Abstract
Optical tweezers use the momentum of photons to trap and manipulate microscopic objects contact-free in three dimensions. Although this technique has been widely used in biology and nanotechnology to study molecular motors, biopolymers and nanostructures, its application in viruses has been very limited largely due to the small size of these nanoparticles. Using optical tweezers that can simultaneously resolve two-photon fluorescence at single-molecule level, here we show that individual HIV-1 can be optically trapped and manipulated, which allows multi-parameter analysis of single virions in culture fluid under native conditions. We show that individual HIV-1 differs in the numbers of envelope glycoproteins by more than one order of magnitude, which implies substantial heterogeneity of these virions in transmission and infection at single-particle level. Analogous to flow cytometry for cells, this fluid-based technique may allow ultrasensitive detection, multi-parameter analysis and sorting of viruses and other nanoparticles in biological fluid with single-molecule resolution.
Optical tweezers (OTs) use the momentum of photons to trap and manipulate microscopic objects contact-free in three dimensions (3D)1–5. Although this technique has been widely used in biology and nanotechnology to study molecular motors, biopolymers3,6–15, and for trapping of metal nanoparticles16–18 and various nanostructures4,19–22, there have been no demonstrations or studies on optical manipulation of a single animal virus23. Several challenges exist besides the issue of biosafety. First, the presence of a permanent or an induced dipole moment in the object is required for optical trapping3. Ashkin and Dziedzic first demonstrated optical trapping of tobacco mosaic virus (TMV) in 198723. TMV is 300 nm long and 15 nm in diameter24. This long cylindrical structure displays a permanent dipole moment25 which may facilitate its trapping by OTs. While majority of animal viruses are spherical in shape26,27, their dipole moments may be intrinsically small as compared to other aspheric objects of similar volume23. Second, animal viruses are typically less than 300 nm in size26,27. The gradient force responsible for trapping of a Rayleigh scatterer scales with the cube of the particle dimension3. The lower viscous drag from the solvent on smaller particles also allows them to easily escape once being trapped. Hence the smaller the particle, the more difficult to trap and manipulate it using light. Third, the refractive indices of animal viruses have never been measured. Although the trapping force increases in general with the refractive index of the object28, how a single animal virus may interact with light rays and generate trapping forces on itself is unknown. All these aspects present challenges to manipulation of an animal virus using light. Can a single animal virus be trapped and manipulated using light? By using an ultrahigh resolution OTs instrument that we constructed recently29, here we show that individual HIV-1 can be trapped and manipulated in culture fluid ‘contact-free’ in 3D under native conditions. We developed this capability into a quantitative technique, ‘virometry’. This technique permits multi-parameter analysis of individual HIV-1 virions in culture fluid under native conditions with single-molecule resolution, and can be applied to other viruses and nanoparticles in solution for their detection and measurement of size, shape and protein compositions with unprecedented sensitivity.
Optical Trapping of Single HIV-1 in Culture Fluid
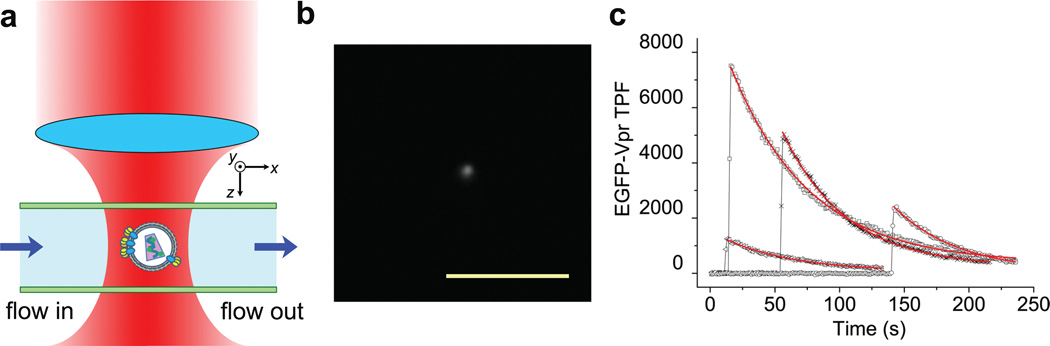
To demonstrate the optical trapping of animal viruses in biological fluid, we have recently prepared the HIV-1 virions derived from the X4-tropic NL4-3 provirus clone30. These viruses were tagged internally with EGFP fused to Vpr31,32, an accessory protein of HIV that physically associates with the virion core33 and thus serves as a fluorescent marker31 for virion manipulation. The infectivity of this virus was 0.1% in TZM-bl cells30, a value that was comparable to wild-type viruses. Control experiments showed that EGFP-Vpr also distinguished virions from microvesicles (Supplementary Note 1). Without any fixation procedure, we diluted the live virus stock in the complete media and injected them into a microfluidic chamber (Fig. 1a). Upon trapping of a virion by the 830 nm infrared laser29 focused at the center of the chamber, the virion was immediately ‘visible’ in the dark background due to simultaneous two-photon excitation of EGFP by the trapping laser29,34 (Fig. 1b). Such events were not observed when complete media only, viruses without EGFP-Vpr labels, or culture supernatants transfected with EGFP-Vpr were used as controls, suggesting that the fluorescent particles resulted from virions tagged with EGFP. Under these conditions, a single fluorescent particle could be trapped in culture fluid and manipulated in various patterns by steering the laser beam without interference from other virions (Supplementary Video). Typical two-photon fluorescence (TPF) time courses from individually-trapped virions are shown in Fig. 1c, where the initial bursts of fluorescence well above background indicated the arrival of HIV-1 virions at the optical trap. The subsequent decay of TPF with time could all be fit by single exponentials with an average time constant of 53.7±8.0 s (N=19), very close to the rate of photobleaching that we measured previously for EGFP either on a surface (50.8±4.2 s)34 or inside a cell (52.7±5.2 s)35. This comparison confirmed the presence of EGFP in these particles.
Figure 1.
