Abstract
Evaluation of myeloid-derived suppressor cells (MDSC), a cell type implicated in T-cell suppression, may inform immune status. However, a uniform methodology is necessary for prospective testing as a biomarker. We report the use of a computational algorithm-driven analysis of whole blood and cryopreserved samples for monocytic MDSC (m-MDSC) quantity that removes variables related to blood processing and user definitions. Applying these methods to samples from melanoma patients identifies differing frequency distribution of m-MDSC relative to that in healthy donors (HD). Patients with a pre-treatment m-MDSC frequency outside a preliminary definition of HD range (<14.9%) were significantly more likely to achieve prolonged overall survival following treatment with ipilimumab, an antibody that promotes T-cell activation and proliferation. m-MDSC frequencies inversely correlated with peripheral CD8+ T-cell expansion following ipilimumab. Algorithm-driven analysis may enable not only development of a novel pre-treatment biomarker for ipilimumab therapy, but also prospective validation of peripheral blood m-MDSC as a biomarker in multiple disease settings.
Introduction
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of granulocyte- and monocyte-like cells that inhibit T-cell function (1, 2). Clinically significant MDSC accumulation has been observed in many challenges to the immune system in humans including chronic infection, transplant and multiple malignancies (3–10). Diversity in phenotype and methods used for analysis creates challenges in prospectively and reproducibly defining the clinical import of this cellular subset. Monocytic MDSC (m-MDSC) are frequently characterized as CD14+/HLA-DRlow/− cells in humans however HLA-DR expression is typically a broad distribution making identification of a specific subset of cells susceptible to inter-user variability. Nevertheless, increased CD14+/HLA-DRlow/− cells in the peripheral blood have been designated m-MDSC in individual datasets based upon this cell population’s ability to suppress lymphocyte function and are prognostic in patients with hematologic cancers (chronic lymphocytic leukemia and multiple myeloma), solid tumors (HCC, non-small cell lung cancer, melanoma, and others), chronic infection (HIV), cirrhosis, and allotransplantation (5, 8, 11–17).
In melanoma, m-MDSCs correlate with melanoma disease activity and are independently prognostic of overall survival in patients with stage IV disease (6, 18–20). Levels of m-MDSC inversely correlate with the presence of NY-ESO-1-specific T cells and appear to be increased in ipilimumab non-responders (20, 21). This suggests a link between m-MDSC and antigen-specific immunity in vivo and provides additional rationale for routinely evaluating m-MDSCs as a biomarker in the context of immunotherapy clinical trials. However, a uniform methodology that corrects for artifacts introduced by cell processing, cryopreservation, and analysis needs to be developed to enable routine measurement of m-MDSC for prospective testing as a biomarker(22).
Immunomodulatory therapy, which has emerged as a promising treatment approach for metastatic melanoma and other cancers, is an area where biomarker development may enable selection of therapy for individuals more likely to achieve prolonged overall survival. Ipilimumab, an antibody that blocks the function of the immune inhibitory molecule cytotoxic T lymphocyte antigen 4 (CTLA-4), was the first immunomodulatory antibody to gain regulatory approval as a cancer therapeutic based on two phase III studies demonstrating significant increases in overall survival (OS) in patients with metastatic melanoma (23, 24). However, only 20–30% of patients achieve long-term survival following therapy (25). This not only supports the need to define biomarkers in this context, but also to identify mechanisms of resistance that could lead to additional therapeutic targets for improved outcomes.
A number of biomarkers examining T-cell proliferation or activation, and antigen-specific immunity have been assessed in the context of ipilimumab therapy. Gene expression profiling on tumor biopsies collected from 45 melanoma patients before and after ipilimumab treatment showed that an immunologically active tumor microenvironment favors clinical response to ipilimumab (26, 27). In peripheral blood, sustained ICOS elevation in CD4+T cells, higher percentage of EOMES+ CD8+ T cells or Ki67+EOMES+CD8+ T cells, and a NY-ESO-1-specific CD8+ T-cell response in NY-ESO-1 seropositive metastatic melanoma patients have all shown an association with clinical benefit and survival following ipilimumab therapy (28, 29).
Absolute lymphocyte count (ALC), the most clinically accessible biomarker, available through a routine complete blood count, has been shown to correlate with overall survival in several single-institution, non-controlled studies (30). More recently, an analysis of almost 2000 ipilimumab-treated patients in multiple studies, including randomized, controlled, phase 3 studies, has demonstrated that an ALC increase is a specific pharmacodynamic biomarker of ipilimumab. In the absence of concomitant chemotherapy, the degree of this pharmacodynamic increase in lymphocyte count at the commercially available ipilimumab dose (3mg/kg) is associated with OS (31) suggesting ALC is worthy of further investigation in the context of risk-adapted clinical trial design.
We report the development of methods to enable uniform analysis of m-MDSC that overcomes issues related to blood processing and inter-user variability. This is achieved by deriving a measure of m-MDSC using coefficient of variance (CV) to assess HLA-DR spread on CD14+/CD11b+ cells and through the evaluation of stabilizers of HLA-DR levels in whole blood. We validate these methods by demonstrating that CD14+/HLA-DRlow/− m-MDSC quantity derived from CV values are both inversely correlated with pharmacodynamics markers of ipilimumab function and also associated with overall survival among patients undergoing treatment with ipilimumab.
