined by immunoprecipitation using multiple nAbs and compared to that of gp120. First, the prototypic nAbs that target a variety of epitopes were tested: 17b (bridging sheet), b12 (CD4BS), 2G12 (carbohydrate) and 447-52D (V3 loop). As shown in Fig. 2A, gp120 was immunoprecipitated by all four nAbs. Among the four antibodies tested, b12 appeared the most efficient. Both gp120-OD and -OD×3 also could be immunoprecipitated by all nAbs except 17b (Fig. 2B and 2C); this was expected since two of the four β-strands that make up the bridging sheet were removed along with the ID. Despite reactivity of gp120-OD and -OD×3 to b12, it was weaker relative to other nAbs when compared to gp120. It should also be noted that both of these proteins failed to bind soluble CD4, as tested by co-immunoprecipation, and that they did not bind to b12 antibody when tested by ELISA (data not shown), suggesting that structural elements on or near the CD4BS are unstable, flexible or prone to misfolding (e.g. when bound to ELISA plate). Compared to gp120 and gp120-OD, the degree of immunoreactivity of gp120-OD×3 to 2G12 and 447-52D was remarkable. This is likely due to the fact that OD×3 has three copies of the epitopes, which should enhance antigen-antibody complex formation.
Fig. 2.
Evaluation of antigenic properties. Immunoprecipitation analyses of gp120 (A), gp120-OD (B) and gp120-OD×3 (C) with mAbs 17b, b12, 2G12 and 447-52D. SPR analyses of interactions between IgG b12 and gp120, gp120-OD and -OD×3. Immunoprecipitation analyses of gp120 (E), gp120-OD (F) and gp120-OD×3 (G) with more recently isolated bnAbs VRC01, 3BNC60, NIH45-46, PG9 and PG16.
To better characterize kinetic parameters of interaction between b12 and the three proteins, surface plasmon resonance (SPR) analyses were conducted. As shown in Fig. 2D and Table 1, the association constant (Ka) between b12 and gp120-OD was about 2-fold higher than that for gp120, suggesting that b12 was able to bind to gp120-OD faster. However, the dissociation constant (Kd) between b12 and gp120-OD was greater than 20-fold higher compared to that for gp120, indicating significantly lower affinity between b12 and gp120-OD. A similar result was reported by Zhou et al. (Zhou et al., 2007). All together, the equilibrium dissociation constant (KD) was about 10 fold lower for gp120 than gp120-OD. The kinetic parameters for gp120-OD×3 were similar to those of gp120, indicating improved antigenic profile of gp120-OD×3 compared to the monomeric gp120-OD.
Table 1.
Kinetic parameters for IgG1 b12 interaction with gp120, gp120-OD and gp120-OD×3.
| Protein | Ka(1/Ms) | Kd(1/S) | KD=(Kd/ Ka) |
|---|---|---|---|
| gp120 | 3.64×104 | 1.83×10−3 | 5.02×10−8 |
| gp120-OD | 8.05×104 | 4.18×10−2 | 5.19×10−7 |
| gp120-OD×3 | 4.08×104 | 1.98×10−3 | 4.85×10−8 |
More recently, bnAbs against HIV-1 with excellent potency and breadth have been isolated from virus-infected patients (Diskin et al., 2011; Scheid et al., 2011; Walker et al., 2009; X. Wu et al., 2010). To further characterize antigenic properties of the three proteins, they were probed with bnAbs that target the CD4BS (VRC01, 3BNC60, NIH45-46), and those that bind to glycans around N160 along with conserved elements on V1/V2 (PG9 and PG16). The intact gp120 bound to all bnAbs, except for PG16 (Fig. 2E). Although CD4BS bnAbs could recognize both gp120-OD and -OD×3, the reactivity seemed to be lower (Fig. 2F, 2G). As expected, they did not bind to PG9 or PG16. SPR analyses were not conducted due to limited availability of these antibodies. Altogether, the results from these studies showed that antigenic structures of epitopes targeted by bnAbs appear better preserved in our gp120-OD and -OD×3 than other OD-based antigens described in past immunogenicity studies.
Antibody responses against gp120, gp120-OD and gp120-OD×3 in rabbits
Many of the epitopes targeted by bnAbs that bind gp120 are non-linear and/or highly conformational. Thus, using adjuvants that can preserve the correct antigenic structure would be critical, especially for proteins like our gp120-OD and gp120-OD×3 that may be more prone to denaturation. Accordingly, we have chosen to use Zn-chitosan as an antigen delivery platform/adjuvant, instead of using adjuvants that contain oil or agents that exhibit chaotropic properties that could potentially disrupt the epitope structure. Chitosan is a cationic polysaccharide (repeating units of P-(1–4)-linked D-glucosamine and N-acetyl-D-glucosamine), derived by the partial deacetylation of chitin. It is nontoxic, biocompatible and biodegradable, and has been used effectively as an adjuvant (Marcinkiewicz et al., 1991; Seferian and Martinez, 2000; Singla and Chawla, 2001; van der Lubben et al., 2001). One property of chitosan is that it can be generated as a metal ion complex with zinc (Zn). Zn-chitosan has a property similar to that of Ni-NTA such that proteins with 6×His residues can be bound (Seferian and Martinez, 2000). Thus, Zn-chitosan could also function as an antigen depot that allows slow release of antigens for extended stimulation of the immune system.
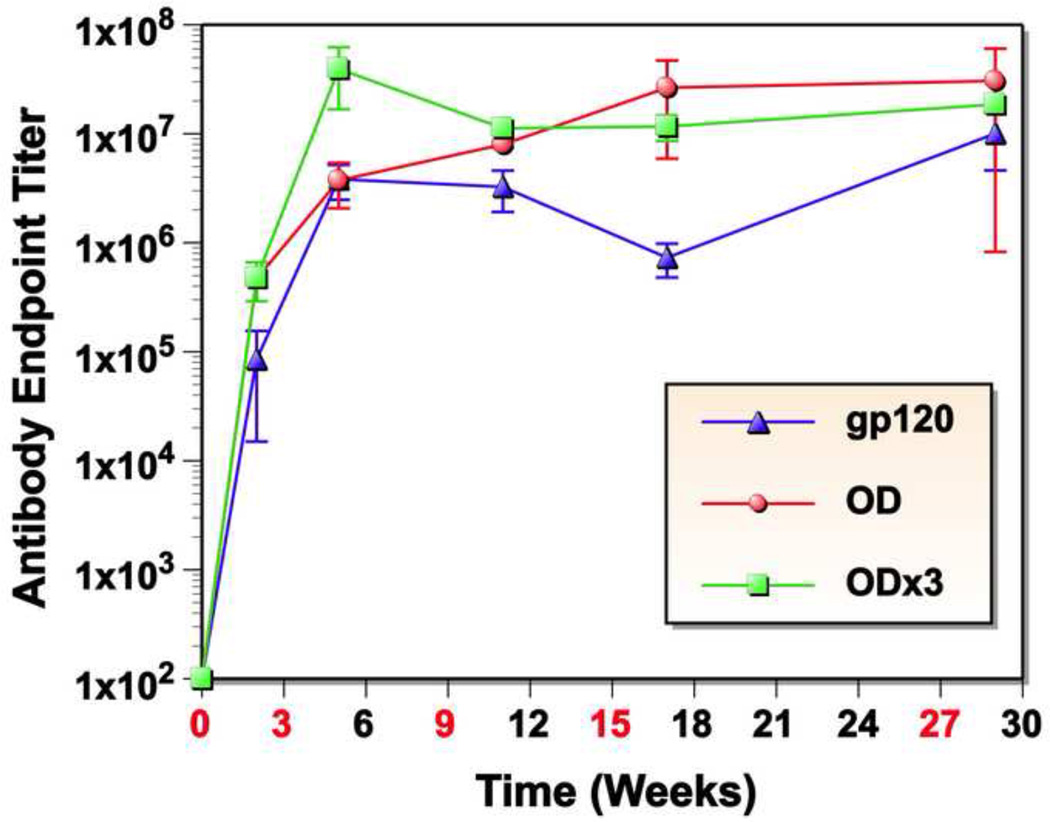
Three rabbits were immunized subcutaneously with each antigen on weeks 0, 3, 9, 15 and 27. Two control animals were inoculated with Zn-chitosan only in PBS. Serum samples were collected before (preimmune) and two weeks after each immunization. Antibody responses were monitored by ELISA using autologous antigens and the end-point antibody titers against each immunogen are shown in Fig. 3. All three protein constructs were highly immunogenic; high end-point titers of antibodies were induced even after a single immunization: 4.7×105 (gp120-OD), 4.8×105 (gp120-OD×3) and 8.5×104 (gp120). Although there were a few minor differences, the antibody induction kinetics were fairly similar for the three proteins. Generally, antibody titers reached the maximal levels after the second immunization, although it continued to increase slightly in gp120-OD-immunized animals. The end-point titers after the final immunization were 3.1×107 (gp120-OD), 1.9×107 (gp120-OD×3) and 1.0×107 (gp120).
