Abstract
Glioblastomas with EGFR amplification represent approximately 50% of newly diagnosed cases and recent studies have revealed frequent coexistence of multiple EGFR aberrations within the same tumor with implications for mutation cooperation and treatment resistance. However, bulk tumor sequencing studies cannot resolve the patterns of how the multiple EGFR aberrations coexist with other mutations within single tumor cells. Here we applied a population-based single-cell whole genome sequencing methodology to characterize genomic heterogeneity in EGFR amplified glioblastomas. Our analysis effectively identified clonal events, including a novel translocation of a super enhancer to the TERT promoter, as well as subclonal loss-of-heterozygosity and multiple EGFR mutational variants within tumors. Correlating the EGFR mutations onto the cellular hierarchy revealed that EGFR truncation variants (EGFRvII and EGFR Carboxyl-terminal deletions) identified in the bulk tumor segregate into non-overlapping subclonal populations. In vitro and in vivo functional studies show EGFRvII is oncogenic and sensitive to EGFR inhibitors currently in clinical trials. Thus the association between diverse activating mutations in EGFR and other subclonal mutations within a single tumor supports an intrinsic mechanism for proliferative and clonal diversification with broad implications in resistance to treatment.
Keywords: single-cell sequencing, single-nucleus sequencing, glioblastoma, EGFR, EGFR variant II
INTRODUCTION
Glioblastoma (GBM), also known as diffuse astrocytoma WHO grade IV, is the most common primary brain tumor in adults and a leading example of a tumor containing a high degree of cellular heterogeneity (1). Recurrent genetic alterations in GBM have been identified (2–4), with frequent amplification of several receptor-tyrosine kinases (RTK) including EGFR, PDGFRA, and MET. Genomic heterogeneity in tumors of the central nervous system, especially GBM, is an emerging problem with substantial ramifications for treatment strategies (5, 6). One of the most prominent examples is the mosaic pattern of different RTK amplifications present in different subpopulations of tumor cells (7, 8).
As the most frequent RTK amplification in glioblastoma, EGFR is focally amplified in about half of all primary glioblastoma and in 95% of the classical subtype (9). Recent reports by The Cancer Genome Atlas (TCGA) revealed unprecedented complexity at the focally amplified EGFR locus within individual tumors, including clusters of chromosomal breakpoints (10, 11), and the expression of multiple EGFR variants and extracellular domain missense mutations (4, 12). However, outside of the variant EGFRvIII and Carboxyl-terminal truncations, it is currently unknown whether many such variants are oncogenic and how such variants co-exist at a single cell level (13–15). Such patterns are important with regards to whether aberrations may functionally cooperate within individual cells or between cells with differing mutations. The functional importance of such distinctions is underscored by recent work showing the differential effects and interactions of variant receptors on EGFR activation within individual cells (16). Furthermore, extrachromosomal amplicons containing EGFRvIII (a constitutively active form of EGFR resulting from the deletion of exons 2–7) was recently shown to be dynamically lost and/or silenced in response to EGFR inhibitor treatment and then to re-emerge following removal of this selective pressure (5). The expression of EGFRvIII is also known to be regionally heterogeneous by immunohistochemistry (IHC) and RT-PCR, and it has been proposed that there may be functional synergy between tumor cells expressing the oncogenic variant (EGFRvIII) and those with the slower growing counterpart (EGFRwt) (5, 14, 17). However, whether the source of this heterogeneity at the single cell level that might support this has not been fully assessed.
Traditional statistical analysis of bulk average sequencing data cannot resolve this complexity at the EGFR locus because both the high copy number and the non-mitotic segregation of EGFR variants present in extrachromosomal amplicons prevent accurate clonality inference (18, 19). While FISH can assess the amplifications of different receptor tyrosine kinase genes (EGFR, PDGFRA and MET), it is difficult to distinguish between multiple variants of these oncogenes by FISH as it is generally limited to the characterization of known alterations spanning large genomic regions, and cannot probe single-nucleotide variants, short deletions, or discover de novo alterations.
Single-cell sequencing has great potential to dissect co-existing genomic alterations. Here we describe a population-based framework for single-cell genomic analysis and demonstrate its potential by dissecting the subclonal populations in two primary glioblastomas with focal EGFR amplifications. Our methodology employs a novel approach for confident inference of loss-of-heterozygosity and enables unambiguous characterization of the subclonal tumor cell populations with implications for EGFR targeted therapies in brain and other cancers.
RESULTS
Multiple somatic events can target EGFR in a single glioblastoma
To characterize the frequency of compound alterations of EGFR in glioblastoma, we examined the RNA-sequencing data of 76 cases of glioblastoma from TCGA with focally amplified EGFR (4, 12) and observed that 71% (54/76) had expression of wild-type EGFR along with at least one aberrant EGFR variant with 30% (23/76) expressing two or more variants. In 34% (28/76) of tumors, missense mutations involving the EGFR extracellular domain coexisted with structural variants. We then performed an exhaustive search of structural rearrangements of EGFR from whole-genome sequencing data in the 25 publicly available cases. Joint analysis of the DNA and RNA-sequencing data not only confirmed the presence of multiple variant EGFR transcripts, but also further revealed that the transcript encoding identical oncogenic EGFR variants could have been generated by distinct alterations at the DNA level.