Optical trapping of HIV virions in culture fluid. (a) HIV-1 virions were delivered into a microfluidic chamber and trapped by the IR laser focused at the center of the chamber. The xyz dimensions are shown as indicated, with y perpendicular to the figure plane. (b) TPF image of a trapped HIV virion. The scale bar is 10 µm. (c) Representative TPF time courses from individually-trapped HIV virions. All traces were fit with single exponential decay (red), with time constants for each trace as follows: 60.7 (square), 49.5 (cross), 51.5 (circle) and 54.1 s (triangle). Time zero started with the onset of TPF collection.
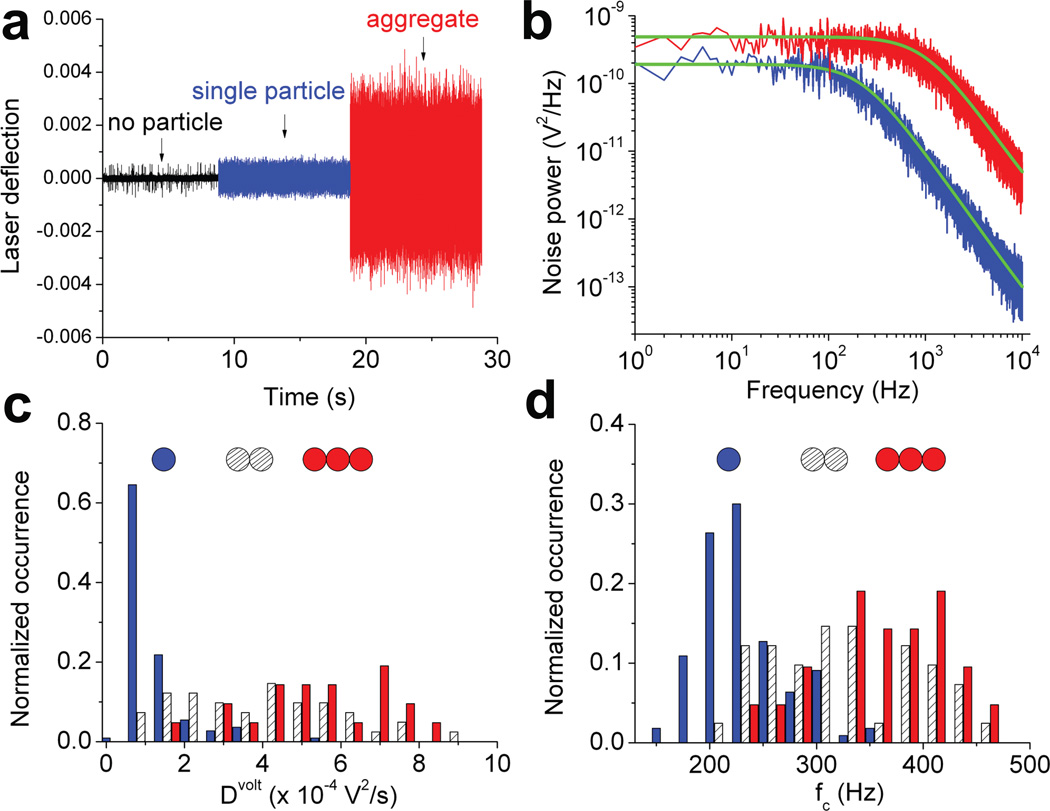
Freely-diffusing particles in culture fluid may undergo concentration-dependent aggregation. How do we know that the trapped particle correspond to a single HIV-1 virion? Independent of EGFP fluorescence, we could also detect the trapping of the virus from changes in laser deflection at the objective’s back focal plane (BFP) (Fig. 2a, blue). The occasional trapping of an apparent viral aggregate produced quantitatively different laser deflection signals29,36 (Fig. 2a, red), suggesting that one could potentially use this signal to distinguish viral particles of different size. We analyzed the time courses of the laser deflection by conversion to power spectra in the frequency domain (Fig. 2b). For both the single particle (blue) and virion aggregate (red), the power spectra could be well described with Lorentzian up to 10 kHz (green curves, Supplementary Methods), consistent with the Brownian motion of a particle in a harmonic photonic field3. Furthermore, this fitting yielded very different parameters for the single particle and virion aggregate: Dvolt, the diffusion coefficient (in the unit of V2/s) and fc, the corner frequency (in Hz). To test the sensitivity of these parameters in differentiating particles of different size, we delivered diluted virions through a microfluidic channel into the complete media followed by trapping of the virion in the vicinity of the channel opening (Supplementary Fig. 1). This scheme allowed us to trap one, two and three particles sequentially using a single laser beam (Supplementary Fig. 1), and measure Dvolt and fc for the apparent single, double and triple particles, respectively. The distributions of these values are plotted in Fig. 2c and d. As shown, both Dvolt and fc conform to statistically different distributions for single versus double or triple particles (Supplementary Table 1), suggesting that the laser deflection is sensitive enough to distinguish a single particle from particle aggregates.
Figure 2.
BFP interferometry to distinguish single HIV-1 particle from aggregates in complete media. (a) The laser deflection signal measured in real time using BFP interferometry. (b) Power spectra calculated from the data shown in (a) and fit to aliased Lorentzian with Dvolt=8.53×10−5 V2/s, fc= 223 Hz (blue) and Dvolt=0.0046 V2/s, fc= 982 Hz (red). Histograms for Dvolt (c) and fc (d) derived from Lorentzian fitting parameters for apparent one (blue, N=110), two (hashed, N=41) and three (red, N=21) particles trapped, with cartoons representing one, two and three particles above each histogram.