MATERIALS AND METHODS
Patients
We identified 83 patients that were treated on a clinical study with ipilimumab between 2/2008 and 3/2012 and had cryopreserved peripheral blood samples in our tissue banks. Peripheral blood from healthy donor volunteers was obtained at the time of the current study and from samples in our institutional tissue bank. MDSC analyses were performed between 12/2011–3/2012. We excluded 4 and 11 samples in the 10mg/kg and 3mg/kg ipilimumab groups, respectively, because of an overnight delay between phlebotomy and processing time, which validation studies confirmed affects levels of HLA-DR (Figure 1E). Patients and healthy donors provided informed consent for the clinical studies and the collection of blood and tumor tissue on a correlative biospecimen protocol. Patients were treated with ipilimumab on Bristol-Myers Squibb studies CA184045 (NCT00495066) or CA184-087 (NCT00920907), with four doses of ipilimumab 10 mg/kg or 3mg/kg IV every 3 weeks during induction therapy, followed by maintenance ipilimumab at the same dose every 12 weeks, starting at week 24. Clinical benefit was determined by investigators at week 24 imaging based upon interpretation of radiographic stable disease or better by mWHO or RECIST criteria. All studies were approved by the Memorial Sloan-Kettering Cancer Center (MSKCC) Institutional Review Board.
Figure 1. Analysis of Myeloid Derived Suppressor Cell Frequency.
Peripheral blood mononuclear cells (PBMC) from advanced melanoma patients and healthy donors were stained with surface antibody and analyzed by multicolor flow cytometry. We defined monocytic myeloid cells based on presence of CD14, CD11b in a CD3, CD16, CD19, CD20, CD56 in lineage (Lin) negative population. Within this monocytic cell population, m-MDSC were isolated based on their low levels of expression of HLA-DR expression.
(A). Gating strategy to isolate myeloid derived cells as CD14+CD11b+Lineage- cells. Based on the 99%ile of healthy donor values, a cut-off for low expression of HLA-DR was set to isolate the population of m-MDSC (shaded in red).
(B) m-MDSC composition by HLA-DR GMFI is subject to fluctuations in staining acquisition and sample handling. CVHLA-DR represents a self-normalizing measurement and is stable among replicate measurements.
(C). Comparison of coefficient of variations (CV) for HLA expression within the myeloid compartment reveals a larger spread for patients pre-treatment, compared to healthy donors and large differences in CV between patients (HD vs. Patients: p<0.05).
(D) Normogram plotting relationship between CV values and m-MDSC frequency.
(E) Evaluation of whole blood collected in standard heparin or Cyto-Chex®tubes (n=9) for m-MDSC frequency and stored at room temperature for the specified interval between analysis and acquisition. Data is expressed as a percentage of total m-MDSC present at baseline. *p=0.002
(F) Correlation between m-MDSC analysis of samples (n=8) cryopreserved using BD Vacutainer® CPT tubes, standard heparin tubes and collected in Cyto-Chex®tubes.
Myeloid-derived suppressor cells staining
Blood was collected and cryopreserved using BD Vacutainer® CPT™ tubes (BD pharmingen) from melanoma patients and healthy donors for the retrospective analysis. Samples were collected from patients and healthy donors in cyto-chex®(Streck), Vacutainer® CPT™, or standard heparin vacutainer tubes for comparative analysis. 5 × 105 peripheral blood mononuclear cells (PBMC) from melanoma patients or healthy donors were washed with 2 mL FACS buffer (PBS containing 2% bovine serum albumin and 0.05 mmol/L EDTA). The following antibodies were then added for 30 minutes at 4 ˚C: Lineage (CD3/CD16/CD19/CD20/CD56) cocktail FITC (BD Pharmingen), CD14-PerCP Cy5.5, CD11b–APC Cy7, CD33-PE-Cy7 (BD Pharmingen), HLA-DR-ECD (Beckman Coulter). Isotype controls included the appropriate fluorochrome-conjugated mouse IgG1, IgG 1k, IgG2a, or IgG2b k (BD Pharmingen, Beckman Coulter, R&D Systems). Whole blood samples were lysed for 10 minutes in BD Phosflow Lyse/Fix after staining (BD Pharmingen). Stained cells were detected using a LSR Fortessa with FACS Diva software (BD Biosciences). Analysis was carried out using FlowJo (Treestar). Monoctyic-MDSCs were quantified as described. Briefly, scale values for HLA-DR within a singlet, live, lineage negative (CD3, CD16, CD19, CD20 CD56) cell population that expressed CD14+CD11b+ were exported from flowjo and analyzed using code written in R software to derive the coefficient of variation (CV), a ratio of standard deviation (σ) and geometric mean fluorescence intensity (GMFI). A % m-MDSC frequency defined as the %HLA-DRlow/− among CD14+CD11b+ cells was derived using a nomogram based on the 99%ile CVHLA-DR among CD14+CD11b+ cells from healthy donors. Absolute number of m-MDSC (/µL) in peripheral blood was estimated using the formula; (%m-MDSC) × (number of monocytes (/µL) from a complete blood count on the same day.
T-cell suppression assay
A T-cell suppression assay was performed as described previously (32). Briefly, CD14+ PBMCs magnetically separated using MACs beads (Miltenyi Biotec, Bergisch Gladbach, Germany) were cultured with 2 × 105 CFSE-labeled autologous CD14− PBMCs in 96-well flat bottom α-CD3-specific Ab-coated plates (OKT3, 100µL at 0.5 µg/mL for 2 hours at 37˚C) in RPMI 1640 medium supplemented with 10% FBS and IL-2 (10 IU/mL; Roche). After 5 days, cells were harvested, stained with CD3-PECy7, CD4-ECD, and CD8-APC Cy7 (BD Pharmingen), and CFSE signal of gated CD8+ T cells (CD3+ CD4−) was measured by flow cytometry. The stimulation index (SI) was calculated by dividing the proliferation measured in the absence of m-MDSC by proliferation measured in the presence of m-MDSC, as previously described (32).