Fig. 3.
Antibody endpoint titers determined by ELISA. Sera obtained before or 2 weeks after each immunization were monitored for autologous antigen-specific antibodies by ELISA. Data are presented as average endpoint titers of three rabbits with standard deviation (SD).
Identification of immunogenic linear epitopes within the OD
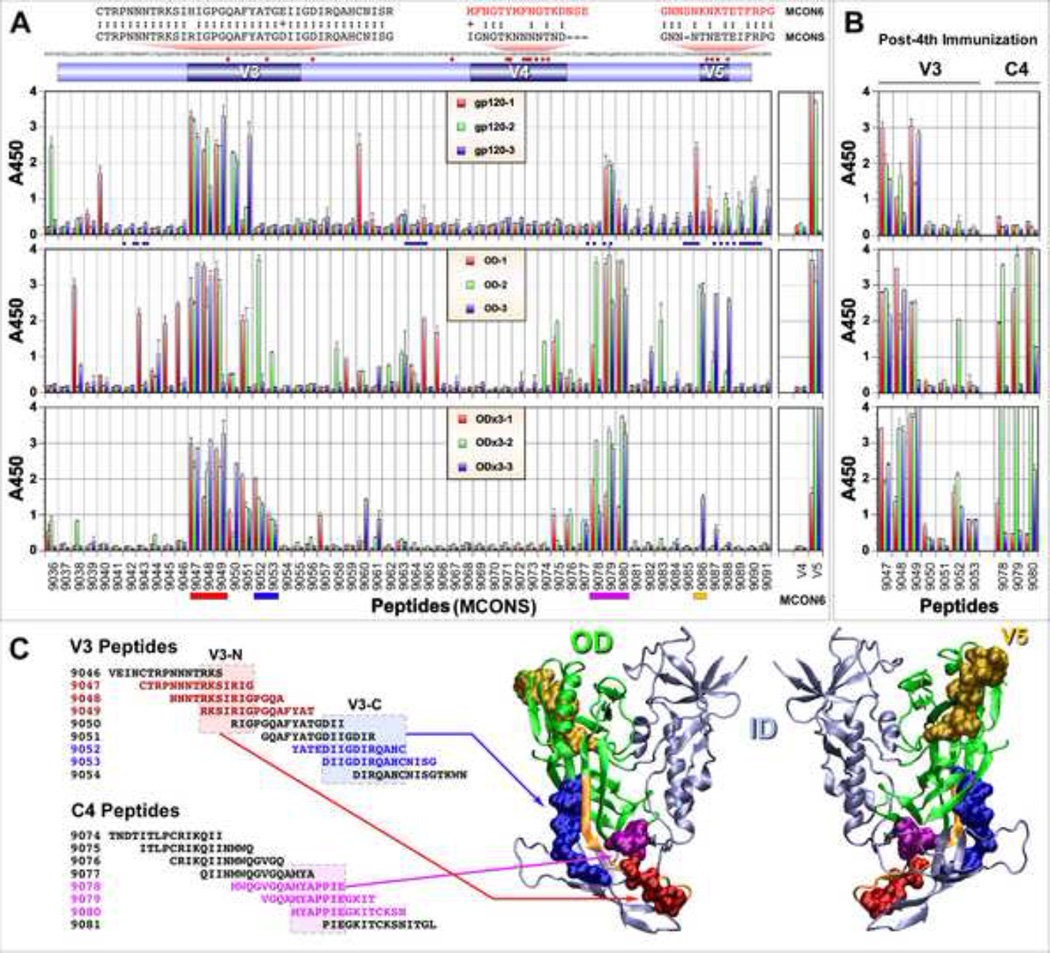
In order to better characterize antibody responses and to compare immunogenic properties of gp120, gp120-OD and -OD×3, we identified immunogenic linear epitopes in the OD region by conducting ELISA with overlapping peptides using immune sera collected after the fifth immunization (Fig. 4). For this analysis, we used peptides available from NIH AIDS RRRP which are based on M group consensus sequence designated as MCONS. Although there are some differences in amino acid sequences between MCONS and the MCON6 we used to generate our immunogens, the variant residues are mostly in the variable loops. Due to the high density of variant residues in the V4 and V5 loops, separate MCON6 peptides were used for these loops. Although there were some differences among the three immunogens and animal-to-animal variations within the group, most of the immunoreactive peptides were within the V3, C4 and V5 regions (Fig. 4A).
Fig. 4.
Comparison of immunogenic linear epitopes by ELISA using overlapping peptides. (A) ELISA using immune sera after the fifth immunization. Peptide numbers represent catalog numbers from the NIH AIDS RRRP. On the top of the figure shows a schematic diagram of gp120-OD, MCONS envelope sequence, identification of amino acid residues different from MCON6 (small red square), and sequence of MCON6 peptides covering the V4 and V5 region (red text). Blue circles below gp120 ELISA data indicates amino acid residues that makes contact with CD4 and/or VRC01. (B) ELISA of select peptides from the V3 and C4 regions with immune sera after the fourth immunization. (C) Plotting of four immunogenic epitopes onto a crystal structure of trimeric gp140 SOSIP (4NCO). Two segments within V3 loop (red and blue), one in C4 (magenta) and one in V5 (gold). Only the likely cores of the V3 and C4 epitopes (boxed in) are plotted on the crystal structure.
Overall, the V3 loop was most immunogenic. The N-terminal half of the loop (peptides 9047-CTRPNNNTRKSIRIG, 9048-NNNTRKSIRIGPGQA and 9049-RKSIRIGPGQAFYAT) was more immunogenic than the C-terminal half (peptides 9051-GQAFYATGDIIGDIR, 9052-YATEDIIGDIRQAHC and 9053-DIIGDIRQAHCNISG) (P<0.0001 for gp120 and gp120-OD; P=0.0039 for gp120-OD×3). This was more evident after the fourth immunization (Fig. 4B). A similar result was observed in a previous study that evaluated immunogenicity of JR-FL gp120 in rabbits (Vaine et al., 2008). The central core of the epitopes recognized in the N-terminal peptides appeared to be “RKSIRIG”, designated as V3-N (Fig. 4C). Interestingly, the C-terminal half of the V3 loop was more immunogenic in the context of gp120-OD×3 than in gp120 (P<0.0001) or gp120-OD (P=0.0028), especially for peptides 9052 and 9053; antibodies were observed consistently in all of the immunized animals even after the fourth immunization. Strong reactivity of these two peptides suggests DIIGDIRQAHC as the core of the epitope being recognized (designated as V3-C; Fig. 4C). These results may suggest some subtle differences in the structure or accessibility of this epitope between gp120-OD×3 and the other two immunogens. Antibodies that recognized upstream of this core were also observed after the fifth immunization (peptides 9050 and 9051).
Three peptides in the C4 region (9078-MWQGVGQAMYAPPIE, 9079-VGQAMYAPPIEGKIT and 9080-MYAPPIEGKITCKSN) were highly reactive to antibodies induced in gp120-OD and -OD×3-immunized rabbits. No anti-C4 antibodies were induced in gp120-immunized animals after the fourth immunization (Fig. 4B) and modest levels of antibodies against only peptides 9079 and 9080 were observed after the fifth immunization, indicating these epitopes are significantly less immunogenic in the intact gp120 (P<0.0001 compared to either gp120-OD or -OD×3). This is not surprising since the likely core of the epitope “MYAPPIE” lies in the interface between the OD and ID of gp120 and becomes exposed only when the ID is removed (Fig. 4C).
Peptides (MCONS) covering the V5 region were reactive to antisera only weakly or sporadically. One possible reason was difference in amino acid sequence between the MCONS-based peptides and MCON6-based immunogens used to immunize rabbits since there are four amino acids different within a stretch of eight amino acids in the V5 region (see top of Fig. 4A). Indeed, when ELISA was performed using a peptide based on MCON6 sequence (GNNSNKNKTETFRPG; underline represents variant residues between MCON6 and MCONS) a strong antibody response was observed for all three immunogens (Fig 4A, right panel). This, however, was not the case for the V4 loop, indicating that V4 loop is not immunogenic in the context of all three immunogens we tested. Alternatively, the peptide we used is unable to mimic the structure that exists in the whole protein.