Measurement of Single HIV-1 Virion Diameter
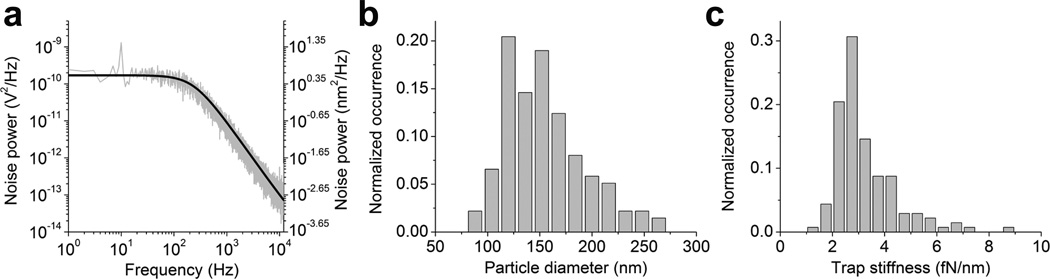
Single HIV-1 particles are heterogeneous in diameter37. To help distinguish individual viral particles of different diameters, we set out to use the sensitive laser deflection signal to directly measure the diameter for each trapped virion in the culture fluid (Methods). This method works by calibration of the laser deflection signal using a distance standard38 and the Dvolt can be converted to the diffusion coefficient of the particle in space (D, in the unit of µm2/s)38. The diameter of the particle, ϕBFP, can then be calculated using the Stokes-Einstein equation (Supplementary Methods). We first tested this method by trapping polystyrene beads of various sizes in water. These results were compared with the diameters of the beads determined from transmission electron microscopy (TEM) images (ϕTEM, Supplementary Fig. 2). Over the range of bead sizes we tested, these two methods yielded good agreement, with ϕBFP slightly larger than ϕTEM by 5.5% on average, which reflected either the error in laser deflection measurement or hydration of particles in water. Fig. 3a shows the power spectrum of a trapped virion obtained from this calibration procedure in complete media, with voltage converted to distance as shown on the right axis, which yields a diameter of 165 nm for this particular virion. Fig. 3b shows the histogram of ϕBFP determined for 137 single HIV-1 particles in complete media at a concentration of 4.0 × 107 virion/ml, which displayed a population with a mean of 154 nm, and a median of 148 nm. The reduced chi-squared statistic from this analysis varied from 1.10 to 1.16 (Supplementary Fig. 3), indicating a good quality of power spectrum fitting. This virion size distribution was quantitatively similar to what was reported previously for authentic, mature HIV-1 virions by cryoelectron microscopy (cEM)37. Our mean diameter was 6.2% larger than 145 ± 25 nm for single HIV virions measured by cEM. This positive deviation was consistent with the trend seen for reference beads, suggesting that these particles indeed corresponded to single virions in complete media. This positive deviation can result from either hydration of the particles in culture media or potential shrinkage of the feature size under EM (Supplementary Note 2). From calibrated power spectra for single HIV-1 virions, we also obtained stiffness of the optical trap (Supplementary Methods), which is shown in Fig. 3c with a mean value of 3.2 fN/nm. This stiffness is about 30-fold lower than typically reported values for bigger particles3 and explains the difficulty in trapping virions of this size. The variation in trap stiffness as compared to variations in particle diameter also suggests that HIV-1 virions deviate from true Rayleigh scatterers. To examine how well the measured particle diameter and trap stiffness can differentiate particles of different size, we conducted sequential trapping of one, two and three particles as illustrated in Supplementary Fig. 1, and measured ϕBFP and trap stiffness for the apparent single, double and triple particles, respectively. The distributions of these values are plotted in Supplementary Fig. 4a and b. As shown, both ϕBFP and trap stiffness conform to statistically different distributions for single versus double or triple particles (Supplementary Table 2), suggesting that these parameters are sensitive enough to distinguish a single particle from particle aggregates.
Figure 3.
Measurement of diameter for single HIV-1. (a) The power spectrum of a trapped virion when the chamber was oscillated at 10 Hz with amplitude of 176 nm along y-axis of the sample plane (Methods). The black curve is fitting of the thermal noise background to aliased Lorentzian with Dvolt=9.37×10−5 V2/s and fc= 233 Hz. (b) Size histogram for single fluorescent particles from chamber oscillation along y-axis (N=137). Concentration of the HIV-1 inside the chamber is 4.0 × 107 virions/ml. (c) Distribution of the optical trap stiffness for each virion trapped (N=137).
Concentration-dependent Aggregation of HIV-1 Virions in the Complete Media
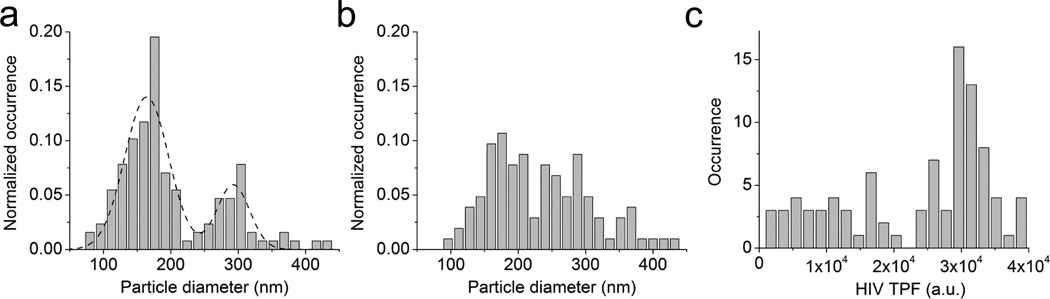
Freely-diffusing particles in the complete media may undergo concentration-dependent aggregation. To examine the aggregation status of HIV-1 virions under native conditions in the complete media, we conducted diameter measurement for single particles trapped at various concentrations in the microfluidic chamber. Fig. 4a shows the histogram of ϕBFP determined for 128 apparent single particles at 1.2 × 108 virions/ml in the complete media (grey bars), which displays two distinct populations as indicated by the double-Gaussian fit (dashed lines). The primary peak, with a mean of 156 nm and a standard deviation of 30 nm, accounts for 72% of the particles. This number is very close to the average diameter of 154 nm for single HIV virions measured at 4.0 × 107 virions/ml, suggesting that these particles indeed correspond to single virions. The secondary peak in this histogram, with a mean of 305 nm and a standard deviation of 44 nm, is about twofold larger than the size of a typical HIV-1 virion. These particles were rarely observed when trapping at a concentration of 4.0 × 107 virions/ml, suggesting that they resulted from a concentration-dependent aggregation. This conclusion is further supported by the same experiment done at even higher concentration of virion particles. Fig. 4b shows the histogram of ϕBFP determined for 103 apparent single particles at 4.0 × 108 virions/ml in the complete media. No clear peaks were present. Particle diameters ranged from 90 to 440 nm, with an average diameter of 232 ± 78 nm (median 219 nm). This feature is reproducible from an independent repeat of the same experiment, suggesting a concentration-dependent aggregation of HIV-1 particles. Fig. 4c shows the distribution of EGFP TPF intensity from single HIV-1 virions upon their initial trapping. This distribution was broad and asymmetric, ranging from 1,000 to 40,000 a.u., with a major peak around 30,000. This variation of TPF intensity exclusively resulted from EGFP-Vpr molecules packaged into virions because control virions without EGFP showed no detectable TPF under these conditions. This substantial variation of the initial TPF amplitudes suggests that one cannot simply use TPF intensity to confirm a single virus. Rather, size or trap stiffness measurement is required. Moreover, virion diameter is a better parameter than Dvolt in resolving subpopulations within a virus pool (Supplementary Fig. 5).