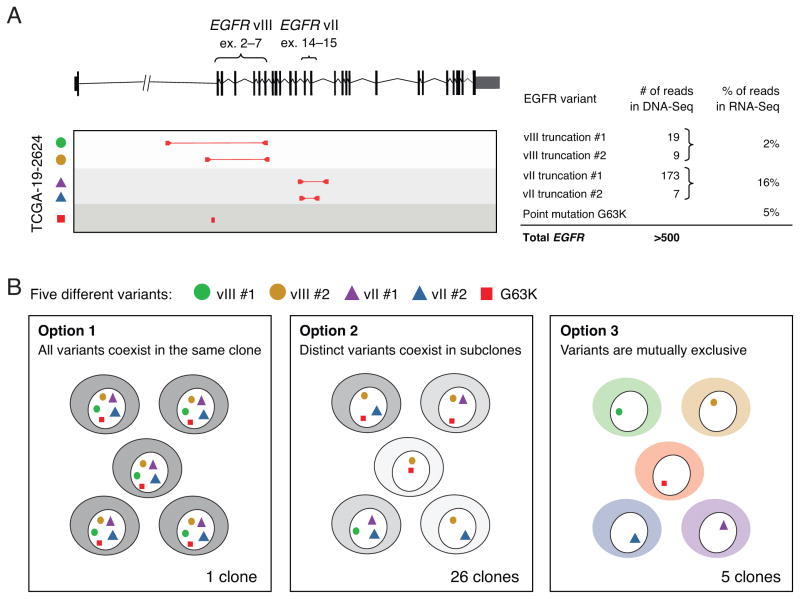
As an example, our analysis suggested that TCGA-19-2624 had expression of wild-type EGFR together with both EGFRvIII (deletion of exons 2–7) and EGFRvII (deletion of exons 14–15). At the DNA level, two distinct genomic rearrangements were found to give rise to EGFRvIII and two other events were found to generate EGFRvII (Fig. 1A). Co-expression of EGFRvII and EGFRvIII was previously described (20), but the frequency of variant reads suggested that EGFRvII is more prevalent in this sample. In addition, an extracellular domain missense mutation, G63K, was also observed at the DNA and RNA level. Based on the five EGFR aberrations alone, TCGA-19-2624 could exist as 32 possible combinations of cellular clones (25). Similar scenarios were discovered in additional samples (Supplementary Fig. 1A–D and Supplementary Table 1) with an overall prevalence of 44% (7 out of 16 with focal EGFR amplifications) harboring multiple EGFR mutations, including both rearrangements and single-nucleotide substitutions. Bulk analyses cannot resolve whether such heterogeneity commonly occurs in a single tumor clone, mutually exclusive clones, or other complex combinations of multiple, yet functionally cooperative variants (Fig. 1B). To answer this question, we sought to assess the clonal structure of EGFR aberrations by single-nucleus sequencing.
Figure 1. Genomic complexity of the EGFR locus in glioblastoma cannot be resolved using bulk tumor sequencing.
(A) RNA-Seq data from GBM patient TCGA-19-2624 revealed co-existence of multiple EGFR aberrations including EGFRvII (16% frequency), EGFRvIII (2% frequency), and a G63K mutation (5% frequency); analysis of whole-genome sequencing further revealed the presence of four distinct intragenic rearrangements producing two variants of EGFRvIII and two variants EGFRvII, each with different allelic frequencies as indicated by read counts. (B) The five distinct EGFR aberrations could result in 25=32 possible clone patterns at the cellular level. Two extreme cases are that all five variants are present in all cells (Option 1) or that the five variants each reside in different cells (Option 3); alternatively, the five variants can exist in up to 26 unique combinations at the cellular level (Option 2).
Single-nucleus sequencing approach
We selected for study two primary glioblastomas (BT325 and BT340) with focal EGFR amplification present as extrachromosomal amplicons based on prior clinical analysis. Bulk tumor and single-nucleus sequencing were performed to examine tumor heterogeneity within a single sampled region taken from each tumor (~1 cm3 in size).
The experimental and analytical framework is summarized in Fig. 2. Single nuclei from tumor resections were isolated by flow cytometry and a limited multiple displacement amplification (MDA) based whole-genome amplification was performed using a protocol that had been optimized for even genome coverage (see Online Methods). Approximately one hundred multiplexed single-nucleus sequencing (SNS) libraries were constructed and sequenced to low depth (~0.01x) for quality evaluation (Online Methods and Supplementary Fig. 2A–B). The quality assessment of single-cell DNA libraries requires an entirely new system of metrics than those utilized for bulk genomic DNA sequencing due to inherent challenges associated with starting with only 2 alleles of template DNA (21–23). Here we described two metrics, both of which can be estimated from low-pass sequencing (median depth ~0.01x): one directly measures the amplitude of over-amplification (whole-genome amplification pileup metric) and the other measures the unevenness of amplification (modified from Zong et al. (21). Both metrics showed consistent results (Supplementary Fig. 2C).
Figure 2.
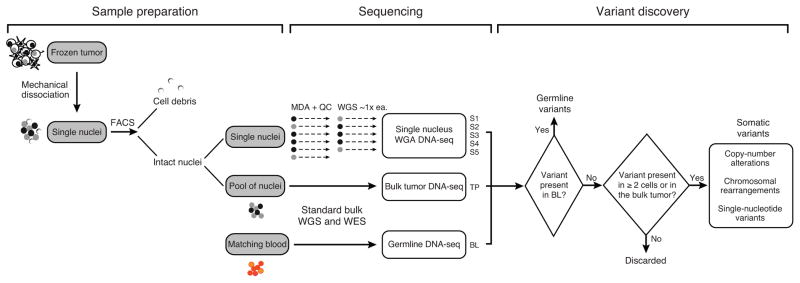
Experimental and analytical workflow of single-nucleus and bulk tumor sequencing for the characterization of tumor heterogeneity
The 50–60 best single-nucleus libraries were selected and sequenced to a combined depth of 30x. In parallel, whole-genome sequencing was performed on dissociated nuclear DNA from the tumor to serve as the bulk tumor reference and on matched blood DNA as germline reference. We performed joint analysis of single-nucleus and bulk DNA sequencing data to infer and guide the clonality estimates of somatic mutations and the subclonal tumor populations. In comparison to prior studies which only examined the single cell genomes, the use of joint analysis allowed us to more rigorously establish and confirm the presence of subclonal alterations using methods that have been previously established for bulk genome sequencing and validate findings from single-nucleus sequencing.