Although immunogenic epitopes on the intact gp120 were also found to be immunogenic in gp120-OD and -OD×3, there were a few subtle differences, especially for gp120-OD. There were multiple peptides that reacted to antisera against gp120-OD, but not at all or less to gp120 or gp120-OD×3. They included peptides 9043–9046 in C2 just upstream of the V3 loop (P<0.0001 for gp120-OD vs. gp120 or gp120-OD×3) and peptides 9063–9066 in C3 (P<0.0001 for gp120-OD vs. -OD×3; P=0.0085 for gp120-OD vs. gp120). These results could be simply due to greater exposure of epitopes that were masked on gp120 or masking of epitopes as a result of trimerization on gp120-OD×3. Interestingly, these two regions are in close proximity to amino acid residues that make up contact residues to CD4 and/or VRC01 (i.e. Loop D and CD4 binding loop; see blue circles under gp120 ELISA data).
Induction of nAbs by gp120, gp120-OD and gp120-OD×3
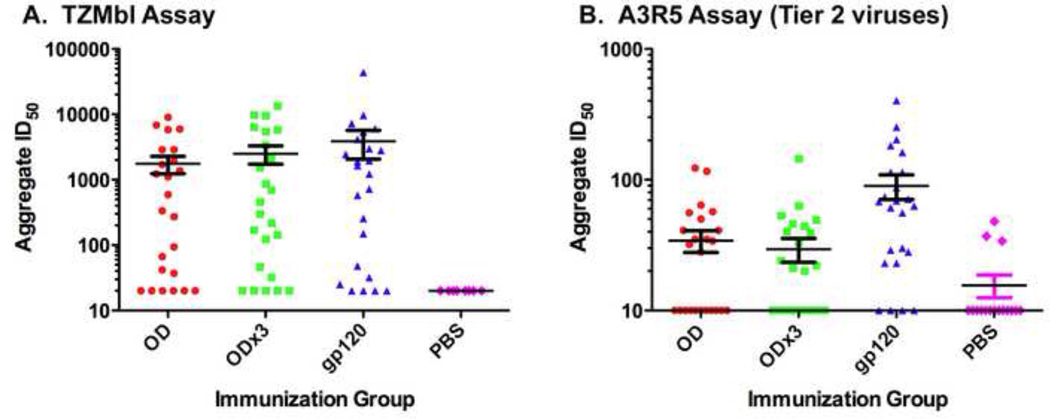
Next, we compared neutralizing activity of antibodies induced by the three immunogens. For the initial analyses, a panel of eight viruses including seven Tier 1 viruses and one Tier 2 virus was tested (Table 2). Antisera collected after the fifth immunization were used. All three immunogens induced nAbs against Tier 1, clade B (MN.3, SF162.LS, W61D-TCLA.71, Bal.26), clade C (MW965.26) and clade AE (TH023.6) pseudoviruses, but not Clade A (Q23.17). Of these six viruses, Bal.26 was least neutralization sensitive. None of the preimmune antisera exhibited neutralizing activity (data not shown). One each of the rabbits immunized with gp120 or gp120-OD×3 showed weak neutralizing activity against Tier 2, clade B isolate (6535.3). Unfortunately, no neutralizing activity was observed against other Tier 2 viruses tested (clade B, PVO.4 and SC422661.8). Analysis of aggregated ID50s from TZM-bl assays showed a general trend of gp120>gp120-OD×3>gp120-OD (Fig. 5A). However, the differences were not statistically significant. Although neutralizing activity was largely limited to Tier 1 viruses, these results are important considering that (1) the activity was induced by a synthetic sequence against heterologous virus isolates, (2) neutralizing activity was observed against virus isolates from different clades, and (3) all three immunogens induced fairly potent nAbs, including OD-based immunogens, which have not been successful in other previously reported studies.
Table 2.
Neutralizing antibody titers of rabbit antisera after 5th immunization in TZMbl.
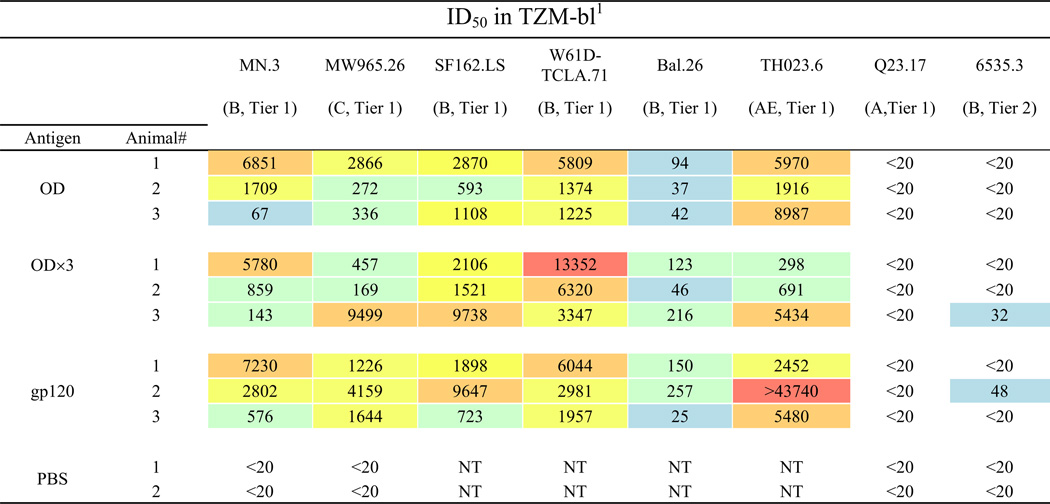 |

Values are the serum dilution at which relative luminescence units (RLUs) were reduced 50% compared to virus control wells (no sera sample). Sera from rabbits after the fifth immunization witheach antigen were tested against the indicated pseudoviruses. All sera failed to neutralize murine leukemia virus pseudovirus at the lowest serum dilution tested (1:20) (data not shown). NT, not tested. Color scale: red, ID50 > 10,000; orange, 5,000 to 9,999; yellow, 1,000–4,999; green 100 to 999; blue, 20 to 99; white, < 20.
Fig. 5.
Aggregate analyses of ID50 values of neutralizing activity. (A) Data from neutralization assay using TZM-bl cells. Data shown in Table 1 are presented. (B). Neutralization assay against Tier 2 pseudoviruses using A3R5 cells. Neutralization assays were done using immune sera collected after the fifth immunization.
Neutralizing activities of the immune sera against Tier 2 viruses (clade B: CH58, THRO and WITO; clade C: Ce1086_B2, DU151.2, Ce1176_A3, Ce2010_F5, and Du422.1) were further analyzed by a more sensitive assay using A3R5.7 cells (41, 42). Analysis showed that neutralizing activity against Tier 2 viruses could be detected in immune sera against all three proteins; however they were relatively weak and sporadic (Fig. 5B). Interestingly, gp120 induced significantly better nAbs against Tier 2 viruses than both gp120-OD and -OD×3 (p=0.0086 and 0.0044, respectively). Results were not statistically different between gp120-OD and -OD×3 (p=0.5955).
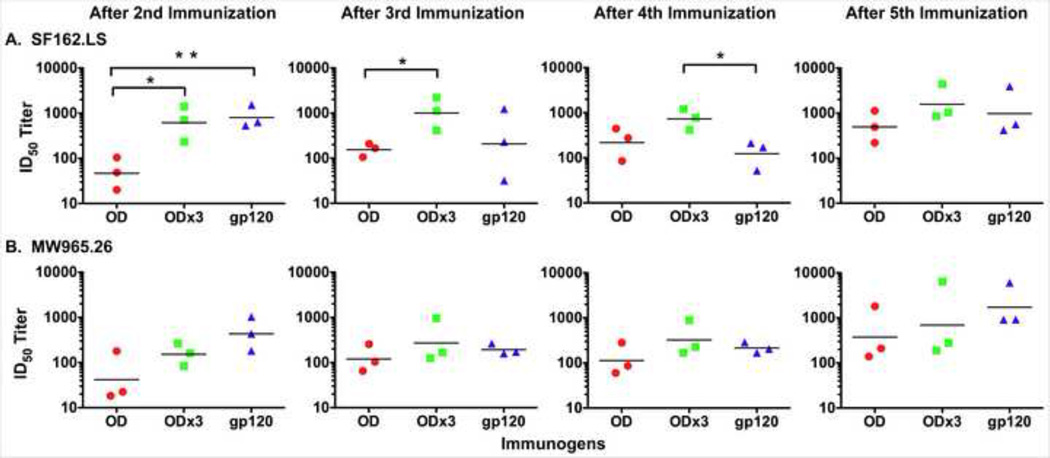
Considering there were some differences in antigenic and immunogenic properties, especially for gp120-OD, we next examined whether there might be any differences in the kinetics of nAb induction among the three immunogens. For these analyses, neutralizing activities of immune sera collected after each immunization were evaluated against Tier 1, clade B SF162.LS (Fig. 6A) and clade C MW965.26 (Fig. 6B). As shown in Fig. 6A, neutralizing activities were detected in all three groups after the second immunization; no neutralizing activities were observed after the first immunization or in mockimmunized animals (data not shown). The activity was significantly lower for gp120-OD group compared to gp120 and gp120-OD×3 (over 12- and 11-fold, respectively). However, the nAb titers for the gp120-OD group increased successively with each immunization. In contrast, the nAb titers for the gp120-OD×3 group reached near plateau after the second immunization and did not increase significantly after additional immunizations. The nAb titers for the gp120 group also seemed to have reached maximum after the second immunization. Unexpectedly, the titer actually decreased after the third and the fourth immunization. The titer increased after the fifth immunization, but only to the level achieved after the second immunization. Together, these results suggest that gp120-OD×3 is superior to gp120-OD and also perhaps to gp120 in that it requires fewer immunizations and is better able to sustain higher nAb titers. The overall pattern of neutralizing activity against MW965.26 was similar although the differences between the groups were less obvious (Fig. 6B).