Figure 4.
Aggregation of HIV-1 observed at high virion concentrations. (a) Size histogram for single fluorescent particles from chamber oscillation along y-axis and its double-Gaussian fit (dashed line) (N=128). Concentration of the HIV-1 inside the chamber is 1.2 × 108 virions/ml. (b) Size histogram for single fluorescent particles from chamber oscillation along y-axis (N=103). Concentration of the HIV-1 inside the chamber is 4.0 × 108 virions/ml. (c) Distribution of EGFP-Vpr TPF intensity from trapped virions that are confirmed to be single based on diameter measurement in (a) (N=92).
Optical Trapping Virometry
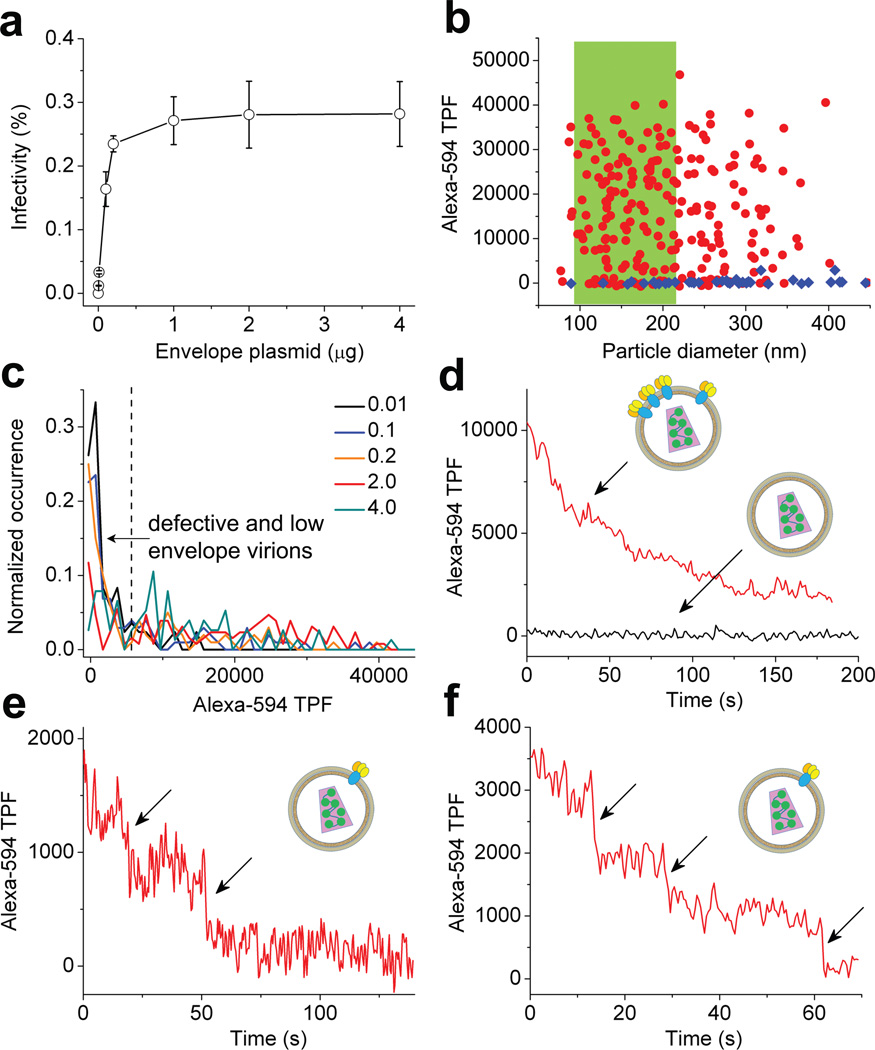
We have recently developed single-molecule TPF imaging capability using our optical trap setup34,35. This imaging uses the optical trapping laser for direct and simultaneous TPF excitation of fluorophores such as fluorescent proteins at the laser focus whose fluorescence is detectable at single-molecule level. Because the optical paths for BFP interferometry operate independently from single-molecule TPF detection, we can thus trap a single HIV-1 in culture fluid and simultaneously monitor viral proteins via TPF by using appropriate fluorophore labels. Because our TPF detection has single-molecule sensitivity, this technique should offer many possibilities for quantitation and potential sorting of virions in culture fluid with exquisite sensitivity. To test this idea, we produced series of HIV-1 virions that were tagged with EGFP-Vpr but might carry different number of envelope glycoproteins as we varied the envelope plasmid input (pEnv, 0–4 µg) during virion production30. The infectivity of these virions increased with increasing pEnv (Supplementary Fig. 6) and reached a plateau at 1 µg pEnv in the presence of 20 µg/ml DEAE-dextran (Fig. 5a). To monitor the envelope content of individual virions, we prepared fluorescent-labeled monoclonal antibody b1239 (Alexa594-b12) that specifically recognizes HIV-1 envelope glycoproteins gp12040,41. We chose Alexa-594 for labeling, which can be excited by the 830 nm trapping laser efficiently and yet has a minimal spectral overlap with EGFP emission (Supplementary Fig. 7). This strategy allowed us to measure EGFP-Vpr and gp120 for the same single virion almost simultaneously, both with single-molecule sensitivity34 (Supplementary Fig. 8). Microvesicles that carried gp120 or antibody aggregates were also excluded from HIV-1 virions by using this two-color strategy. We incubated the EGFP-tagged viruses with Alexa594-b12 at an antibody concentration (15 µg/ml) that was sufficient to neutralize >95% HIV infectivity (Supplementary Fig. 9) to ensure the saturation binding of all functional gp120. Control experiments showed that gp120 shedding was negligible at this concentration of b12 (Supplementary Fig. 10). We thus expect the level of Alexa594-b12 fluorescence associated with individual virions to correlate with gp120 content on virion surface. We measured HIV virions prepared with 0, 0.01, 0.1, 0.2, 2 and 4 µg pEnv by trapping them in complete media. A typical result is shown in Fig. 5b using 2 µg pEnv as an example, where the intensity of Alexa-594 TPF for each particle was plotted as a function of particle diameter in red circles (N=231). For 2 µg pEnv, 55% of the particles had diameters that were consistent with single virions and were highlighted in green. The intensity of Alexa-594 TPF associated with these particles displayed a broad distribution, varying from zero to 40,000 a.u.. Control experiments using virions without envelope glycoproteins (pEnv=0) showed no Alexa-594 fluorescence in >95% cases (Fig. 5b blue diamonds), confirming that the different levels of Alexa-594 TPF resulted from specific gp120 binding by Alexa594-b12. Fig. 5c shows a compendium of Alexa-594 TPF histograms from single virions prepared with varied pEnv. Throughout, all the distributions were broad even when the infectivity for the corresponding