EGFR copy number is highly variable between single cells
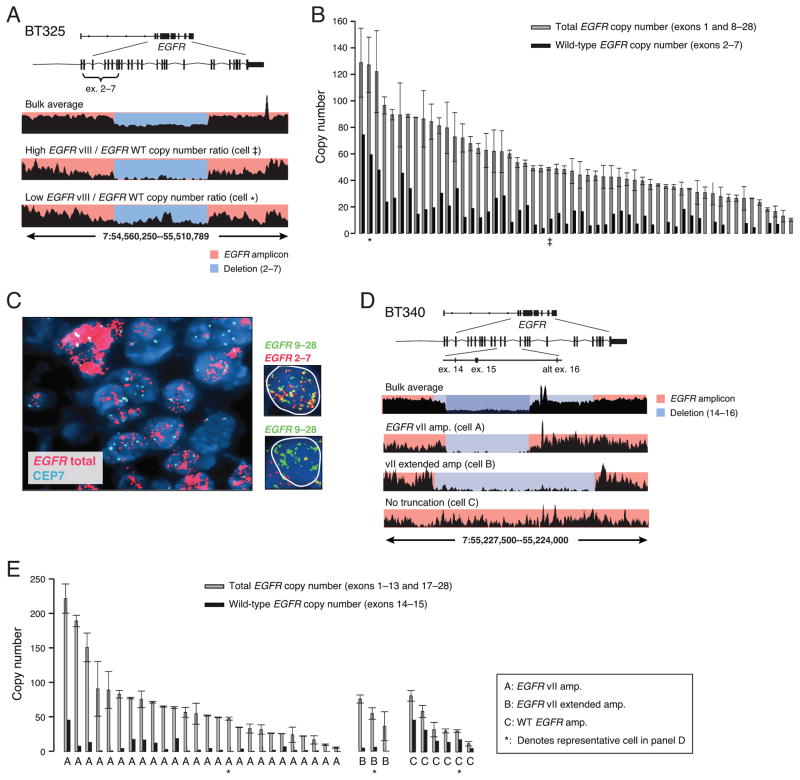
Sequencing of the bulk BT325 tumor revealed the presence of a 935kb amplicon containing four genes including EGFR, with an additional deletion corresponding to EGFRvIII (Fig. 3A and Supplementary Fig. 3A). Bulk tumor analysis suggested an approximate 50% reduction in read density within this region, consistent with a mixture of amplified wild-type EGFR and EGFRvIII deletion. A total of 60 single nucleus libraries from BT325 were sequenced and all the tumor-derived nuclei harbored the EGFR amplification as well as the vIII deletion (Fig. 3B). The common EGFR amplicon boundaries and EGFRvIII deletion breakpoints shared among tumor nuclei suggested that the deletion occurred after the wild-type EGFR amplification. The total EGFR copy number and the wild-type EGFR copy number estimated from the read depth demonstrated that each individual cell harbored amplifications of both wild-type EGFR as well as EGFRvIII, albeit at varying levels. To further validate our findings, we designed an oligonucleotide based multi-probe FISH assay that specifically discriminates between the exons deleted in the EGFRvIII variant and those retained. Application of this novel FISH approach to BT325 validated the large differences in total EGFR copy number including the presence of variation in levels of EGFRvIII amongst individual cells (Fig. 3C). Our observation of highly variable levels of EGFRvIII at the single-cell DNA level with respect to wild-type EGFR further supports a mechanism for local selective pressure of EGFR variants that may be associated with nutrient availability and diversification.
Figure 3. Heterogeneities in the copy number and mutation status of focally amplified EGFR in two glioblastomas.
(A) Read coverage plots of BT325 showing concurrent amplification of EGFR wild type and EGFRvIII alleles (bulk average). Single-nucleus sequencing plots show the same features as bulk but each cell varies in the ratio of EGFR wild-type and EGFRvIII amplification. (B) Quantified levels of total EGFR (exons 1, 8–28) and wild-type EGFR (exons 2–7) across all tumor nuclei using read depths from these regions. EGFRvIII (total minus wild-type) is co-amplified with wild-type in all tumor nuclei but to different degrees. The copy numbers of representative single-nucleus (SN) libraries shown in (A) are denoted by ‡ and ★. (C) Fluorescence in situ hybridization (FISH) for total EGFR and the chromosome 7 centromere (CEP7) confirming varying total EGFR copy number levels in BT325. SureFISH assay (far right nuclei) designed to specifically detect retained and deleted exons shows that individual cells range from low (top) to high levels of vIII deletion (bottom) (D) Read coverage plots of BT340 showing two overlapping deletions encompassing exons 14 and 15 (bulk average). Three representative SN libraries (lower tracks) show different mutation patterns. Cell A contained only EGFRvII truncation, cell B contained only the longer EGFRvII (vII-extended), and Cell C contained only wild-type EGFR. (E) Quantified levels of total EGFR (exons 1–13, 17–28) and wild-type EGFR (exons14–15) read depth within single tumor nuclei demonstrate three distinct populations containing amplified EGFRvII, EGFRvII-extended or wild-type EGFR. Discordant read pair signatures confirmed the presence of vII or vII-extended variants in population A and B; no discordant pairs were found in population C. Asterisks denote the representative nuclei displayed in (D).
Multiple EGFRvII variants exist in non-overlapping cell populations
We next performed sequencing of the BT340 tumor. This tumor harbored a 535kb amplicon containing the EGFR locus (Supplementary Fig. 3B). Bulk sequencing showed a marked decrease in read density in the region of exons 14 and 15 indicative of EGFRvII (Fig. 3D, top panel). An additional region of decreased read density indicative of a 7.6kb deletion that included alternative exon 16 was also found. Discordant read pairs in the bulk tumor sequencing data corroborated the presence of these deletions and defined their precise boundaries as being distinct (Supplementary Fig. 3C). We denoted this novel deletion variant as EGFRvII-extended.