Fig. 6.
Kinetic analyses of neutralizing antibodies induced by gp120, gp120-OD, and -OD×3. (A) Neutralizing activity against clade B SF162.LS with immune sera collected after the second, third, fourth, and fifth immunization. The geometric mean titer for each group of sera is shown as a horizontal bar. Statistical analysis was done with Prism GraphPad 5 using a two-tailed t-test. **P<0.01; *0.01<P<0.05. (B) Neutralizing activity against clade C MW965.26. There were no statistical differences between the groups.
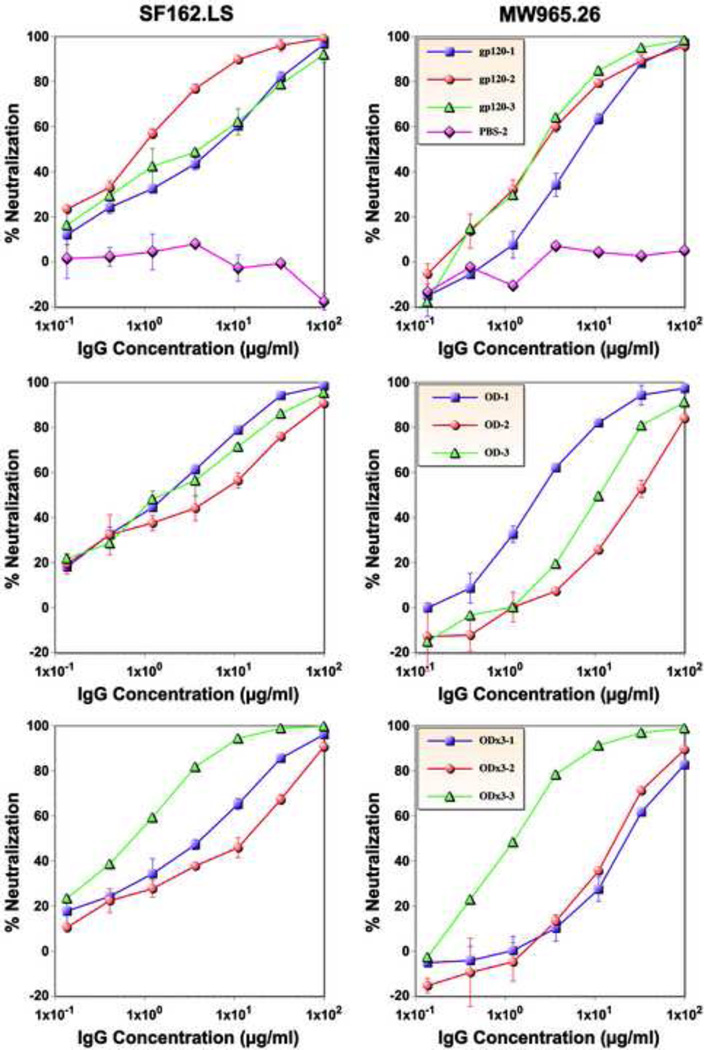
Neutralizing activity of purified IgG
To confirm neutralizing activities are indeed conferred by antibodies, IgG was purified from immune sera of all immunized rabbits and evaluated against Tier 1 clade B (SF162.LS) and clade C (MW965.26) viruses. As shown in Fig. 7, purified IgG exhibited potent neutralizing activity against both viruses. ID50 of serum neutralization inversely correlated with IC50 of IgG concentration, confirming that IgG is responsible for the neutralizing activity. For example, the ID50s for gp120-OD×3 rabbits 2 and 3 against MW965.26 were 169 and 9499, respectively. The IC50s of purified IgG from the corresponding samples were 16.1 and 1.1 µg/ml, respectively. IgG purified from one of the two mock-immunized animals (PBS-2) exhibited no neutralizing activity.
Fig. 7.
Neutralizing activity of purified serum IgG. Serial dilutions of purified IgG from gp120, gp120-OD, and -OD×3 immunized rabbits and negative control (PBS only) were tested for neutralizing activity against the SF162.LS and MW965.26 viruses.
Defining neutralization epitope(s)
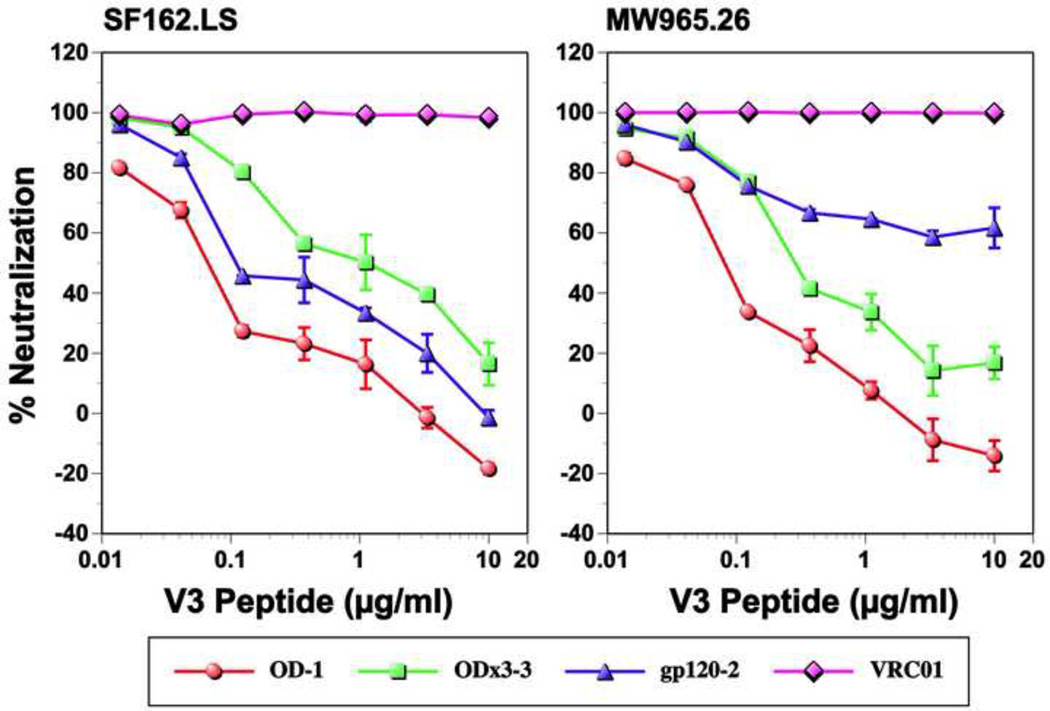
Since the V3 loop was highly immunogenic for all three immunogens (Fig. 4), it seemed likely that the V3 loop could be a major neutralizing epitope. To confirm this, we conducted a neutralization inhibition assay using the MCON6 V3 loop peptide (TRPNNNTRKSIHIGPGQAFYATGEIIGDIRQAH). For this analysis, neutralizing activity of purified IgG (50 µg/ml) from one rabbit from each immunogen group that exhibited the highest neutralizing activity was evaluated (i.e. gp120-OD-1, -OD×3-3, and gp120-2) in the presence or the absence of various concentrations of the peptide. The IC50 values of these IgG samples were fairly comparable (1.9, 0.6, and 0.9 µg/ml against SF162.LS, and 2.3, 1.1, and 1.6 µg/ml against MW965.26, respectively). VRC01 (5 µg/ml) was used as a negative control. As shown in Fig. 8, neutralization of both SF162.LS and MW965.26 viruses by gp120-OD-induced nAbs was efficiently inhibited by the peptide. In contrast, neutralizing activity of VRC01 was not affected, as expected. Neutralization of both viruses by gp120-OD×3-induced nAbs was also inhibited, but higher concentrations of the peptide were required. This could be simply due to higher anti-V3 loop antibody response induced by gp120-OD×3. Neutralization of SF162.LS by gp120-induced nAbs was also similarly inhibited. Interestingly, however, inhibition of neutralizing activity against MW965.26 was less efficient, reaching maximal inhibition of only about 40%. One plausible explanation is that this could be due to the presence of nAbs that targets epitopes outside of the V3 loop. In fact, neutralizing activity also could not be completely inhibited when using the entire gp120-OD protein as an inhibitor (data not shown), indicating that neutralizing epitope(s) could lie within the ID (e.g. V1/V2 loops), or requires structural elements of both the inner and the outer domains. Not surprisingly, immunoreactive C4 peptides (peptides #9078, 9079 and 9080) did not inhibit neutralizing activity (data not shown).