batch of HIV-1 was at plateau. The single-molecule sensitivity of current setup allowed us to roughly estimate the number of envelope trimers in single virions based on the average TPF intensity of a single Alexa-594 molecule, which yielded 0–18 envelope trimers per virion if assuming each Alexa594-b12 bound two gp120 molecules or 0–9 envelope trimers per virion if each Alexa594-b12 bound only one gp120 (Supplementary Note 3). Indeed, as shown in Fig. 5d, we could clearly distinguish particles that carried gp120 (red curve) from those that didn’t due to single-molecule sensitivity (black curve). 12% of the single particles from 2 µg pEnv did not show any Alexa-594 fluorescence (Fig. 5c), indicating that no gp120 was present on the virion surface. This fraction increased to 26% as we lowered pEnv to 0.01 µg (Fig. 5c). Because envelope glycoproteins are required for virion infectivity (Fig. 5a), these data suggest that defective HIV-1 virions can be produced during virion budding, the fraction of which increases when envelope glycoproteins are supplied under limiting conditions (Supplementary Table 3). Furthermore, there were particles that had gp120 but clearly showed only two or three steps of Alexa-594 photobleaching (Fig. 5e and f). The fraction of these virions was 5% for 2 µg pEnv and increased to 33% for virions prepared with 0.01 µg pEnv. These data indicate that at most a single envelope trimer is present on these virions. The number of envelope trimers per virion was estimated previously using various methods42,43. However, as low as a single trimer on virion surface has not been reliably reported. Our technique allows us to clearly detect the presence of a single trimer on HIV-1 virion surface in culture fluid without ambiguity.
Figure 5.
Optical trapping virometry. (a) Infectivity of HIV-1 virions as a function of pEnv in the presence of 20 µg/ml DEAE-dextran. The error bars are standard deviations from three independent replicates. (b) Alexa-594 TPF as a function of particle diameter. Red circles are for HIV-1 from 2 µg pEnv (N=231) while blue diamonds are for virions without envelope (N=42). (c) TPF intensity histograms for Alexa-594 from single HIV-1 particles, with black, blue, orange, red and olive curves for virions from 0.01 (N=84), 0.1 (N=102), 0.2 (N=100), 2.0 (N=128) and 4.0 µg pEnv (N=76), respectively. (d)–(f) representative Alexa-594 TPF time courses from single virions bound with Alexa594-b12, where individual photobleaching steps in (e) and (f) are indicated with arrows. Insets, cartoons of HIV-1 virions with varied numbers of envelope glycoproteins.
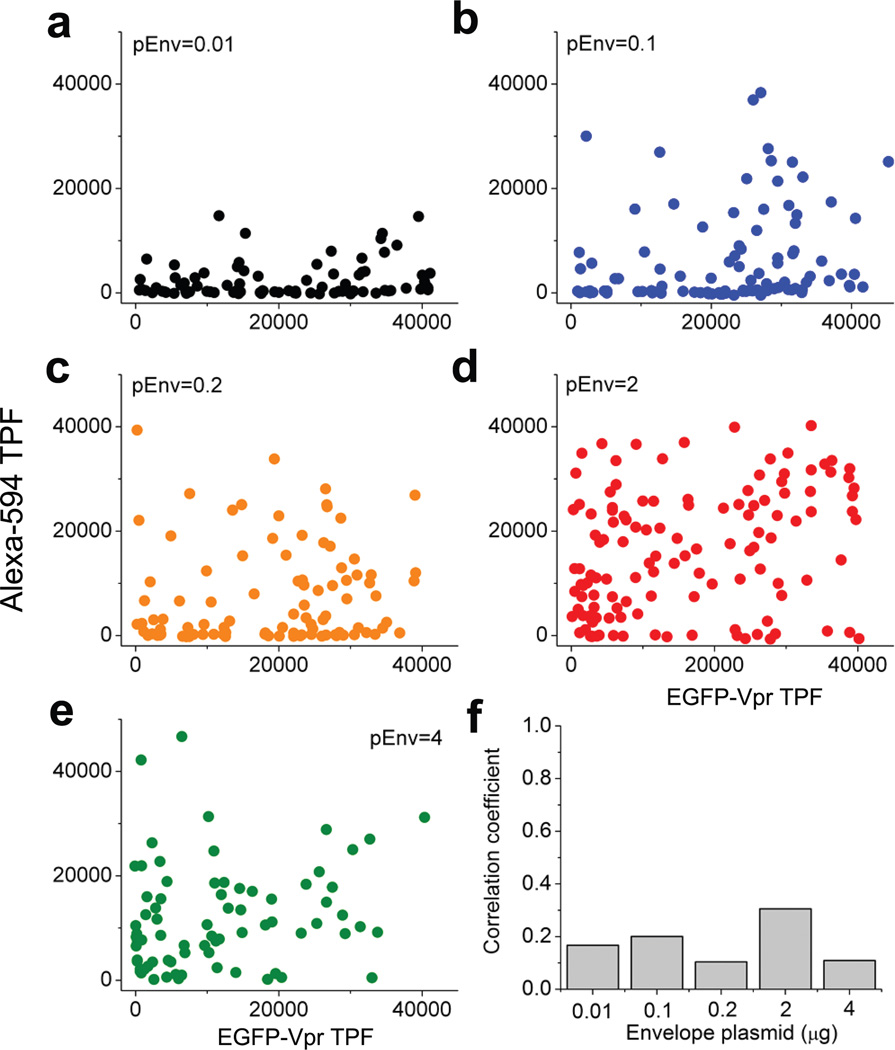
Lastly, the ability to use a single 830 nm laser to excite both EGFP and Alexa-594 simultaneously offers a unique advantage to our technique. Because EGFP and Alexa-594 have minimal emission spectrum overlap (Supplementary Fig. 7), we can resolve both fluorescence for a single HIV-1 particle and thus examine the potential correlation between the two kinds of proteins within a single particle. To this end, we plotted Alexa-594 TPF as a function of EGFP-Vpr TPF for each single HIV-1 virions in Fig. 6a–e for the five batches of viruses that we have prepared with different pEnv inputs. As shown, there was little correlation between Alexa-594 and EGFP-Vpr in their TPF intensities at single-particle level. This conclusion is true regardless of the input quantity of pEnv. The correlation coefficients between Alexa-594 TPF and EGFP-Vpr TPF are 0.1668, 0.2002, 0.1035, 0.3054, 0.1087 for 0.01, 0.1, 0.2, 2 and 4 µg pEnv (Fig. 6f), respectively, suggesting that Vpr and envelope glycoprotein incorporation into individual HIV-1 virions are two independent processes during HIV-1 virion budding.
Figure 6.
Two-color correlation analysis for EGFP-Vpr and envelope glycoproteins in single HIV-1 virions. (a)–(e) Alexa-594 TPF was plotted as a function of EGFP-Vpr TPF for virions prepared with 0.01, 0.1, 0.2, 2 and 4 µg pEnv, respectively as indicated (the same color code as Figure 5c). (f) The correlation coefficients calculated for each panel from a to e as a function of the pEnv inputs during virion production.