Statistics
Patient characteristics were described using median and range for continuous variables and frequency and percentages for categorical variables. The primary endpoint for this retrospective analysis was overall survival, which is defined as the time from pre-treatment %m-MDSC assessment to death or last follow-up. Landmark analysis from week 6 was also performed. Patients alive at last follow-up are censored. Maximally selected log-rank statistics was used to find a cutoff value for %m-MDSC. The Kaplan-Meier method and log-rank test were used to compare differences in survival for categorical variables. Univariate and multivariate Cox proportional hazards regression was used to assess the association between clinical variables and overall survival. A student’s T test with Welch’s correction was used for comparisons of %m-MDSC frequency in patient and healthy donor groups. Pearson correlation was used to evaluate for relationships between %m-MDSC and lymphocyte subsets.
Results
Measuring HLA-DR spread using a computational algorithm removes user bias and inter-replicate variability in m-MDSC assessment
Published reports of m-MDSC frequency have evaluated this cellular subset by gating on lineage- CD14+CD11b+ HLA-DRlow/− cells in the peripheral circulation. We similarly developed a flow cytometric strategy to define m-MDSC based on high abundance of CD14, CD11b, and low or absent HLA-DR expression in a CD3, CD16, CD19, CD20 CD56, lineage (Lin) negative population (Supp. Figure 1). HLA-DR expression on myeloid cells displayed a wide continuous distribution rather than distinct populations. Log rank tests based on different gating cutoffs resulted in a broad range of m-MDSC cutoff values and highly variable survival curves. Thus, selection of an accurate gate for a low or negative HLA-DR fraction is challenging and prone to user bias and experimental unreliability. However, we observed distinct spreads for the HLA-DR distribution between individual patients suggesting that evaluating this parameter on CD11b+CD14+ cells could serve as a measure of m-MDSC. Thus, we gated on CD11b+CD14+ cells and measured HLA-DR GMFI, standard deviation (SD), and the coefficient of variation (CV), a ratio between GMFI and SD (Figure 1A). Evaluating CV corrects for shifting GMFI due to staining protocol and nearly eliminates inter-replicate variability (Figure 1B) enabling measurement of HLA-DR distribution on myeloid cells objectively and independently of staining fluctuations (33). Measurements across a cohort of healthy donors (n=20) and melanoma patients (n=68) revealed a higher value of CVHLA-DR among patient myeloid cells (Figure 1C). Furthermore, we found a cohort of patient samples with CVHLA-DR levels above the range for healthy donors (defined by the 99th%ile in CV values among healthy donors). for these patients, the higher CV value indicates a higher HLA-DR spread, representative of abnormal elevations in the number of m-MDSC (HLA-DRlow cells). To explicitly quantify the number of m-MDSC, we utilize the upper limit of CVs for healthy donors (again, the 99th%ile, = X) as a "cutoff" and generate a nomogram to calculate an ad hoc quantitative measure of MDSC frequency (%m-MDSC). By translating the mean-normalized variance in the data to a concrete percentage of the population, we relate CVHLA-DR to a classical immunophenotyping measurement.
Given the potential for changes in HLA-DR expression that may occur during blood processing or transport to significantly alter m-MDSC evaluation we evaluated our methods in whole blood stored at room temperature as well as cryopreserved ficoll purified PBMCs. We noted CVHLA-DR was significantly reduced as the interval between phlebotomy time and analysis increased: a 48h delay until processing demonstrated a nearly 50% reduction from baseline. Levels of CVHLA-DR were however consistent over time in cyto-chex® blood collection tubes even if whole blood was stored at room temperature prior to processing for up to 8 days after phlebotomy (Figure 1E). Actual CVHLA-DR values were different but clearly correlated between cyto-chex® BCT, vacutainer® CPT™ cell preparation tubes (r=0.83), and standard heparin tubes (r=0.87) (Figure 1F).
m-MDSCs occur with relative higher frequency among patients with metastatic melanoma than in healthy donor controls
Using our CVHLA-DR/%m-MDSC conversion nomogram, we determined the relative frequency of m-MDSC for 68 melanoma patients treated with ipilimumab at either 10mg/kg (n= 28) or 3mg/kg (n= 40) for whom pre-treatment and week 6 PBMC samples were processed the same day as phlebotomy and stored in our tissue repository. We again used healthy donors as controls. The baseline characteristics of the patients and healthy donors are described in Table 1. Overall median time from initial m-MDSC measurement to last recorded follow-up was 13.6 months (range 0.66–63.9).
Table 1.