Fig. 8.
Inhibition of neutralizing activity by V3 loop peptide. Neutralizing activity of purified IgG (50 µg/ml) from one rabbit from each immunogen group was evaluated in the absence or the presence of various concentrations of MCON6 V3 peptide (TRPNNNTRKSIHIGPGQAFYATGEIIGDIRQAH). VRC01 bnAb (5 µg/ml) that targets the CD4BS was used as a negative control.
Discussion
In this report, we described construction, expression and purification of three different immunogens (gp120, gp120-OD and -OD×3) based on an M group consensus sequence (MCON6). We evaluated their antigenic properties using variety of nAbs and assessed their potential to induce nAbs in rabbits. All three immunogens induced potent nAbs that were able to neutralize viruses from three different clades including clade B (MN, SF162.LS, W61D-TCLA.71 and Bal.26), clade C (MW965.26), and clade AE (TH023.6). Although neutralizing activity was limited largely to Tier 1 viruses, results are still meaningful considering that previous OD-based immunogens reported by other investigators have either failed to induce or induced nAbs with lessor breadth and/or modest potency (Bhattacharyya et al., 2010; Chen et al., 2007; Yang et al., 2004).
One possible reason why we were able to induce potent nAbs with our OD-based constructs when others failed to do so is that we used an antigen-delivery platform/adjuvant that may have been better at preserving the native structure of the proteins, yet had potent immunostimulating properties. In the study conducted by Yang et al. (Yang et al., 2004), rabbits were immunized with their OD1 construct using 1xRibi as an adjuvant, which is an oil-in-water emulsion. Considering that OD-based proteins might already be structurally unstable with many hydrophobic amino acid residues exposed on the surface (on the interface between the inner and the outer domains), exposure to oil likely denatured the protein. Similarly, Bhattacharyya et al. (Bhattacharyya et al., 2010) used Freund’s adjuvant, which is a water-in-oil emulsion. Chen et al. (Chen et al., 2007) used an OD construct fused with the human IgG1 Fc domain to function as an adjuvant. Immunization of mice yielded end-point antibody titers of only about 400, and no neutralizing activity was detected. Low antibody titer suggests ineffectiveness of IgG1 Fc as an adjuvant. In our study we used Zn-chitosan, which is a cationic polysaccharide chelated with Zn. Our immunogens could be loaded onto the matrix simply by gently mixing the two components in a nearphysiological buffer (PBS, pH 8.0). As such, our methodology likely preserved the native antigenic conformation of potential neutralizing epitopes on gp120. Yet, chitosan exhibited high immunostimulatory properties, inducing 105–106 end-point antibody titers after the first immunization and 106-107 after the second immunization.
Another possible reason why we were able to induce fairly potent nAbs with our immunogens is the immunogens themselves (e.g. cellular origins of recombinant proteins, primary amino acid sequences, extent of glycosylation, three-dimensional structure, etc.). Our immunogens were derived from an M group consensus sequence (MCON6), and they were produced in HEK 293 cells, a human embryonic kidney cell line. In contrast, immunogens evaluated by other groups were derived from a specific HIV-1 isolate: The OD1 construct reported by Yang et al. (Yang et al., 2004), is based on clade B YU2 strain and was produced in S2 Drosophila cells. The construct studied by Chen et al. (Chen et al., 2008; 2007)is based on a clade C virus, CN54, and was produced in Sf9 Spodoptera frugiperda cells using a recombinant baculovirus. In the study described by Bhattacharyya et al. (Bhattacharyya et al., 2010), the immunogen was based on the clade B HxBc2 strain and was produced in E. coli. The proteins expressed in E. coli are non-glycosylated, and it is well known that proteins expressed in mammalian and insect cells are glycosylated differently (Marchal et al., 2001) . These glycosylation variances can have a significant effect on the biochemical, structural, functional and immunological properties of proteins. Additional studies will be needed to determine full implication of differential glycosylation on the ability to induce nAbs against HIV-1.
Construction of envelope glycoproteins based on MCON6 sequence and their immunogenic properties were first described by Gao et al. (Gao et al., 2005). In that study, soluble gp120 was produced from 293T cells infected with a recombinant vaccinia virus. Four guinea pigs were immunized subcutaneously using RIBI-CWS (cell wall skeleton from a tubercule bacillus) as an adjuvant. In these animals, the average neutralization titer against SF162.LS was about 737, and no neutralizing antibodies were detected against BAL. In our rabbits, the average neutralization titers against the two viruses were 4089 and 144, respectively. Considering that the gp120 immunogen used in the two studies are practically the same, and were produced in the same cell line, the use of different adjuvants and/or the animal model system used could be responsible for the different levels of nAbs induced. It is also possible that antigenic dosage and immunization schedule may also influence antibody titers.
The ID of gp120 is highly immunogenic, but the antibodies induced against this domain are non-neutralizing. As such, we and others have hypothesized that immunogens based solely on OD of gp120 would be better at inducing bnAbs (e.g. VRC01) than the intact gp120 because they would be able to focus the immune response toward the OD. Although our immunogen/vaccine formulation was able to induce fairly potent nAbs that other groups failed to achieve in the past, they were not any better than what could be induced with the intact gp120. In fact, the kinetics of inducing nAbs were slower for gp120-OD than for gp120, requiring more boosts. Interestingly, our gp120-OD×3 was able to induce higher nAb titers than gp120-OD. Not only that, it was better able to induce nAb response more consistently than gp120 throughout the immunization study. It is not clear why nAb titers decreased after the third or fourth immunization for gp120.
Although all three immunogens we evaluated were able to neutralize multiple virus isolates from different clades, they were limited to Tier 1 viruses. Based on V3 loop peptide inhibition data, it is most likely that nAbs are targeting conserved elements on the V3 loop. Presently, the structural parameters of the V3 loop on Tier 1 and Tier 2 viruses have not been clearly delineated. It is possible that V3 loops on Tier 2 viruses are less exposed, thus avoiding recognition by antibodies. In fact, recently determined crystal and cryo-EM structures of a soluble gp140 trimer (SOSIP gp140) of a Tier 2, clade A virus isolate (BG505) suggested that V3 loop could be shielded by intra- and intertrimeric V1/V2 loops (Julien et al., 2013; Lyumkis et al., 2013). At present, we do not know whether the V3 loop on our MCON6 gp120 resembles those on Tier 1 or Tier 2 viruses. Regardless, better understanding of common structural and immunogenic properties of the V3 loop on Tier 2 viruses might allow better immunogen design that could induce bnAbs against the V3 loop.
Based on the observation that V3 loop peptide was not able to fully inhibit neutralizing activity of gp120-induced nAbs against MW965.26, we hypothesize that some of the nAbs are targeting non-V3 loop epitopes. Thus, enhancing immunogenicity of these neutralizing epitopes and minimizing immunogenicity of the V3 loop could allow inducing higher titers of nAbs with greater breadth. Perhaps combining our gp120 with other immunogens that lack V3 loop (e.g. eOD-GT6) (Jardine et al., 2013) in a heterologous prime-boost vaccine strategy could specifically enhance antibody responses towards non-V3 neutralizing epitopes. To date, the immunogenicity of eOD-GT6 has not been reported and will need to be evaluated before an effective prime-boost strategy can be formulated.
In this study, we were able to determine only the linear immunogenic epitopes. Identification of non-linear epitopes would better enhance our understanding of immunogenic properties of gp120 and facilitate designing future immunogens. Unfortunately, there is no simple, efficient way to identify nonlinear epitopes. Development of a high throughput methodology capable of identifying non-linear epitopes could allow for more thorough characterization of B cell responses and facilitate AIDS vaccine development.