Optical Trapping of a Single Animal Virus
Optical trapping technique has been widely used in biology and nanotechnology to study molecular motors, biopolymers, and various nanostructures. However, there have been no demonstrations or studies on optical manipulation of any animal virus, largely due to the small size of these particles. Here we show that it is feasible to optically manipulate a single HIV-1 virion in culture fluid. Our technique combines the sensitive BFP interferometry36,44 and simultaneous TPF at single-molecule level in a single experimental setup, which allows multi-parameter analysis of single HIV-1 virion in culture fluid with single-molecule resolution (Supplementary Note 4). Analogous to flow cytometry for cells, this technique may well be applied to a broad spectrum of animal viruses26 for ultrasensitive detection, multi-parameter analysis of particle heterogeneity, and potential sorting in biological fluid.
Heterogeneity of HIV-1 Virions
Much is known about the heterogeneity of RNA viruses at nucleic acids level thanks to the advances in DNA sequencing technology. However little is known about the heterogeneity of RNA viruses in their protein compositions due to the lack of sensitive techniques to measure viral protein contents at single-molecule level per virion basis. Virometry directly reveals the heterogeneity of HIV-1 in their protein compositions with single-molecule sensitivity. For gp120, we found that its copy number varied over one order of magnitude despite the fact that all these virions were derived from a single clone. This property is a feature of single HIV-1 virions, as demonstrated by our analysis using different cutoff values for single virion diameters (Supplementary Fig. 11). The consequence of this variation might be a broad spectrum of infectivity for individual HIV-1 virions as shown in Fig. 5a and hypothesized in literature45. Indeed, recent studies have shown that the transmitted ‘founder’ virus that established infection in AIDS patients corresponded to virions that had higher envelope glycoprotein content46. Future experiments are necessary to determine if the heterogeneity observed in the spike number of NL4-3 virions is also relevant to R5-tropic viruses, which are generally believed to establish primary infections47. The substantial heterogeneity of envelope content in virions from both low and high pEnv inputs indicates a stochastic component in envelope glycoprotein incorporation during virion budding48–50. It will be of future interest to dissect the mechanisms behind this apparent heterogeneity in viral envelope content, whether it results from a passive incorporation process or an active recruitment mechanism through protein-protein interactions48,49,51.
Methods
Production of HIV-1 virions
Various HIV-1 virions were generated and assayed as described recently30. Briefly, 293T cells were transfected with 1.0 µg pNL4-3R−E− plasmid, various amounts of pEnv (NL4-3 envelope expression plasmid, the same as pcDNA3.1REC in Kim et al.) and pEGFP-Vpr using Mirus LT-1 transfection reagents in 2-ml culture volume in a 35-mm dish. 0.1 µg pEnv and 0.3 µg pEGFP-Vpr plasmids were used as default unless otherwise noted. Media was changed six hours post transfection and virions were harvested at 24 hours post transfection30. Infectious virion concentrations were measured using TZM-bl indicator cell line and the physical concentrations of the virion particles were determined using p24 ELISA assay as described30. The infectivity of the virions was calculated by taking the ratio between infectious virion concentration and physical particle concentration. More than 90% of the infectious units were retained in the complete media over the time frame of the experiments (Supplementary Fig. 12).