Patient and Healthy Donor Characteristics.
| Characteristics | Ipilimumab 10mg/kg | Ipilimumab 3mg/kg | Healthy Donors* |
|---|---|---|---|
| Number of Patients | 28 | 40 | 20 |
| Median Age (range) | 62 (34–83) | 60 (34–80) | 38(26–58) |
| Sex (%) | |||
| Male | 17(61) | 29 (73) | 10 (50) |
| Female | 11(39) | 11(27) | 7 (35) |
| Stage of disease (%) | |||
| III (unresectable) | 0 | 1 | - |
| M1a | 3 | 0 | - |
| M1b | 4 | 5 | - |
| M1c | 21 | 34 | - |
| Median number of prior therapies (range) | 1 (0–3) | 1 (0–5) | - |
| Median LDH (range) | 209 (113–968) | 211(117–816) | - |
| ≥Upper limit of normal (% of available LDH) | 13 (46) | 28(70) | - |
| <Upper limit of normal (% of available LDH) | 15 (54) | 12(30) | - |
| MDSC Frequency %HLA-DR low/− in Lin"CD14+CDllb+ (range)** | 11.4 (3.9–24.5) | 11.2(5.8–20.9) | 10.3(6.4–14.3) |
| ≥ 14.9 (%) | 7(25) | 7(18) | 0(0) |
| < 14.9 (%) | 21(75) | 33 (82) | 20 (100) |
| Median baseline ALC (range) | 1250 (500–5100) | 1100(600–8100) | - |
| ≥ 1000/µl (%) | 19(68) | 25 (63) | - |
| < 1000/µl (%) | 9(32) | 15 (37) | - |
Data for anonymously donated blood bank samples is unavailable
Baseline values
We found that the relative frequency of peripheral blood m-MDSC was increased among patients with metastatic melanoma (p=0.05) when compared with a group of healthy individuals (Figure 2A). Pre-treatment m-MDSC frequency did not differ significantly in our cohort between patients that were treated with different doses of ipilimumab (Figure 2B).
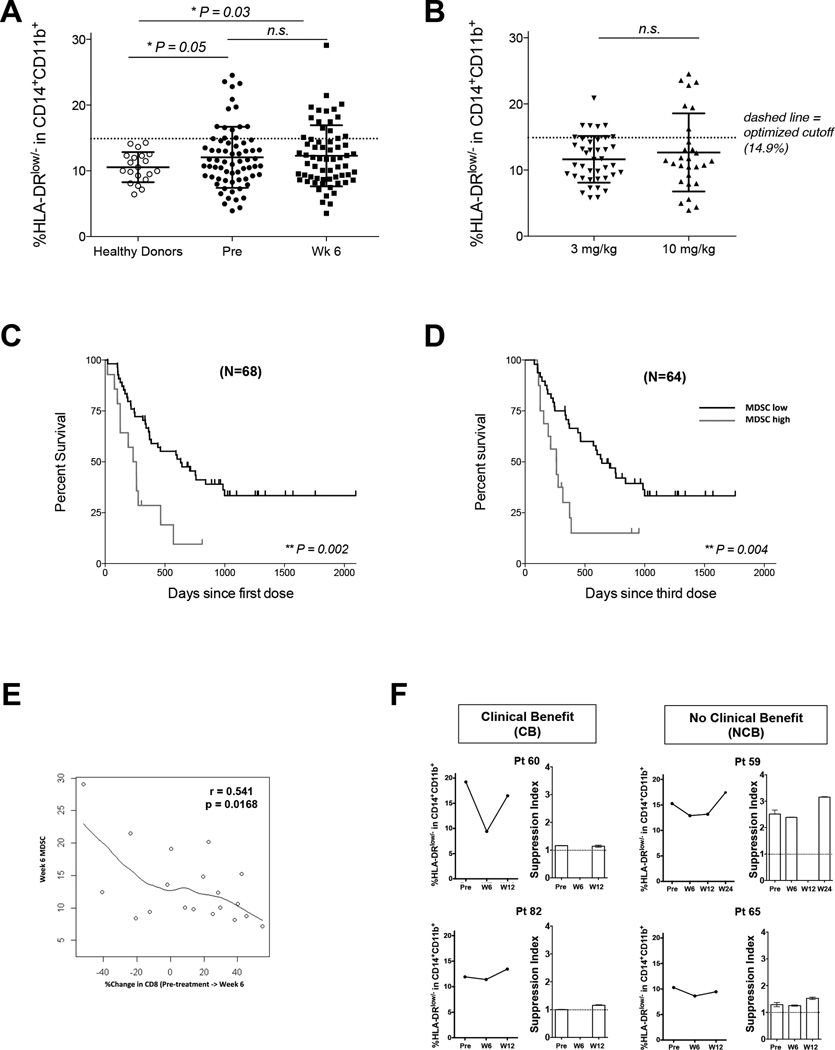
Figure 2. Functionally Suppressive m-MDSC are increased in patients with metastatic melanoma less likely to achieve prolonged overall survival following ipilimumab.
(A) Peripheral blood mononuclear cell (PBMC) from advanced melanoma patients and healthy donors analyzed for %m-MDSC based on CVHLA-DR. The frequency of m-MDSC in healthy donors (n=20), melanoma patients analyzed at pre-treatment baseline and week 6 (Healthy donor vs pretreatment p=0.05; healthy donor vs week 6, p=0.03),
(B) Pre-treatment values for subsets of patients treated with ipilimumab 10mg/kg (n=28) or 3mg/kg (n=40).
(C) Overall survival based on m-MDSC quantity at pre-treatment baseline.
(D) Overall survival from 6 weeks after start of ipilimumab treatment.
(E) The correlation between %change in CD8 T cells and wk 6 m-MDSC frequency (r = − 0.541, p = 0.02). % change in CD8 T cells = [(wk6 absolute CD8 – baseline absolute CD8) ÷ (baseline absolute CD8)].
(F) The average stimulation index is graphed for 2 melanoma patients with clinical benefit and 2 melanoma patients with non-clinical benefit assessed at week 24. Stimulation index = (% proliferated CD3+ T cells in CD14-depleted PBMCs) / (% proliferated CD3+ T cell in CD14-PBMCs with CD14+ cells added back).