Materials and methods
Construction of gp120, gp120-OD and -OD×3
All of the constructs used in the study were derived from pcDNA-MCON6gp160 (kindly provided by Dr. Beatrice Hahn (Gao et al., 2005)). For a purpose unrelated to this study, MluI and EcoRV restriction enzyme sites in the vector were eliminated first by mutating EcoRV (by digesting the plasmid with EcoRV and XhoI, followed by Klenow fill-in and ligation). The resulting plasmid was digested with MluI followed by Klenow fill-in and ligation to eliminate the site. The resulting plasmid was renamed pcDNA*MCON6gp160. A segment of DNA encoding gp160 was cloned into pTM-NdeI (Cho et al., 1994) using EcoRI and BamHI. Subsequently, a short DNA fragment between NdeI and EcoRI was removed by digesting with both enzymes followed by Klenow fill-in and ligation, to generate pTM-MCON6gp160. Restriction enzyme sites (MluI and EcoRV) were inserted at the base of the V3 loop by introducing silent mutations using site directed mutagenesis. For the MluI site, sense primer 5'-GAGATCAACTGC-ACGCGTCCCAACAACAAC-3', and antisense primer 5'-GTTGTTGTTGGGACGCGTGCAGTT-GATCTC-3' were used with QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene) according to manufacturers protocol. EcoRV site was introduced using sense primer 5'-CGAGATCATCGGC-GATATCCGCCAGGCCCAC-3' and antisense primer 5'-GTGGGCCTGGCGGATATCGCCGATGA-TCTCG-3'. Underlined sequences signify restriction enzyme sites, and bold letters represent mutagenesis sites. Restriction enzyme digestion and DNA sequencing confirmed correct cloning. The plasmid is referred to as pTM-MCON6gp160-V3.
To clone gp120 with C-terminal 6xHis tag, gp120 coding region was PCR amplified from pTM-MCON6gp160-V3 using sense primer 5'-GACCATCACCCTGCCCTGCC-3' and antisense primer 5'-GGCCCCTCGAGTTAATGGTGATGATGGTGATGGCGCTTCTCGCGCTCCACCACGCG-3'. The PCR fragment was digested with BsrG1 and XhoI, and cloned into the corresponding sites in pTM-MCON6gp160-V3 to generate pTM-MCON6gp120-V3. Subsequently, the BstEII2-HincII fragment from this construct was inserted into BstEII-HindIII (after Klenow fill-in) sites of pcDNA*MCON6gp160 to generate pcDNA*MCON6gp120.
The OD of gp120 was PCR amplified from 249STQLLL… to …RPGGGD477 with forward primer 5'-CCATGGTGACCGGATCCACCCAGCTGCTGCTGAACGGC-3' and reverse primer 5'-GGCCT-CGAGAAGCTTAATGGTGATGATGGTGATGGTCGCCGCCGCCGGGGCG-3'. The amplified PCR fragment was then cloned into the BstEII and HindIII sites of pcDNA*MCON6gp120 to form pcDNA*MCON6gp120-OD. To express gp120-OD×3, three copies of OD were covalently linked by a multistep process. First, an AgeI site was introduced in pcDNA*MCON6gp120-OD at the C-terminal end by site directed mutagenesis. The OD insert was prepared by BstEII digestion, followed by Klenow fill-in, and then HindIII digestion. The pcDNA*MCON6gp120-OD vector backbone was prepared in a similar manner, with an initial AgeI digestion, followed by blunting and HindIII digestion. The resulting fragments were ligated to form pcDNA*MCON6gp120-ODx2. Subsequently, the OD×2 fragment was removed with partial BstEII digestion, end blunting and then HindIII digestion. The ODx2 fragment was ligated into the pcDNA*MCON6gp120-OD vector, as previously described, to construct the final pcDNA*MCON6gp120-OD×3. Ultimately, the OD×3 contained three copies of OD covalently joined by a four amino acid linker (STQL…GGGD-RVTG-STQL…GGGD-RVTG-STQL…GGGD) encoded by 5'-CGGGTGACCGGATCC-3' (BstEII and BamHI sites are underlined; codons encoding RVTG are indicated by boldface). All three proteins were fused to the original MCON6gp160 signal sequence, and all included a C-terminal 6xHis tag to facilitate protein purification.
Expression and purification of gp120, gp120-OD and -OD×3
All three constructs were stably transfected into HEK 293 cells. Cell culture supernatants containing the recombinant proteins were collected for purification using tandem affinity chromatography as previously described (Wang et al., 2010) with few modifications. First, recombinant proteins were enriched using Con A Sepharose column (GE Healthcare Life Science). The proteins were eluted from the column using elution buffer (20 mM Tris-HCl, pH 7.4, 0.5 M NaCl, and 0.5 M methyl α-dmannopyranoside). The recombinant proteins were further purified using a Ni-NTA column (Qiagen). After binding, column was washed with wash buffer (20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, and 5 mM imidazole). Subsequently, the proteins were eluted using the same buffer containing 250 mM imidazole. Remaining protein contaminants were removed through a Q-Sepharose column (GE Healthcare Life Science). The purified proteins, which were in the flow-through fraction, were concentrated with an Amicon Ultra concentrator (Millipore) and stored at −80°C. The protein purity was evaluated by SDSPAGE and silver staining.
Deglycosylation analyses of immunogens
2 µg of purified gp120, gp120-OD or -OD×3 were deglycosylated using either EndoH or PNGaseF (New England Biolabs) for partial or complete deglycosylation, respectively. Proteins were first denatured in a denaturation buffer (5% SDS and 0.4M DTT), then incubated with 500 U of PNGaseF or EndoH for 3 hrs at 37°C. Proteins were subjected to SDS-PAGE using a Novex 4–20% Tris-glycine Mini gel (Invitrogen), followed by Western blot analysis with an anti-gp120 MCON6-specific mouse monoclonal antibody (S7.5, hybridoma generated from gp120 immunized mouse, recognizing gp120 outer domain, from unpublished work). Alexa Fluor® 680 R-Phycoerythrin conjugated goat anti-mouse IgG (H+L) was used as a secondary antibody. The blot was scanned with an Odyssey infrared imager (Li-Cor)
Immunoprecipitation (IP) analyses
Purified gp120, gp120-OD or -OD×3 were immunoprecipitated with mAbs 17b (Kwong et al., 1998; Sullivan et al., 1998; Thali et al., 1993; Trkola et al., 1996a; Wyatt et al., 1998; 1995), 2G12 (Buchacher et al., 1994; Crawford et al., 1999; Etemad-Moghadam et al., 1999; Mascola et al., 1999; Trkola et al., 1996b), b12 (Barbas et al., 1992; Burton et al., 1991; 1994; Roben et al., 1994), 447-52D (Conley et al., 1994; Gorny et al., 1992; 1997; 1993; Nyambi et al., 1998; Zolla-Pazner et al., 1995) NIH45-46 (Diskin et al., 2011), VRC01 (X. Wu et al., 2010), PG9 and PG16 (Walker et al., 2009), all of which were obtained through the NIH AIDS Research and Reference Reagent Program (RRRP). MAb 3BNC60 heavy chain and light chain variable regions were synthesized by Genscript according to the published sequence (Scheid et al., 2011). The synthesized heavy chain and light chain variable regions were cloned into Ab expression vectors pFUSE-CHIg-hGI and pFUSE-CLIg-hK (Invivogen), respectively. The 3BNC60 Ab was purified from the supernatant of 293F cells co-transfected with the expression vectors using a protein A column (Pierce). IP was done by incubating the proteins, mAbs and protein A-agarose in IP buffer (10mM Tris, pH 7.5, 200mM NaCl, 1mM EDTA, 0.5% Triton X-100). After end-to-end rotation at 4°C overnight, the bound antigen-antibody complex was washed three times with IP buffer. Proteins were denatured and eluted from the resin by boiling for 5 min in 2×SDS-PAGE sample buffer. Immunoprecipitated proteins were subjected to SDS-PAGE using a Novex 4–20% Tris-glycine Mini gel (Invitrogen), followed by Western blot analysis using mouse anti-gp120 outer domain mAb (S7.5, unpublished) and goat anti-mouse IgG-HRP (Pierce). Protein bands were detected by chemiluminescence on X-ray films.
Surface plasmon resonance (SPR) analyses with IgG1 b12
IgG1 b12 was diluted in 10mM glycine-HCl buffer (pH 3.0) to a final concentration of 10 µg/ml and then coated onto the CM5 sensor chip (Biacore) via carboxyl moieties on the dextran using the standard primary amine coupling method until the resonance signal reached 1300 resonance units (RUs). A reference surface was prepared by activating and blocking a flow cell in the absence of b12. SPR analyses were performed with the Biacore 3000 (Biacore) at room temperature in HBS-P running buffer (10mM HEPES, pH 7.4, 150mM NaCl, and 0.005% (v/v) of the surfactant P20). To determine kinetic parameters for binding of immunogens (gp120, gp120-OD and -OD×3) to immobilized b12, sensorgrams were obtained by passing various concentrations of analytes (5–200 nM for gp120 and gp120-OD×3; 10–500 nM for gp120-OD) over the sensor surface at a flow rate of 10 µL/min using a 2 min association phase and a 2 min dissociation phase. The specific binding profiles of analytes to immobilized b12 were obtained after subtraction of the response signal from the reference flow cell. Protein binding kinetics were evaluated using BiaEvaluation (Biacore) based on 1:1 Langmuir binding model.