Examination of SNS libraries showed three distinct cell populations with respect to exons 14–16 of the EGFR locus (Fig. 3D). EGFRvII and EGFRvII-extended segregated into two populations based on read depth and discordant read pairs corresponding to the rearrangements (Fig. 3D, Cell A and Cell B). Cells that did not harbor any discordant read pairs supporting either EGFRvII variant were inferred to retain these exons (Fig. 3D, Cell C). The initial amplicon was derived from a single copy of the wild-type allele (allele A) as assessed by germline SNVs; however, the exact number of copies per amplicon cannot be determined.
We further characterized the copy number of different EGFR variants within each individual cell. The total EGFR copy number was estimated from the read depths in exons 1–13 and in exons 17–28, and the wild-type EGFR copy number was estimated from the read depth in exons 14–15. In 35 of the 48 single nuclei libraries we were able to confidently assess the absolute number of EGFR copies. The total EGFR copy number showed an extremely broad range from 5 copies to more than 200 copies in different nuclei and was consistent with our FISH analysis (Fig. 3E, gray bars and Supplemental Fig. 3D). From 35 nuclei, we identified 26 containing EGFRvII and three EGFRvII-extended with no overlap. Further examination of the other six nuclei within the non-vII cluster revealed amplified EGFR, with three nuclei showing discordant read pairs spanning c-terminal breakpoints that occur in the presence of wild-type EGFR.
Determination of EGFRvII evolutionary hierarchy based on tumor subclones
The common genomic boundaries of the overall EGFR amplicon in all three populations in BT340 suggested that the clonal diversification occurred after the common ancestral EGFR amplification. Although the vII-extended and the vII deletion existed in distinct cell populations, their clonal association could not be unambiguously determined (e.g. the larger vII-extended deletion may theoretically be derived from the smaller vII). We therefore sought to characterize the clonal hierarchy based on co-existing alterations unrelated to EGFR in order to infer the clonal origin of the two EGFR variants.
Single-nucleus sequencing allows one to reconstruct the lineage evolution history in tumors using copy-number profiling and single-nucleotide variant clustering (24–27). However, the generic clustering approach previously employed in these studies cannot identify the exact clonal or subclonal alterations that differentiate subclonal populations as in bulk analysis (19, 28) and can be confounded by artifacts that are introduced during whole-genome amplification (22). To circumvent this limitation, we developed and applied an integrative approach to identify distinct tumor populations based on joint detection of clonal and subclonal events from the bulk tumor and single-nucleus genomes. Of particular importance, by requiring true variants to be present in at least 2 single-nucleus libraries, or with evidence in the bulk tumor DNA, we were able to exclude the majority of random MDA artifacts generated within individual libraries, and at the same time, increase detection sensitivity for subclonal events that are present in the bulk library at low allelic frequencies.
Profiling clonal aberrations increases sensitivity for detection of subclonal alterations
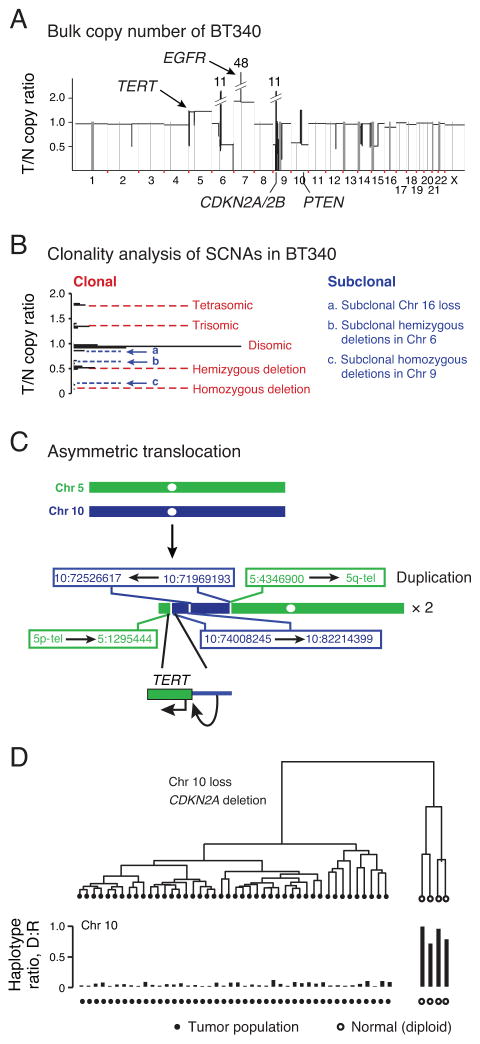
We first focused on clonal events inferred from bulk tumor analysis that could be used as markers for SNS analysis. In both BT340 and BT325 we identified characteristic glioblastoma somatic copy number alterations (SCNAs), including homozygous loss of CDKN2A/CDKN2B, hemizygous loss of chromosome 10, as well as gains of chromosome 7 (Fig. 4A–B and Supplementary Figure 4A–B). BT325 was found to have a common activating mutation within the promoter region of TERT (5:1295228, G->A) that is observed in >80% of glioblastomas (4, 29–31). In BT340, we observed a non-reciprocal translocation of a putative super enhancer within chromosome 10 (32) to the TERT promoter that could be an alternative mechanism for activating TERT in glioblastoma (Fig. 4C).
Figure 4. Characterization of clonal genomic alterations in single tumor nuclei from BT340.