OTs and TPF experiments
A home-made OTs instrument using tapered amplifier diode laser at 830 nm (SYS-420-830-1000, Sacher LaserTechnik LLC, Germany) was used for optical trapping and simultaneous TPF measurement of HIV-1 virions29. Briefly, the trapping laser was focused to a diffraction limited spot using a 60× water immersion microscope objective (Nikon) with a numerical aperture of 1.2. We diluted the live virus stock in complete media to a concentration of 0.4~4.0 × 108 virions/ml and injected them into a microfluidic chamber for optical trapping. Brownian motion of the trapped virions was recorded using BFP interferometry44 at 62.5 kHz for 10 s36. To measure virion diameter in culture fluid, a closed-loop nanopositioning stage (3D200, Mad City Labs) was used to oscillate the microfluidic chamber, the amplitude of which was measured directly using video microscopy (Supplementary Fig. 13). For trapping of virions, control experiments showed that oscillation did not induce any change in thermal background. An electron-multiplying charge-coupled device (EMCCD) camera (Evolve, Photometrics) was used for all fluorescence detection. To collect TPF with single-molecule sensitivity, a motorized filter wheel was installed in front of the EMCCD camera, in which the TPF from EGFP was monitored using an emission filter (HQ525/50m, Chroma) and the TPF of Alexa-594 was monitored using an emission filter (ET645/75, Chroma). In addition, a short pass filter (E700sp-2p, Chroma) was installed in front of the EMCCD camera to block the 830 nm trapping laser. A laser power of 130.8 mW at the focus was used throughout for optical trapping and simultaneous TPF excitation. The laser power was monitored using a position-sensitive detector and kept within 1% variation throughout all the experiments. The fluorescence averaged over the initial 2 s exposure time was recorded as the initial fluorescence of the virion. For the two-color experiments, the viruses bound with Alexa594-b12 were injected into the microfluidic chamber. Upon trapping of individual particles in culture media, the TPF emission from Alexa-594 was measured first and then filter was switched to EGFP channel to measure EGFP TPF. The residue leakage of EGFP channel fluorescence into Alexa-594 channel was experimentally measured and quantified by linear regression (Supplementary Fig. 7) and this information was used to correct for Alexa-594 TPF throughout. All the TPF was collected as described using Nis-Element (Nikon), with an EM gain of 200 and exposure time of 400 ms, except the TPF time courses in Fig. 1, where an exposure time of 1 s without EM gain was used. All the trapping and TPF experiments were conducted at a constant temperature of 20.0 ± 0.2 °C in complete media, which consisted of DMEM supplemented with 10% Fetal Bovine Serum (HyClone). The step-finding algorithm that we developed recently52 was used to identify steps from all single-molecule traces. All the correlation analysis was done using custom-written MATLAB scripts and the correlation coefficients were calculated using MATLAB built-in functions. Throughout this work, all errors are standard deviations unless otherwise noted.
Supplementary Material
Acknowledgments
This work was supported by NIH Director’s New Innovator Award 1DP2OD008693-01 to WC, NSF CAREER Award CHE1149670 to WC and also in part by Research Grant No. 5-FY10-490 to WC from the March of Dimes Foundation. We thank Dr. Akira Ono and Alice Telesnitsky for helpful discussions and Cheng Lab members, especially Mike DeSantis for critical reading of the manuscript. The MATLAB code for analysis of TEM images of polystyrene beads was provided by Dr. Mike DeSantis. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH): pNL4-3 from Dr. Malcolm Martin; pNL4-3.Luc.R-E- from Dr. Nathaniel Landau; pEGFP-Vpr from Warner C. Greene; TZM-bl cells from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc; b12 antibody from Dr. Dennis Burton and Carlos Barbas.
Footnotes
W. C. conceived and directed the project; H. S. and J. H. K. prepared experimental materials; Y. P., H. S., J. H. K., X. H. and W. C. conducted the experiments; Y. P., H. S., J. H. K., X. H. and W.C. performed the analysis; Y. P. and W.C. wrote the paper.
Additional information
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
The authors declare no competing financial interests.
References
- 1.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Observation of a Single-Beam Gradient Force Optical Trap for Dielectric Particles. Optics Letters. 1986;11:288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 2.Grier DG. A revolution in optical manipulation. Nature. 2003;424:810–816. doi: 10.1038/nature01935. [DOI] [PubMed] [Google Scholar]
- 3.Neuman KC, Block SM. Optical trapping. Review of Scientific Instruments. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marago OM, Jones PH, Gucciardi PG, Volpe G, Ferrari AC. Optical trapping and manipulation of nanostructures. Nat Nanotechnol. 2013;8:807–819. doi: 10.1038/nnano.2013.208. [DOI] [PubMed] [Google Scholar]
- 5.Qian B, Montiel D, Bregulla A, Cichos F, Yang H. Harnessing thermal fluctuations for purposeful activities: the manipulation of single micro-swimmers by adaptive photon nudging. Chemical Science. 2013;4:1420–1429. [Google Scholar]
- 6.Wang MD. Manipulation of single molecules in biology. Curr Opin Biotechnol. 1999;10:81–86. doi: 10.1016/s0958-1669(99)80015-9. [DOI] [PubMed] [Google Scholar]
- 7.Perkins TT, Quake SR, Smith DE, Chu S. Relaxation of a Single DNA Molecule Observed by Optical Microscopy. Science. 1994;264:822–826. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 8.Heller I, et al. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat Methods. 2013;10:910–916. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- 9.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bormuth V, Varga V, Howard J, Schaffer E. Protein friction limits diffusive and directed movements of kinesin motors on microtubules. Science. 2009;325:870–873. doi: 10.1126/science.1174923. [DOI] [PubMed] [Google Scholar]
- 11.Gross SP. Application of optical traps in vivo. Methods Enzymol. 2003;361:162–174. doi: 10.1016/s0076-6879(03)61010-4. [DOI] [PubMed] [Google Scholar]