Preparation of zinc (Zn)-chitosan and immunization
Zn-chitosan was prepared as previously described (Seferian and Martinez, 2000) with a few minor modifications. Briefly, 2 g of chitosan (Sigma, C3646) was dissolved in 100 ml of 2% acetic acid followed by autoclave sterilization. The 2% (w/v) chitosan solution was diluted 1:1 using deionized water, and 4 mL of the resulting 1% chitosan solution was added to 10 ml of 0.2 M zinc acetate. The suspension was then mixed on an end-to-end rocker for 4 hrs at room temperature (RT). The mixture was then sonicated using a Branson Digital Sonifier for 5 min (70% Amplitude), and the pH was adjusted to 12.0–12.5 with 10 N NaOH. The resulting precipitate was centrifuged at 1000×g for 10 min. The pellet was washed twice with PBS (pH 7.4) by resuspension followed by centrifugation at 1000×g for 10 min, and finally resuspended in PBS (pH 8). Prior to immunization, antigens were loaded onto Zn-chitosan by mixing the two (1:1000 w/w) for 3 hrs. Female New Zealand white rabbits (2.5-3 kg; Charles River) were immunized subcutaneously with 200 µg of protein with 200 mg of Zn-chitosan in 1 ml (injected into 6–8 sites) on weeks 0, 3, 9, 15 and 27. Three animals were immunized with each immunogen and two animals were mock-immunized with Zn-chitosan in PBS. Blood was collected from the central ear artery prior to (pre-immune) and two weeks after each immunization and serum samples were prepared. All of the studies conducted were approved by IACUC.
Enzyme-linked immunosorbent assay (ELISA)
Antigen-specific antibody levels were monitored by ELISA. Purified immunogens were coated onto 96-well Nunc-Immuno Plates overnight at 4oC at 30 ng per well using an antigen coating buffer (15 mM Na2CO3, 35 mM NaHCO3, 3 mM NaN3, pH 9.6). Uncoated surface of the wells were blocked with 200 µl of a blocking buffer (PBS, pH 7.4, containing 2.5% skim milk and 25% Fetal Bovine Serum) for 1 hr at 37°C Wells were subsequently washed five times with a wash buffer (PBS containing 0.1% Tween 20) using a Biotek automated plate washer. Rabbit sera were diluted in the blocking buffer (at indicated dilutions or 3-fold serial dilutions for end-point titration), and 100 µl was added to each well and incubated for 2 hrs at 37°C. Wells were washed five times, and further incubated for 1 hr at 37°C with a secondary antibody (horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L), Pierce; 1:3000 dilution in the blocking buffer). Wells were washed again five times and HRP reaction was initiated by adding 100 µl TMB HRP-substrate (Bio-Rad). The reaction was stopped after 10 min by adding 50 µl of 2 N H2SO4. Plates were read on a microplate reader (Versamax by Molecular Devices) at 450 nm. All assays were done in duplicate. The endpoint ELISA titers were defined as serum dilution factor that gave readings of average ±2× SD of the background (i.e. sera from animals mock immunized with PBS).
The 15-mer overlapping peptide set for gp120 based on the M group consensus sequence (CON-S) was obtained from the NIH AIDS RRRP (Cat# 9487). V3, V4 and V5 peptides based on the MCON6 sequence (H-TRPNNNTRKSIHIGPGQAFYATGEIIGDIRQAH-OH, H-MFNGTYMFNGTKDNSE-OH and H-GNNSNKNKTETFRPG-OH, respectively) were synthesized commercially by CHI Scientific (Maynard, MA). Peptides were coated onto wells at 20 pmol per well.
Purification of rabbit IgG
Rabbit IgG was purified from sera with Protein A column (Pierce). Briefly, 1 ml of rabbit serum was diluted 1:1 with Protein A Binding buffer (Pierce) and then applied to a protein A column. After washing, IgG was eluted with elution buffer (0.1M glycine pH 3.0), then was immediately adjusted to physiologic pH by adding 100 µl of neutralization buffer (1M Tris, pH 8.5) per 1 ml of eluate. The eluted antibody was pooled and dialyzed in PBS. The purity of purified IgG was confirmed by SDS-PAGE. Antibodies were kept at −80°C.
Neutralization assay
Virus neutralization assays were done using the single cycle HIV-1 pseudovirus infections of TZM-bl cells as previously described (Li et al., 2005; Wei et al., 2002). Briefly, heat inactivated rabbit sera (56°C for 1 hr) or purified IgG were diluted in 100 µl of Dulbecco’s modified Eagle’s medium supplemented with 10% heat inactivated FBS (DMEM/FBS). Serum/antibody was mixed with 200 TCID50 (50% tissue culture infectious dose) of viruses in 50 µl and incubated for 1 hr at 37°C. Into the mixture, 1×104 TZM-bl cells in DMEM/FBS containing DEAE-dextran (Sigma; 10 µg/ml final concentration) were added. Plates were incubated at 37°C for 48 hrs. Cells were harvested and processed using Bright-Glo Luciferase Assay kit (Promega) as per manufacturer’s recommended protocol, and luciferase activity was measured with Synerge 2 luminometer (Biotek). The 50% inhibitory dose (ID50) or concentration (IC50) was defined as either the inverse of serum dilution or IgG concentration at which relative luminescence units (RLU) were reduced by 50% compared to virus control wells after subtraction of background RLU in cell-only control wells. For the protein/peptide inhibition neutralizing assays, the same procedure was followed except that antibodies were preincubated with gp120-OD (50µg/ml) or peptides (in varying concentrations) for 1 hr at 37°C. Neutralization assays against a larger panel of pseudoviruses (in TZM-bl and A3R5 cells) were assayed in Montefiori lab as previously described (Li et al., 2005; McLinden et al., 2013; Montefiori, 2005)
Statistical analyses
Statistical analyses were performed using SAS 9.3 and GraphPad Prism 5. ELISA values were log-transformed and analyzed using linear mixed models. Peptide, group, and their interaction were used as fixed effects, whereas animal and animal-peptide interactions were random effects. Differences among groups were assessed by F-tests followed by post-hoc Tukey’s pairwise t-tests for each group and region. P-values ≤0.05 were considered significant.
Two novel immunogens based on HIV-1 gp120 outer domain were generated.
Critical neutralizing epitopes were preserved in both immunogens.
Potent cross-clade neutralizing antibodies were induced in rabbits.
These prototypic immunogens are good candidates for further improvements.