Pre-treatment m-MDSC quantity correlates with overall survival (OS) in patients treated with ipilimumab
In order to evaluate the hypothesis that lower frequency of m-MDSC was associated with overall survival, we parsed our patients according to their %m-MDSC at baseline and after 2 doses of ipilimumab (week 6). Based on logrank statistics within our ipilimumab-treated cohort we defined 14.9% as the cutoff between “high” and “low” %m-MDSC. The distribution of m-MDSC frequencies among analyzed patients is summarized in Table 1.
Having less than 14.9% m-MDSC pre-treatment was associated with improved OS among 68 patients treated with ipilimumab (Figure 2C, Table 2) with a HR of 0.35 (95%CI: 0.18 – 0.70, p=0.003). When analyzed by individual dose groups, the difference was seen in patients treated at 10 mg/kg, but not at 3 mg/kg (Table S1). We performed univariate (Table 2) and multivariate analysis (Table 3) to evaluate the impact of ALC, LDH, and monocyte counts on survival in our patient cohort. %m-MDSC < 14.9% correlated with superior OS on both univariate and multivariate analysis. Monocyte quantity was not predictive suggesting %m-MDSC represents a relative activation state within the monocyte compartment and is not a direct reflection of monocyte numbers.
Table 2.
Univariate analysis of relationship between m-MDSC and overall survival at pre-treatment baseline and week 6 after ipilimumab.
| Ipilimumab Treated | ||||||
|---|---|---|---|---|---|---|
| Pre-treatment | Week 6 | |||||
| n | HR (95% CI) | p-value | n | HR (95% CI) | p-value | |
| MDSC < 14.9% | 68 | 0.35 (0.18 – 0.70) | 0.002 | 64 | 0.38 (0.19 – 0.75) | 0.004 |
| ALC ≥1000 cells/µl | 68 | 0.73 (0.41 – 1.33) | 0.303 | 64 | 0.22 (0.11 – 0.45) | < 0.001 |
| LDH < 250 | 68 | 0.33 (0.18 – 0.59) | <0.001 | 65 | 0.37 (0.20 – 0.68) | 0.001 |
| Monocytes < 300 cells/µl | 68 | 0.70 (0.25 – 1.95) | 0.495 | 64 | 1.77 (0.69 – 4.51) | 0.233 |
Table 3.
Multivariate analysis of relationship between m-MDSC and overall survival at pre-treatment baseline and week 6 after ipilimumab treatment.
| Ipilimumab | ||||||
|---|---|---|---|---|---|---|
| Pre-treatment | Week 6 | |||||
| n | HR (95% CI) | p-value | n | HR (95% CI) | p-value | |
| MDSC ≤ 14.9% | 68 | 0.47 (0.23 – 0.94) | 0.033 | 63 | 0.38 (0.18 – 0.81) | 0.012 |
| ALC ≥1000 cells/µl | - | - | - | 63 | 0.21 (0.10 – 0.46) | < 0.001 |
| LDH < 250 | 68 | 0.38 (0.21 – 0.69) | 0.002 | 63 | 0.29 (0.15 – 0.56) | < 0.001 |
On Treatment (week 6), the frequency of m-MDSC correlates with overall survival (OS) in patients treated with ipilimumab
We also evaluated associations between %m-MDSC at week 6 and OS similarly to what has been evaluated previously for ALC (Table 2, Table S1) (30, 34). %m-MDSC below 14.9% at week 6 was associated with superior OS (Figure 2D) in patients receiving ipilimumab treatment with a HR of 0.38 (95%CI: 0.19 – 0.75, p=0.005). As expected ALC greater than or equal to 1000 at week 6 was associated with improved OS in our cohort and normal LDH (<250) at week 6 correlated with improved OS in patients treated with ipilimumab. To address potential confounding by ALC and LDH, a multivariate analysis was performed and week 6 %m-MDSC remained significantly associated with OS, even when accounting for both LDH and week 6 ALC. (Table 3).
%m-MDSC is inversely correlated with CD8+ T-cell rise on therapy and suppresses T-cell proliferation in vitro
Ipilimumab has a specific pharmacodynamic effect on absolute lymphocyte counts, but data on the specific subset of cells affected is limited. Our group previously reported on 35 patients treated with ipilimumab 10mg/kg in which the relationships between increases in CD8+ T cells, CD4+ T cells, and CD4+CD25+ regulatory T cells, and clinical outcome were analyzed. In this analysis the majority of patients had increases in all three lymphocyte subsets, but only the mean increase in CD8+ T cells was significantly associated with clinical benefit (35).
Since m-MDSCs are defined by the ability to suppress CD8+ T-cell proliferation, we examined whether m-MDSC frequency impacts T cells in vivo or in vitro. We first sought to explore if relationships between ALC and m-MDSC were observed consistent with m-MDSC suppressive function in vivo. Based on the known biologic functions of m-MDSC, we reasoned that a greater frequency of m-MDSC would limit the T-cell proliferative response to ipilimumab. However, we did not find correlations between the %change in total ALC ([week 6 – pretreatment] ÷ [pre-treatment]) and pre-treatment or week 6 m-MDSC frequency. Data on CD4+ and CD8+ T-cell subsets were available for 19 of the 40 patients treated with ipilimumab at 3mg/kg. We observed a statistically significant inverse correlation only between %change in absolute CD8+ T-cell number and m-MDSC frequency at week 6 (r = − 0.54, p = 0.0168) (Figure 2E) and no correlation was observed between that and %change in CD4+ T-cell numbers on therapy.