- 12.Vale RD. Microscopes for fluorimeters: the era of single molecule measurements. Cell. 2008;135:779–785. doi: 10.1016/j.cell.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagida T, Iwaki M, Ishii Y. Single molecule measurements and molecular motors. Philos Trans R Soc Lond B Biol Sci. 2008;363:2123–2134. doi: 10.1098/rstb.2008.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The Complex Folding Network of Single Calmodulin Molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 15.Tinoco I., Jr Force as a useful variable in reactions: unfolding RNA. Annu Rev Biophys Biomol Struct. 2004;33:363–385. doi: 10.1146/annurev.biophys.33.110502.140418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosanac L, Aabo T, Bendix PM, Oddershede LB. Efficient optical trapping and visualization of silver nanoparticles. Nano Lett. 2008;8:1486–1491. doi: 10.1021/nl080490+. [DOI] [PubMed] [Google Scholar]
- 17.Ploschner M, Cizmar T, Mazilu M, Di Falco A, Dholakia K. Bidirectional optical sorting of gold nanoparticles. Nano Lett. 2012;12:1923–1927. doi: 10.1021/nl204378r. [DOI] [PubMed] [Google Scholar]
- 18.Tong L, Miljkovic VD, Kall M. Alignment, rotation, and spinning of single plasmonic nanoparticles and nanowires using polarization dependent optical forces. Nano Lett. 2010;10:268–273. doi: 10.1021/nl9034434. [DOI] [PubMed] [Google Scholar]
- 19.Geiselmann M, et al. Three-dimensional optical manipulation of a single electron spin. Nat Nanotechnol. 2013;8:175–179. doi: 10.1038/nnano.2012.259. [DOI] [PubMed] [Google Scholar]
- 20.Reece PJ, et al. Characterization of semiconductor nanowires using optical tweezers. Nano Lett. 2011;11:2375–2381. doi: 10.1021/nl200720m. [DOI] [PubMed] [Google Scholar]
- 21.Chen YF, Serey X, Sarkar R, Chen P, Erickson D. Controlled photonic manipulation of proteins and other nanomaterials. Nano Lett. 2012;12:1633–1637. doi: 10.1021/nl204561r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauzauskie PJ, et al. Optical trapping and integration of semiconductor nanowire assemblies in water. Nat Mater. 2006;5:97–101. doi: 10.1038/nmat1563. [DOI] [PubMed] [Google Scholar]
- 23.Ashkin A, Dziedzic JM. Optical trapping and manipulation of viruses and bacteria. Science. 1987;235:1517–1520. doi: 10.1126/science.3547653. [DOI] [PubMed] [Google Scholar]
- 24.Lauffer MA, Stevens CL. Structure of the tobacco mosaic virus particle; polymerization of tobacco mosaic virus protein. Adv Virus Res. 1968;13:1–63. doi: 10.1016/s0065-3527(08)60250-x. [DOI] [PubMed] [Google Scholar]
- 25.Newman J, Swinney HL. Length and dipole moment of TMV by laser signal-averaging transient electric birefringence. Biopolymers. 1976;15:301–315. doi: 10.1002/bip.1976.360150208. [DOI] [PubMed] [Google Scholar]
- 26.Knipe D, et al. Fields Virology. Lippincott Williams & Wilkins. 2007 [Google Scholar]
- 27.Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendix PM, Oddershede LB. Expanding the optical trapping range of lipid vesicles to the nanoscale. Nano Lett. 2011;11:5431–5437. doi: 10.1021/nl203200g. [DOI] [PubMed] [Google Scholar]
- 29.Cheng W, Hou X, Ye F. Use of tapered amplifier diode laser for biological-friendly high-resolution optical trapping. Opt Lett. 2010;35:2988–2990. doi: 10.1364/OL.35.002988. [DOI] [PubMed] [Google Scholar]
- 30.Kim JH, Song H, Austin JL, Cheng W. Optimized Infectivity of the Cell-Free Single-Cycle Human Immunodeficiency Viruses Type 1 (HIV-1) and its Restriction by Host Cells. PLOS One. 2013;8:e67170. doi: 10.1371/journal.pone.0067170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald D, et al. Visualization of the intracellular behavior of HIV in living cells. Journal of Cell Biology. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaeffer E, Geleziunas R, Greene WC. Human immunodeficiency virus type 1 Nef functions at the level of virus entry by enhancing cytoplasmic delivery of virions. J Virol. 2001;75:2993–3000. doi: 10.1128/JVI.75.6.2993-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Accola MA, Ohagen A, Gottlinger HG. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6(gag) J Virol. 2000;74:6198–6202. doi: 10.1128/jvi.74.13.6198-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou X, Cheng W. Single-molecule detection using continuous wave excitation of two-photon fluorescence. Opt Lett. 2011;36:3185–3187. doi: 10.1364/OL.36.003185. [DOI] [PubMed] [Google Scholar]
- 35.Hou X, Cheng W. Detection of single fluorescent proteins inside eukaryotic cells using two-photon fluorescence. Biomed Opt Express. 2012;3:340–353. doi: 10.1364/BOE.3.000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng W, Arunajadai SG, Moffitt JR, Tinoco I, Jr, Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briggs JAG, Wilk T, Welker R, Krausslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. Embo Journal. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolic-Norrelykke SF, et al. Calibration of optical tweezers with positional detection in the back focal plane. Review of Scientific Instruments. 2006;77:103101. [Google Scholar]
- 39.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 40.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chertova E, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 44.Gittes F, Schmidt CF. Interference model for back-focal-plane displacement detection in optical tweezers. Opt Lett. 1998;23:7–9. doi: 10.1364/ol.23.000007. [DOI] [PubMed] [Google Scholar]
- 45.Klasse PJ. The molecular basis of HIV entry. Cell Microbiol. 2012;14:1183–1192. doi: 10.1111/j.1462-5822.2012.01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parrish NF, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger EA, et al. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 48.Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson MC. Mechanisms for Env glycoprotein acquisition by retroviruses. AIDS Res Hum Retroviruses. 2011;27:239–247. doi: 10.1089/aid.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muranyi W, Malkusch S, Muller B, Heilemann M, Krausslich HG. Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail. PLoS Pathog. 2013;9:e1003198. doi: 10.1371/journal.ppat.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arunajadai SG, Cheng W. Step detection in single-molecule real time trajectories embedded in correlated noise. PLoS One. 2013;8:e59279. doi: 10.1371/journal.pone.0059279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.