Acknowledgements
We are grateful to Drs. Beatrice Hahn for providing pcDNA-MCON6gp160. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 gp120 MAb (VRC01) from Dr. John Mascola, IgG1 b12 from Dr. Dennis Burton and Carlos Barbas, 2G12 from Dr. Hermann Katinger, 17b from Dr. James E. Robinson, 447-52D Dr. Susan Zolla-Pazner, PG9 and PG16 from Dr. Dennis Burton, NIH45-46 G54W IgG from Dr. Pamela J. Bjorkman, HIV-1 Consensus Group M Env peptides, pCAGGS SF162 gp160 from Drs. L. Stamatatos and C. Cheng-Mayer, pSVIII-93MW965.26 from Dr. Beatrice Hahn. This work was supported by the NIH grants AI074286 and HHSN27201100016C. MWC has an equity interest in NeoVaxSyn Inc., and serves as CEO/President. NeoVaxSyn Inc. did not contribute to this work or the interpretation of the data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barbas CF, Björling E, Chiodi F, Dunlop N, Cababa D, Jones TM, Zebedee SL, Persson MA, Nara PL, Norrby E. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc Natl Acad Sci USA. 1992;89:9339–9343. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Rajan RE, Swarupa Y, Rathore U, Verma A, Udaykumar R, Varadarajan R. Design of a non-glycosylated outer domain-derived HIV-1 gp120 immunogen that binds to CD4 and induces neutralizing antibodies. J Biol Chem. 2010;285:27100–27110. doi: 10.1074/jbc.M110.152272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, la Peña de AT, Cupo A, Julien J-P, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. Structural Delineation of a Quaternary, Cleavage-Dependent Epitope at the gp41-gp120 Interface on Intact HIV-1 Env Trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Burton DR, Barbas CF, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Center RJ, Leapman RD, Lebowitz J, Arthur LO, Earl PL, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J Virol. 2002;76:7863–7867. doi: 10.1128/JVI.76.15.7863-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center RJ, Schuck P, Leapman RD, Arthur LO, Earl PL, Moss B, Lebowitz J. Oligomeric structure of virion-associated and soluble forms of the simian immunodeficiency virus envelope protein in the prefusion activated conformation. Proc Natl Acad Sci USA. 2001;98:14877–14882. doi: 10.1073/pnas.261573898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu X, Jones IM. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirology. 2007;4:33. doi: 10.1186/1742-4690-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu X, Lin H-H, Chen S-H, Forsman A, Aasa-Chapman M, Jones IM. Mapping the immune response to the outer domain of a human immunodeficiency virus-1 clade C gp120. J Gen Virol. 2008;89:2597–2604. doi: 10.1099/vir.0.2008/003491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MW, Teterina N, Egger D, Bienz K, Ehrenfeld E. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology. 1994;202:129–145. doi: 10.1006/viro.1994.1329. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Gorny MK, Kessler JA, Boots LJ, Ossorio-Castro M, Koenig S, Lineberger DW, Emini EA, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J Virol. 1994;68:6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Earl PL, Moss B, Reimann KA, Wyand MS, Manson KH, Bilska M, Zhou JT, Pauza CD, Parren PW, Burton DR, Sodroski JG, Letvin NL, Montefiori DC. Characterization of primary isolate-like variants of simian-human immunodeficiency virus. J Virol. 1999;73:10199–10207. doi: 10.1128/jvi.73.12.10199-10207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Klein F, Gao H, Gnanapragasam PNP, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Sun Y, Nicholson EK, Karlsson GB, Schenten D, Sodroski J. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J Virol. 1999;73:8873–8879. doi: 10.1128/jvi.73.10.8873-8879.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang C-H, McBride R, Bredow von B, Shivatare SS, Wu C-Y, Chan-Hui P-Y, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong C-H, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. Broadly Neutralizing HIV Antibodies Define a Glycan-Dependent Epitope on the Prefusion Conformation of gp41 on Cleaved Envelope Trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Korber BT, Weaver E, Liao H-X, Hahn BH, Haynes BF. Centralized immunogens as a vaccine strategy to overcome HIV-1 diversity. Expert Rev Vaccines. 2004;3:S161–S168. doi: 10.1586/14760584.3.4.s161. [DOI] [PubMed] [Google Scholar]
- Gao F, Weaver EA, Lu Z, Li Y, Liao H-X, Ma B, Alam SM, Scearce RM, Sutherland LL, Yu J-S, Decker JM, Shaw GM, Montefiori DC, Korber BT, Hahn BH, Haynes BF. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol. 2005;79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Hioe C, Israel ZR, Michael NL, Conley AJ, Williams C, Kessler JA, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. The Journal of Immunology. 1997;159:5114–5122. [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006;5:579–595. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- Hoxie JA. Toward an antibody-based HIV-1 vaccine. Annu Rev Med. 2010;61:135–152. doi: 10.1146/annurev.med.60.042507.164323. [DOI] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. NATURE. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien J-P, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang P-S, Macpherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MG, Kanekiyo M, Xu L, Biertümpfel C, Boyington JC, Moquin S, Shi W, Wu X, Yang Y, Yang Z-Y, Zhang B, Zheng A, Zhou T, Zhu J, Mascola JR, Kwong PD, Nabel GJ. Outer domain of HIV-1 gp120: antigenic optimization, structural malleability, and crystal structure with antibody VRC-PG04. J Virol. 2013;87:2294–2306. doi: 10.1128/JVI.02717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Hedestam GB, Fouchier RAM, Phogat S, Burton DR, Sodroski J, Wyatt RT. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat Rev Micro. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- Khayat R, Lee JH, Julien J-P, Cupo A, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. Structural characterization of cleaved, soluble HIV-1 envelope glycoprotein trimers. J Virol. 2013;87:9865–9872. doi: 10.1128/JVI.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, Kraft Z, Liu Y, Pietzsch J, Hurley A, Poignard P, Feizi T, Morris L, Walker BD, Fätkenheuer G, Seaman MS, Stamatatos L, Nussenzweig MC. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med. 2012;209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Bjorkman PJ. Few and Far Between: How HIV May Be Evading Antibody Avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Gnanapragasam PNP, Galimidi RP, Foglesong CP, West AP, Bjorkman PJ. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. PNAS. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR, Nabel GJ. Rational Design of Vaccines to Elicit Broadly Neutralizing Antibodies to HIV-1. Cold Spring Harb Perspect Med. 2011;1:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. NATURE. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche CC, Galasso G, Moore JP, Bolognesi DP, Hirsch MS, Hammer SM. HIV fusion and its inhibition. Antiviral Res. 2001;50:95–115. doi: 10.1016/s0166-3542(01)00130-9. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal I, Jarvis DL, Cacan R, Verbert A. Glycoproteins from insect cells: sialylated or not? Biol. Chem. 2001;382:151–159. doi: 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewicz J, Polewska A, Knapczyk J. Immunoadjuvant properties of chitosan. Arch. Immunol. Ther. Exp. (Warsz.) 1991;39:127–132. [PubMed] [Google Scholar]
- Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, Louder MK, Brown CR, Sapan CV, Frankel SS, Lu Y, Robb ML, Katinger H, Birx DL. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- McLinden RJ, Labranche CC, Chenine A-L, Polonis VR, Eller MA, Wieczorek L, Ochsenbauer C, Kappes JC, Perfetto S, Montefiori DC, Michael NL, Kim JH. Detection of HIV-1 neutralizing antibodies in a human CD4 /CXCR4 /CCR5 T-lymphoblastoid cell assay system. PLoS ONE. 2013;8:e77756. doi: 10.1371/journal.pone.0077756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter. 2005;12 doi: 10.1002/0471142735.im1211s64. pUnit 12.11. [DOI] [PubMed] [Google Scholar]
- Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P, Katinger H. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel GJ, Kwong PD, Mascola JR. Progress in the rational design of an AIDS vaccine. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2011;366:2759–2765. doi: 10.1098/rstb.2011.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle DC. Consensus and Ancestral State HIV Vaccines. Science. 2003;299:1515c–1518c. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J Virol. 1998;72:9384–9391. doi: 10.1128/jvi.72.11.9384-9391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Burton DR. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, Tauer C, Berger R, Barrett N, Jungbauer A. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1651–1658. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AL, Bråve A, Scarlatti G, Manrique A, Buonaguro L. Progress towards development of an HIV vaccine: report of the AIDS Vaccine 2009 Conference; Presented at the The Lancet infectious diseases; 2010. pp. 305–316. [DOI] [PubMed] [Google Scholar]
- Scharf L, Scheid JF, Lee JH, West AP, Chen C, Gao H, Gnanapragasam PNP, Mares R, Seaman MS, Ward AB, Nussenzweig MC, Bjorkman PJ. Antibody 8ANC195 Reveals a Site of Broad Vulnerability on the HIV-1 Envelope Spike. Cell Rep. 2014;7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferian PG, Martinez ML. Immune stimulating activity of two new chitosan containing adjuvant formulations. Vaccine. 2000;19:661–668. doi: 10.1016/s0264-410x(00)00248-6. [DOI] [PubMed] [Google Scholar]
- Singla AK, Chawla M. Chitosan: some pharmaceutical and biological aspects--an update. J. Pharm. Pharmacol. 2001;53:1047–1067. doi: 10.1211/0022357011776441. [DOI] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. NATURE. 1996a;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996b;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaine M, Wang S, Crooks ET, Jiang P, Montefiori DC, Binley J, Lu S. Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. J Virol. 2008;82:7369–7378. doi: 10.1128/JVI.00562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lubben IM, Verhoef JC, Borchard G, Junginger HE. Chitosan and its derivatives in mucosal drug and vaccine delivery. Eur J Pharm Sci. 2001;14:201–207. doi: 10.1016/s0928-0987(01)00172-5. [DOI] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, Protocol G, Principal Investigators Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. NATURE. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G, Principal Investigators s, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Qin Y, Ilchenko S, Bohon J, Shi W, Cho MW, Takamoto K, Chance MR. Structural Analysis of a Highly Glycosylated and Unliganded gp120-Based Antigen Using Mass Spectrometry. Biochemistry. 2010;49:9032–9045. doi: 10.1021/bi1011332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou T, Yang Z-Y, Svehla K, O'Dell S, Louder MK, Xu L, Mascola JR, Burton DR, Hoxie JA, Doms RW, Kwong PD, Nabel GJ. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol. 2009;83:5077–5086. doi: 10.1128/JVI.02600-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. NATURE. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol. 2004;78:12975–12986. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang S-H, Yang X, Zhang M-Y, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. NATURE. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Chertova E, Bess J, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, O'Leary J, Burda S, Gorny MK, Kim M, Mascola J, McCutchan F. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J Virol. 1995;69:3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]