We next assessed for suppressive function by measuring T-cell proliferation in PBMCs in the presence or absence of CD14+ cells. We inferred that suppressive function is present if enhanced proliferation was observed among PBMCs stimulated with anti-CD3 and IL-2 in the absence of CD14-expressing cells (Supplemental Figure 2). Proliferation of CD3+ T cells was increased to a greater extent in the absence of CD14-expressing cells only in PBMC samples taken from patients that did not achieve clinical benefit as measured at week 24 imaging (Figure 2F). These data suggest that higher frequency of m-MDSC in patients with inferior outcomes correlated with diminished T-cell proliferation in vitro.
Discussion
We developed an objective methodology to evaluate m-MDSC frequency in the peripheral blood of metastatic melanoma patients receiving immunotherapy with ipilimumab at our center. In our single institution cohort, we found that patients with metastatic melanoma have a greater frequency of m-MDSC than a group of healthy donors. An m-MDSC quantity prior to treatment and at week 6 that was outside the healthy donor range that we defined was significantly associated with inferior overall survival, independent of LDH (at baseline and week 6) and ALC (at week 6) in a multivariate model. Our observations suggest that m-MDSC frequency is a novel prognostic indicator of overall survival in metastatic melanoma patients treated with ipilimumab.
The CV-based cutoff presented here represents an objective methodology for determining m-MDSC composition independent of fluorescence variability in FACS analysis. The cutoff level derived here was consistent with a level greater than the 99%ile of a preliminary cohort of healthy donor m-MDSC, suggesting that our method enables distinction of normal versus abnormal CVHLA-DR and m-MDSC evaluation in a prospective fashion. Thus, we suggest that using healthy donors as a calibration can lead to an easily implementable, automated and objective tool for monitoring the frequency of m-MDSC within patients’ blood samples, and distinguish between “normal” and “high” ranges. However, it is important to note the preliminary nature of our healthy donor range and that further study of the effects of age, gender, BMI, and non-malignant comorbid conditions on CVHLA-DR are necessary to propose a “cutoff” value capable of prospectively segregating melanoma patients more or less likely to benefit from ipilimumab.
As the coefficient of variation (CV) is defined as the ratio of the standard deviation to the mean, we obtained a metric independent of non-biologically meaningful fluctuations in sample handling, FACS protocol and fluorescence intensity. While using “non-normalized” metrics for HLA-DRlow/− populations (GMFI, standard deviation; Supplemental Figure 3) replicate the reported observations, the survival-based cutoffs determined here by GMFI or standard deviation do not represent universal standards, and would be expected to be unstable differentiating factors with subsequent validation. Utilizing CV effectively captures either the decreasing GMFI and/or increasing standard deviation of the HLA-DR fluorescence intensity characteristic of cellular populations with higher numbers of m-MDSCs and eliminates replicate variability. By establishing a protocol in which healthy donor CV provides the necessary threshold for a “normal” CV range, we can achieve a robust identification of patients with high m-MDSC composition.
ALC rise on therapy has been associated with improved OS following ipilimumab therapy. In a small cohort of patients treated with ipilimumab 10mg/kg at our center we found that CD4+ lymphocytes, CD4+CD25+ regulatory T cells, and CD8+ lymphocytes all increased with therapy. However, increases in the absolute number of CD8+ lymphocytes were significantly greater among patients that achieved clinical benefit from ipilimumab when compared with patients that did not benefit (35). In the current analysis, we observed inverse correlations between %change in CD8+ T cells with m-MDSC frequency in vivo. These findings are consistent with an in vitro suppression assay where higher frequencies of m-MDSC were associated with greater suppressive activity. We propose that in melanoma patients receiving ipilimumab, the pharmacodynamic effects on lymphocyte subset increase and the quantity of m-MDSC are interrelated and worthy of further study as pharmacodynamic markers of therapeutic efficacy perhaps sufficient to guide risk-adapted clinical trial design. Furthermore, taken together the observations reported here suggest the hypothesis that m-MDSC suppression of lymphocytes may be limiting the therapeutic benefit of ipilimumab. A larger cohort of patients will need to be studied to confirm our findings and to assess if escalating ipilimumab dose or combination therapies including m-MDSC-directed therapies can modulate CD8+ and m-MDSC interactions.
In our study we developed an objective method to evaluate pre-treatment Lin−CD14+HLA-DRlow/− m-MDSC frequency building on the phenotype reported in the literature by other research groups (5, 6, 21). Similar to results from Gros et al and Meyer et al, we found a greater frequency of these cells in the peripheral blood of melanoma patients in comparison to healthy donors (19, 21). These cells co-express CD11b (Supplemental Figure 1) and in most cases also co-express CD33 (data not shown). Similar to others’ finding we also found that disease course paralleled m-MDSC frequency, i.e. patients without radiographic benefit following ipilimumab tended to have increasing frequencies of m-MDSC over time (data not shown). Nevertheless, the prognostic significance of m-MDSC frequency in our analysis was independent of LDH, a known prognostic marker associated with disease burden in melanoma patients (36).
Clinically significant m-MDSC accumulation characterized with diverse myeloid phenotypic markers has been observed in a number of malignancies in humans (4–10). Young et al. measured CD34+ natural suppressor cells and found that excess of CD34+ cells at the tumor site was associated with relapse of head and neck cancer. Solito et al and Gabitass et al have reported that the quantity of m-MDSC with an immature myeloid phenotype (lineage negative, HLA-DR−, CD11b+, CD33+) are prognostic markers in breast and colorectal cancer or gastric, esophageal, and pancreatic cancer, respectively (37, 38). In renal cell carinoma, Zea et al. have described a CD33+,CD15+ granulocytic MDSC(10) while in melanoma, both Filipazzi et al. and Poschke et al have found that m-MDSC function is within the monocytic CD14+, HLA-DRlow/− cell population. Our report adds to this emerging literature with the first description of statistically significant associations between m-MDSC accumulation, survival outcomes, and specific lymphocyte parameters following an immunomodulatory antibody therapy.