(A) Bulk average somatic copy-number profile derived from whole-genome sequencing of GBM BT340 showing characteristic glioblastoma alterations. T/N= tumor vs. normal. (B) Clonality of somatic copy-number alterations inferred from the copy ratios (18). Clonal SCNAs generate integral copy-number alterations with peaks of copy ratios separated by equal spacing (red dashed lines); off-peak copy ratios corresponding to non-integral copy-number alterations suggest subclonal SCNA events (blue dashed lines). The clonality of subclonal SCNA events was later resolved at the single-cell level (see Fig. 5). (C) Schematic drawing of the inter-chromosomal rearrangements between Chr. 5 and Chr. 10 leading to TERT rearrangement and duplication. Translocation fuses a super enhancer element in Chr. 10 to the 5′ promoter region of TERT. (D) Hierarchical clustering of 48 nuclei from BT340 based on the haplotypes in chromosome 10. Forty-four nuclei (black circles) harbored loss-of-heterozygosity in Chr. 10 (with almost none of the deleted haplotype) and four nuclei (open circles) as heterozygous in Chr. 10 (1:1 ratio between the deleted and the retained haplotypes). The same two groups were also validated by clustering using other clonal aberrations (Supplementary Fig. 6).
We then characterized the SNS libraries of BT340 according to the clonal deletion events. In regions of hemizygous deletion or loss-of-heterozygosity (LOH), single-nuclei should only harbor one consensus haplotype. We utilized this feature to cluster individual nuclei based on the differences in their observed genotypes in LOH regions and infer the consensus haplotype within these regions (Supplementary Fig. 5A–H and Online Methods). Using the germline heterozygous sites to infer the haplotype information from chromosome 10, we clustered the 48 nuclei derived from BT340 into two groups (Fig. 4D). The 44 tumor-derived nuclei (black circles) showed minimal presence of the deleted haplotype (whole-genome amplification and sequencing errors) and the four normal diploid nuclei (open circles) exhibited roughly 1:1 ratio between the deleted and the retained haplotypes. The clustering of nuclei was further confirmed by haplotype analysis in hemizygously deleted regions in 9p and from the average copy ratio of chromosome 7, which is polysomic in tumor but disomic in normal (Supplementary Fig. 6A–C).
Haplotyping of 60 SNS libraries derived from BT325 based on deleted regions in chromosome 10 and chromosome 9p identified 56 libraries as tumor derived (Supplementary Fig. 7A–7C, middle panel). Of the other four libraries, two showed an approximate 1:1 ratio between the deleted and the retained haplotypes, consistent with having been derived from normal diploid cells (open circles); the other two exhibited a 1:2 ratio between the deleted and the retained haplotype, implying a 1:1 mixture of tumor and normal nuclei (Supplementary Fig. 7D and 7E).
The haplotype based method for clustering SNS libraries allowed for unprecedented sensitivity and specificity by utilizing a common event in cancer (loss-of-heterogeneity) rather than sequencing read depths that can be grossly distorted by whole-genome amplification. Moreover, haplotype-based clustering utilizes the digital nature of single-cell genotypes (in contrast with allele frequencies in bulk sequencing data) and the clonal structure of tumor populations to achieve high accuracy at low sequencing depths (~1x) even for libraries with high amplification bias.
Complex chromosomal rearrangements drive diversification
Chromothripsis has been observed to be a frequent event in glioblastoma and often results in the disruption of regions that undergo recurrent somatic copy-number alterations (10, 33). In both BT325 and BT340, we observed that homozygous inactivation of CDKN2A/CDKN2B resulted from two events, one a simple deletion and the other a complex genomic rearrangement event resembling chromothripsis. In BT325, the chromothripsis event involved chromosomes 1, 3, 8 and 9 and resulted in three derivative chromosomes, one of which was subsequently amplified (Supplementary Fig. 7F). In a 1.25Mb region encompassing the CDKN2A locus we observed alternating copy-number events between three states (0, 1, and 2) from the bulk read depth characteristic of chromothripsis. The single-nucleus sequencing coverage captured the retained regions after chromothripsis and the signal was not influenced by the background contribution of contaminating normal cells. In BT340, a complex double-minute amplification involving multiple segments of Chr. 6 and Chr. 9 resulted in homozygous inactivation of CDKN2A/2B (Supplementary Fig. 8A–E).
Subpopulations of BT325 are distinguished by a chromothriptic event
Clonality analysis of SCNA events from bulk tumor sequencing of BT325 suggested three subclonal deletions within chromosomes 1, 4 and 6 at copy ratio ~0.61 (Supplementary Fig. 4A–B). Zooming into these regions we observed alternating copy-number alterations involving chromosomes 1, 4, and 6 that is characteristic of chromothripsis (Supplementary Fig. 9A–E). Of the 56 tumor-derived nuclei, 46 nuclei (red circles) harbored hemizygous deletions in all the chromothriptic regions while six nuclei (blue circles) were heterozygous at these loci (Supplementary Fig. 9B). Four additional SNS libraries had a haplotype ratio suggesting a 1:1 mixture of the tumor clones (magenta circles). This “all-or-none” scenario of all deletions clearly established that these events had been acquired within a single event as implied in chromothripsis.
Based on the presence of shared clonal features such as EGFRvIII, TERT promoter mutations and loss of chromosome 10, but the absence of the chromosome 1, 4 and 6 chromothriptic event in six nuclei, we infer that the minor subclone split from the major subclone at this point (Supplementary Fig. 9F).