In summary, we have developed a method that enables accurate measurement of Lin-CD14+HLA-DRlow/− m-MDSC independent of technical variables related to sample processing time and flow cytometry. Using this method, we demonstrate for the first time higher pretreatment quantities of Lin-CD14+HLA-DRlow/− m-MDSC are associated with inferior OS in metastatic melanoma patients treated with ipilimumab. Inverse correlations between CD8+ T-cell increases and m-MDSC frequency in patients treated with ipilimumab suggest a role for m-MDSC-mediated lymphocyte suppression in overall survival following ipilimumab therapy. Further prospective studies are needed to validate m-MDSC measurement as a prognostic biomarker for melanoma and other disease states. Uniform analysis methods and the use of cyto-chex® tubes enable these to proceed across multiple labs.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health (RC2 CA148468 and R01 CA056821 to JDW), the American Cancer Society (MRSG-11-054-01-LIB, to AML), the Melanoma Research Alliance (to AML and JDW), Swim Across America (to JDW), the Cancer Research Institute (to JDW), the Lita Annenberg Hazen Foundation (to JDW), and the Commonwealth Foundation for Cancer Research (to JDW).
Footnotes
Author’s Contribution:
S.K., J.D.W., and A.M.L. conceived, designed, and performed experiments, analyzed data, and wrote the manuscript. M.A.P., C.Z., and G.A.B. analyzed data and wrote the manuscript, D.K. and K.S.P performed statistical analysis and edited manuscript. C.C. and T.R. performed experiments. J.Y. analyzed data and edited manuscript.
Conflicts of Interest Statement:
A.M.L. and J.D.W. have a patent pending entitled "Prediction of Responsiveness to Treatment with Immunomodulatory Therapeutics and Method of Monitoring Abscopal Effects During Such Treatment". A.M.L, M.A.P., and J.D.W. receive research funding on behalf of their institution and consult for Bristol Myers-Squibb.
References and Notes
- 1.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poschke I, Kiessling R. On the armament and appearances of human myeloid-derived suppressor cells. Clin Immunol. 2012;144:250–268. doi: 10.1016/j.clim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young MR, Wright MA, Lozano Y, Prechel MM, Benefield J, Leonetti JP, et al. Increased recurrence and metastasis in patients whose primary head and neck squamous cell carcinomas secreted granulocyte-macrophage colony-stimulating factor and contained CD34+ natural suppressor cells. Int J Cancer. 1997;74:69–74. doi: 10.1002/(sici)1097-0215(19970220)74:1<69::aid-ijc12>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 5.Filipazzi P, Valenti R, Huber V, Pilla L, Canese P, Iero M, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25:2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 6.Poschke I, Mougiakakos D, Hansson J, Masucci GV, Kiessling R. Immature Immunosuppressive CD14+HLA-DR-/low Cells in Melanoma Patients Are Stat3hi and Overexpress CD80, CD83, and DC-Sign. Cancer Res. 2010;70:4335–4345. doi: 10.1158/0008-5472.CAN-09-3767. [DOI] [PubMed] [Google Scholar]
- 7.Diaz-Montero CM, Salem ML, Nishimura MI, Garrett-Mayer E, Cole DJ, Montero AJ. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chikamatsu K, Sakakura K, Toyoda M, Takahashi K, Yamamoto T, Masuyama K. Immunosuppressive activity of CD14+ HLA-DR- cells in squamous cell carcinoma of the head and neck. Cancer Science. 2012;103:976–983. doi: 10.1111/j.1349-7006.2012.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serafini P, Meckel K, Kelso M, Noonan K, Califano J, Koch W, et al. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med. 2006;203:2691–2702. doi: 10.1084/jem.20061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 11.Qin A, Cai W, Pan T, Wu K, Yang Q, Wang N, et al. Expansion of monocytic myeloid-derived suppressor cells dampens T cell function in HIV-1-seropositive individuals. J Virol. 2013;87:1477–1490. doi: 10.1128/JVI.01759-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mougiakakos D, Jitschin R, von Bahr L, Poschke I, Gary R, Sundberg B, et al. Immunosuppressive CD14+HLA-DRlow/neg IDO+ myeloid cells in patients following allogeneic hematopoietic stem cell transplantation. Leukemia. 2013;27:377–388. doi: 10.1038/leu.2012.215. [DOI] [PubMed] [Google Scholar]
- 13.Luan Y, Mosheir E, Menon MC, Wilson D, Woytovich C, Ochando J, et al. Monocytic Myeloid-Derived Suppressor Cells Accumulate in Renal Transplant Patients and Mediate CD4(+) Foxp3(+) Treg Expansion. Am J Transplant. 2013;13:3123–3131. doi: 10.1111/ajt.12461. [DOI] [PubMed] [Google Scholar]
- 14.Huang A, Zhang B, Wang B, Zhang F, Fan KX, Guo YJ. Increased CD14(+)HLA-DR (-/low) myeloid-derived suppressor cells correlate with extrathoracic metastasis and poor response to chemotherapy in non-small cell lung cancer patients. Cancer Immunol Immunother. 2013;62:1439–1451. doi: 10.1007/s00262-013-1450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arihara F, Mizukoshi E, Kitahara M, Takata Y, Arai K, Yamashita T, et al. Increase in CD14+HLA-DR −/low myeloid-derived suppressor cells in hepatocellular carcinoma patients and its impact on prognosis. Cancer Immunol Immunother. 2013;62:1421–1430. doi: 10.1007/s00262-013-1447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafson MP, Abraham RS, Lin Y, Wu W, Gastineau DA, Zent CS, et al. Association of an increased frequency of CD14+ HLA-DR lo/neg monocytes with decreased time to progression in chronic lymphocytic leukaemia (CLL) Br J Haematol. 2012;156:674–676. doi: 10.1111/j.1365-2141.2011.08902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan XK, Zhao XK, Xia YC, Zhu X, Xiao P. Increased circulating immunosuppressive CD14(+)HLA-DR(-/low) cells correlate with clinical cancer stage and pathological grade in patients with bladder carcinoma. J Int Med Res. 2011;39:1381–1391. doi: 10.1177/147323001103900424. [DOI] [PubMed] [Google Scholar]
- 18.Postow MA, Callahan MK, Barker CA, Yamada Y, Yuan J, Kitano S, et al. Immunologic correlates of the abscopal effect in a patient with melanoma. N Engl J Med. 2012;366:925–931. doi: 10.1056/NEJMoa1112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gros A, Turcotte S, Wunderlich JR, Ahmadzadeh M, Dudley ME, Rosenberg SA. Myeloid cells obtained from the blood but not from the tumor can suppress T-cell proliferation in patients with melanoma. Clin Cancer Res. 2012;18:5212–5223. doi: 10.1158/1078-0432.CCR-12-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weide B, Martens A, Zelba H, Stutz C, Derhovanessian E, Di Giacomo AM, et al. Myeloid-derived suppressor cells predict survival of advanced melanoma patients: comparison with regulatory T cells and NY-ESO-1- or Melan-A-specific T cells. Clin Cancer Res. 2014;20:1601–1609. doi: 10.1158/1078-0432.CCR-13-2508. [DOI] [PubMed] [Google Scholar]
- 21.Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, et al. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother. 2014;63:247–257. doi: 10.1007/s00262-013-1508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biomarkers CoQo, Disease SEiC, Medicine Io. Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease. The National Academies Press; 2010. [PubMed] [Google Scholar]
- 23.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 2010363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robert C, Thomas L, Bondarenko I, O'Day S, M DJ, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 25.Wolchok JD, Weber JS, Maio M, Neyns B, Harmankaya K, Chin K, et al. Four-year survival rates for patients with metastatic melanoma who received ipilimumab in phase II clinical trials. Ann Oncol. 2013;24:2174–2180. doi: 10.1093/annonc/mdt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamid O, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med. 2011;9:204. doi: 10.1186/1479-5876-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carthon BC, Wolchok JD, Yuan J, Kamat A, Ng Tang DS, Sun J, et al. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin Cancer Res. 2010;16:2861–2871. doi: 10.1158/1078-0432.CCR-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J, Adamow M, Ginsberg BA, Rasalan TS, Ritter E, Gallardo HF, et al. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical benefit in advanced melanoma patients treated with ipilimumab. Proc Natl Acad Sci U S A. 2011;108:16723–16728. doi: 10.1073/pnas.1110814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ku GY, Yuan J, Page DB, Schroeder SE, Panageas KS, Carvajal RD, et al. Single-institution experience with ipilimumab in advanced melanoma patients in the compassionate use setting: lymphocyte count after 2 doses correlates with survival. Cancer. 2010;116:1767–1775. doi: 10.1002/cncr.24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michael A. Postow, Scott D. Chasalow, et al. submitted. [Google Scholar]
- 32.Mandruzzato S, Solito S, Falisi E, Francescato S, Chiarion-Sileni V, Mocellin S, et al. IL4Ralpha+ myeloid-derived suppressor cell expansion in cancer patients. J Immunol. 2009;182:6562–6568. doi: 10.4049/jimmunol.0803831. [DOI] [PubMed] [Google Scholar]
- 33.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolchok JD, Neyns B, Linette G, Negrier S, Lutzky J, Thomas L, et al. Ipilimumab monotherapy in patients with pretreated advanced melanoma: a randomised, double-blind, multicentre, phase 2, dose-ranging study. The lancet oncology. 2010;11:155–164. doi: 10.1016/S1470-2045(09)70334-1. [DOI] [PubMed] [Google Scholar]
- 35.Yang A, Kendle RF, Ginsberg BA, Roman RA, Heine AI, Pogoriler E, et al. CTLA-4 blockade with ipilimumab increases peripheral CD8+ T cells: Correlation with clinical outcomes. J Clin Oncol. 2010;28 (suppl; abstr 2555) [Google Scholar]
- 36.Deichmann M, Benner A, Bock M, Jackel A, Uhl K, Waldmann V, et al. S100-Beta, melanoma-inhibiting activity, and lactate dehydrogenase discriminate progressive from nonprogressive American Joint Committee on Cancer stage IV melanoma. J Clin Oncol. 1999;17:1891–1896. doi: 10.1200/JCO.1999.17.6.1891. [DOI] [PubMed] [Google Scholar]
- 37.Solito S, Falisi E, Diaz-Montero CM, Doni A, Pinton L, Rosato A, et al. A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood. 2011;118:2254–2265. doi: 10.1182/blood-2010-12-325753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabitass RF, Annels NE, Stocken DD, Pandha HA, Middleton GW. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60:1419–1430. doi: 10.1007/s00262-011-1028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.