Subclonal deletions define two subpopulations in BT340
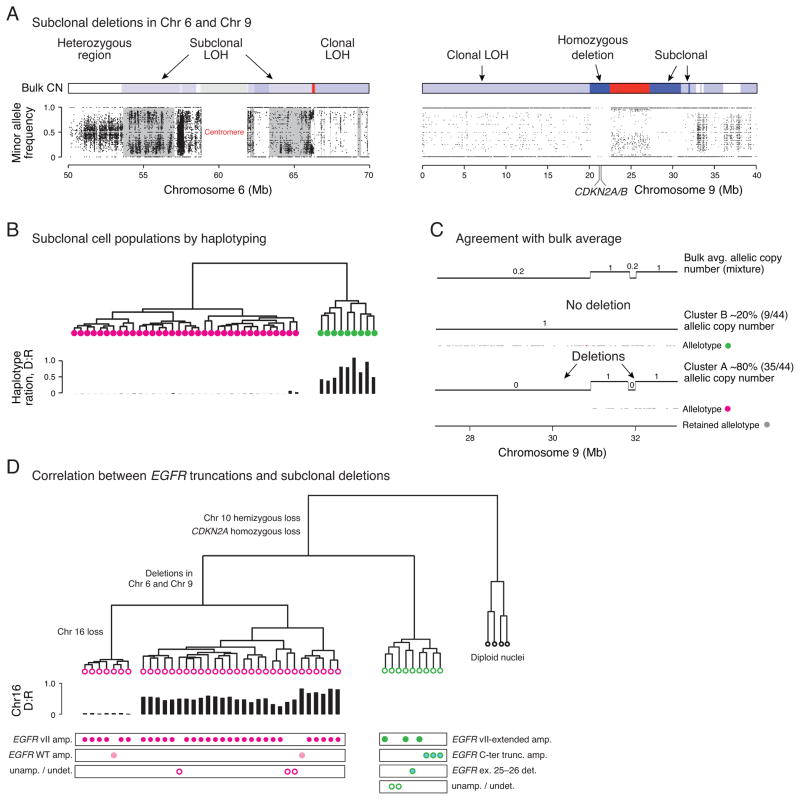
Analysis of SCNAs from the BT340 bulk tumor also showed copy ratios of approximately 0.20, 0.64, and 0.86 corresponding to non-integer copy numbers, indicating that such deletions were likely subclonal (Fig. 4A–B).
Regions of subclonal LOH were observed in chromosome 6p11.2 from 53.6 to 57 Mb, in 6q12 from 63.3 to 66.1 Mb, and in 9p21.1 from 27.1 to 30.9 Mb in the bulk tumor sequencing (Fig. 5A and Supplementary Fig. 6D). Clustering analysis of the 44 tumor-derived nuclei at these regions of chromosomes 6 and 9 resulted in two unique populations (Fig. 5B and Supplementary Fig. 6D). Cluster 1 (pink circles), consisting of 35 nuclei, was characterized by hemizygous deletions in 6p11.2, 6q12 and homozygous deletions in 9p21.1 (27.1–30.9 Mb, 31.8–32 Mb). The second cluster of nine nuclei (green circles) was characterized by the retention of segments from 6p11.2 and 6q12 and hemizygous deletion of segments from 9p21.1. The haplotype ratio between the deleted (D) and the retained (R) in hemizygously deleted regions in 6p11.2 confirmed the hemizygous loss of these regions in cluster 1.
Figure 5. Subclonal deletions delineate two tumor populations harboring distinct EGFR variants.
(A) Subclonal LOH in chromosomes 6 and 9 of BT340 is reflected by incomplete segregation of SNVs from the population of single tumor nuclei. Gray masking on the left panel indicates regions of subclonal LOH flanked by regions with clonal LOH in chromosome 6. Regions of subclonal homozygous deletions in chromosome 9 have a few heterozygous sites but there are none in regions of clonal homozygous deletion. (B) Hierarchical clustering of 44 tumor nuclei from BT340 based on the single-nucleus haplotypes in regions of subclonal LOH in Chr. 6 identifies two clusters with either loss or preservation of heterozygosity. (C) Two segments in 9p21 with bulk average copy number ~0.20 (top track) are resolved as a mixture of one subclone with 80% of tumor nuclei (35/44) having deletions in both regions. Gray dots show the retained haplotype that is only present in regions that are not deleted. (D) Correlation between subclonal populations as delineated by deletions in 6p11.2, 6q12 and 9p21.1 and distinct EGFR truncations suggest that the two distinct EGFRvII truncations independently emerged after clonal segregation determined by the deletion events.
We then compared the subclonal copy numbers statistically inferred from the bulk analysis to the clonal fractions from single-cell analysis. The bulk average allelic copy number of the subclonal deletion in 9p21.1 is ~0.2 after correcting for tumor purity (90%) and ploidy (2.1). Clustering of single tumor nuclei found 20% (9/44) retained these regions and 80% (35/44) harbored the deletion (Fig. 5C). The same agreement was observed for regions in chromosome 6 where the bulk average copy number is ~0.70. Finally, the copy ratio of 0.86 from the bulk analysis, corresponding to a subclonal deletion of chromosome 16 at ~19% clonal fraction, was associated to 7 out of 35 nuclei within cluster 1 with hemizygous loss (Supplementary Fig. 6E). The excellent concordance between the bulk clonality analysis and the single-cell analysis highlights the power of our population-based approach to robustly and efficiently resolve heterogeneous subclonal populations even at low clonal frequencies. The consistency between the clonality analysis of bulk sequencing and population analysis of single nuclei not only validates the statistical rationale behind bulk tumor analysis (34) but further sets a standard paradigm for single-cell based tumor analysis. Furthermore, single-cell analysis clearly resolved that the subclonal Chr. 16 loss occurred in a different population than the subclone that retained segments from Chr. 6 and 9, although their clonal fractions are similar (~19% and ~20%). Thus single-cell sequencing provides an unambiguous solution to associate independent subclonal alterations with distinct subclonal populations.
EGFRvII variants reside within distinct clones
Another application of single-cell sequencing in dissecting heterogeneous tumors is demonstrated by correlating alterations in amplified loci with other subclonal events, which is impossible to be inferred from bulk tumor sequencing data. Nucleus-by-nucleus comparison of EGFR mutation status to the clonal hierarchy constructed from subclonal deletions above revealed that EGFRvII and EGFRvII-extended were exclusively associated with only one of the two subclonal populations: EGFRvII was present only in cluster 1 with 6p11.2 and 6q12 deletions, while EGFRvII-extended was present only in cluster 2 that retained these chromosomal segments (Fig. 5D). In addition, cluster 2 also contained cells with a known oncogenic C-terminal truncation (ex. 25–28) (15) and a second C-terminal deletion (ex. 26–28), all of which occur in the context of wild-type EGFR amplification. Three cells from cluster 1 and two cells from cluster 2 were found to not contain focally amplified EGFR and thus could not be assigned as being a variant. In summary, 89% (39/44) of tumor cells were found to contain EGFR variants, with EGFRvII being the most common, followed by EGFRvII-ext., EGFR (del25–28), and EGFR (del25–26). Remarkably, all EGFR variants were mutually exclusive, suggesting that the EGFR mutations were most likely independently derived and expanded. In particular, the convergent evolution of EGFRvII variants suggested its oncogenic potential.
EGFRvII is oncogenic in the absence of EGFRvIII
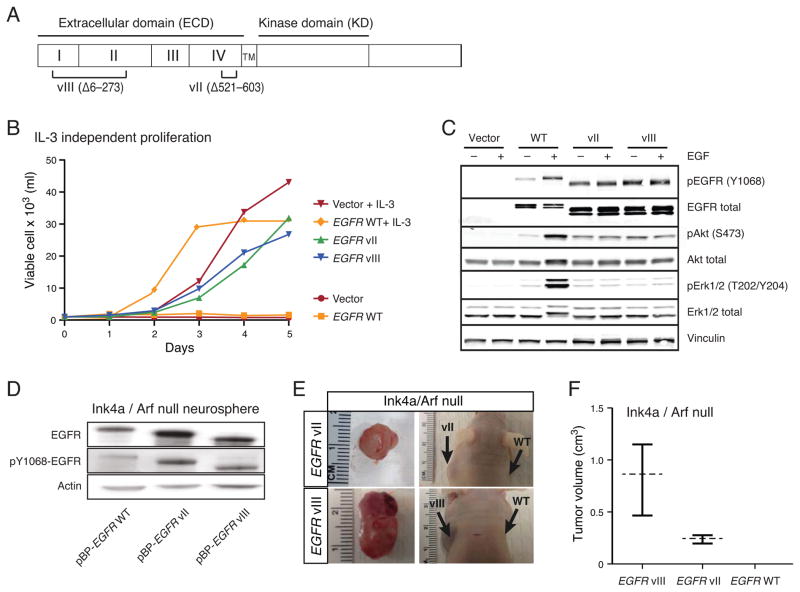
The EGFR variant II was initially described in GBM two decades ago, but was reported to be non-oncogenic (35, 36). Subsequent studies identified the presence of EGFRvII coexisting with EGFRvIII and thus EGFRvIII was inferred to be the driving oncogenic variant (20). However, in TCGA-19-2624, the EGFRvII variant was found to be more prevalent than EGFRvIII at both DNA and RNA levels, arguing in favor of its functional relevance. Furthermore, the convergence of independent clonal diversifications in BT340 resulting in EGFRvII and EGFRvII-extended further underscored its oncogenic potential. We therefore assessed the transformation potential of EGFRvII and the previously characterized EGFRvIII using two model systems (Fig. 6A).
Figure 6. EGFRvII transforms Ba/F3 and Ink4a/Arf−/− neural stem cells.
(A) Schematic representation of the EGFR vII and vIII mutants. (B) Proliferation of Ba/F3 cells stably containing empty vector or expressing EGFR wild-type, EGFRvII, or EGFRvIII were plated in the presence or absence of IL-3. Proliferation was measured over 5 days. (C) Immunoblot of Ba/F3 cells stably expressing empty vector, EGFR wild-type, EGFRvII or EGFRvIII. Cells were serum starved for 3 hours and then stimulated with 25 ng/ml of EGF for 10 min. (D) Immunoblot of Ink4a/Arf null neurospheres stably expressing EGFR wild-type, EGFRvII or EGFRvIII. (E) Xenografted Ink4a/Arf null neurospheres stably expressing EGFRvII or EGFRvIII form subcutaneous tumors after 6- or 11-weeks, respectively. (F) Measurement of the tumor volumes of subcutaneous xenografts stably expressing EGFRvII or EGFRvIII after 6- or 11-weeks respectively. Cells expressing wild-type EGFR failed to form tumors.
Ba/F3 cells are dependent upon supplemental IL-3 for survival but can be transformed to ligand independence by expression of oncogenic EGFR mutants (37, 38). Ectopic expression of EGFRvII and EGFRvIII permitted Ba/F3 cells to grow in the absence of IL-3, while cells containing the empty parental vector or wild-type EGFR did not survive (Fig. 6B). Constitutive phosphorylation of EGFRY1068 and AktS473 was observed in serum starved EGFRvII- and EGFRvIII-expressing cells demonstrating ligand independent activation (Fig. 6C).
To evaluate the oncogenic potential of the EGFRvII variant in a genetically relevant model of GBM, we expressed EGFR variants in E14.5 neural stem cells derived from Ink4a/Arf−/− mice (39). All variants demonstrated constitutive phosphorylation of EGFR as compared to wild-type EGFR expressing cells (Fig. 6D). Consistent with published reports, the expression of EGFRvIII induced tumor growth when subcutaneously grafted in Nude mice (Fig. 6E) (13). Expression of EGFRvII was also tumorigenic, albeit tumors developed at a slower rate (11 weeks for EGFRvII compared to 6 weeks for the EGFRvIII) (Fig. 6E–F). Histologically EGFRvIII and EGFRvII both generated high-grade gliomas, but we observed that the EGFRvII tumors exhibited oligodendroglial features and a corresponding lower rate of proliferation. These findings suggest that these similar mutations produce different functional effects.
To examine the drug sensitivities of EGFRvII and EGFRvIII, transformed Ba/F3 cells were tested for sensitivity to a panel of EGFR inhibitors. Ba/F3 cells dependent on EGFRvII were killed by erlotinib, lapatinib, afatinib and neratinib (Supplementary Fig. 10A–D), with irreversible inhibitors afatinib and neratinib exhibiting the lowest IC50 values for both EGFRvII and EGFRvIII expressing cells (Supplementary Fig. 10C–D). These results are similar to previous reports for EGFRvIII expressing Ba/F3 cells (38) and suggested that EGFRvII may be a therapeutic target for kinase inhibition.
DISCUSSION
Our identification of subclone-specific rearrangements within the EGFR gene expands upon the recent discoveries of heterogeneous genomic alterations in oncogenic kinase drivers in glioblastoma (7, 8, 14, 40). These results further suggest that a substantial fraction of the multiple EGFR variants reported in the TCGA glioblastoma collection may represent distinct subclonal populations within the tumor.
High-resolution genomic approaches have begun to reveal genomic complexity and heterogeneity to a degree not previously understood. These methods include clonality analysis of bulk tumor sequencing (18, 28, 41), regional sampling of tumors (6, 42, 43), and single-nucleus sequencing (24–26). Among these techniques, single-nucleus sequencing has the unique power to resolve cellular heterogeneity but can be technically challenging and difficult to interpret. Our methods for loss-of-heterozygosity detection by allele-specific delineation of haplotypes address limitations in genome-coverage due to MDA and expand upon the applications of single cell DNA analysis. As single-cell sequencing studies expand in size and scope, our methods provide the basis for maximizing sequencing information given the numbers of cells sequenced. Clonality inference by combining bulk tumor analysis with single-cell population analysis further provides a unique strategy for resolving clonal evolution and co-association of independent genomic alterations at the single cell level. The combination of SNS and joint calling with bulk sequencing allowed us to detect minor subclonal populations that were present in as low as 10% of the tumor at relatively low sequencing depth.
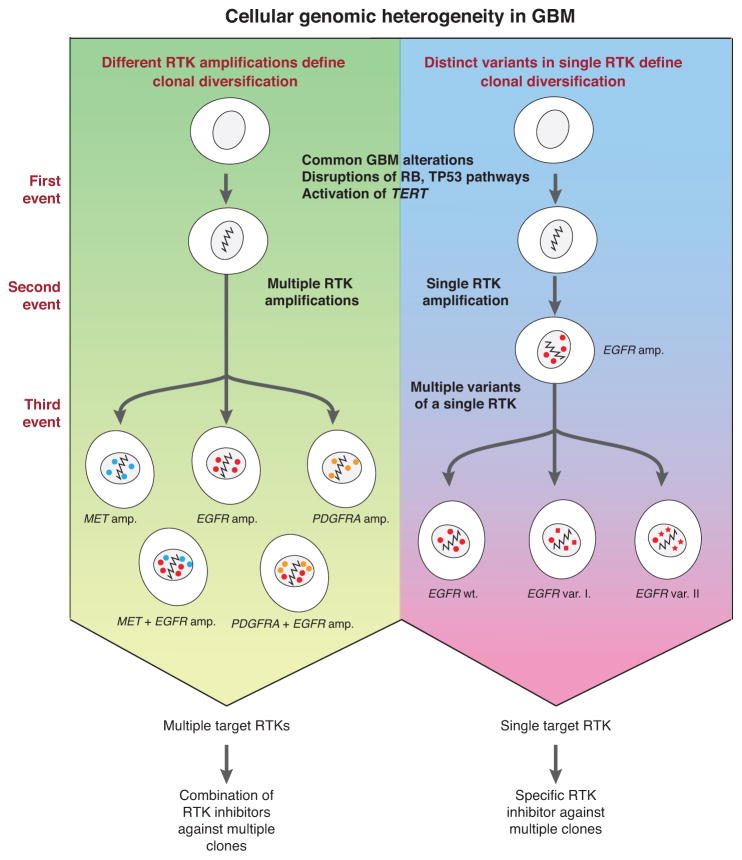
Genomic diversification in cancer is known to contribute to targeted and cytotoxic therapy failure and emergence of resistance (5, 44, 45). Large-scale genomic studies of cancer have outlined sets of alterations that impact growth promoting and tumor suppressing pathways that are necessary to drive tumorigenesis. In glioblastoma, the RB, TP53 and PTEN tumor suppressors are frequently inactivated directly or indirectly through aberrations in pathway molecules such CDKN2A/CDKN2B or MDM2 (4). These genomic alterations serve as the foundation upon which receptor tyrosine kinases like EGFR, MET, and PDGFRA can become amplified and evolve as the tumor cells begin to compete with each other for growth advantage. As FISH studies have demonstrated, RTKs within a tumor can be amplified alone or in combination (Fig. 7, left panel) (7, 8). Presumably clones with diverse growth signaling pathways will gain an advantage and expand as competition ensues.
Figure 7. Summary of cellular genomic heterogeneity in glioblastoma.
(Left panel) Different receptor-tyrosine kinase amplifications can lead to clonal diversification after common clonal mutations. (Right panel) Mutations in a single receptor-tyrosine kinase can also lead to clonal diversification through multiple variants, SNVs, and amplification of these different alleles. The presence of multiple concurrent RTK amplifications/mutations implies potential need for multiple drugs targeting the same RTK via different mechanisms.
Here our study reveals an additional paradigm for the diversification of growth signaling pathways through the acquisition of multiple variants of a single receptor tyrosine kinase gene (Fig. 7, right panel). The convergence of subclone-specific EGFRvII deletions reflects the dynamic evolution of amplified EGFR copies and pinpoints its oncogenic potential. We further demonstrated that over-expression of EGFRvII does cause transformation in both Ba/F3 and Ink4a/Arf−/− neural stem cell models, a variant previously thought not to be oncogenic because it co-occurred with EGFRvIII (20, 35). TCGA RNA-sequencing even revealed the expression of EGFRvII to be present in 9% (7/76) of focally amplified EGFR cases (4). Our functional characterizations found that constitutive expression of EGFRvII results in downstream activation of AKT signaling, consistent with that of EGFRvIII, but not with enhanced ERK activation. It is possible that EGFRvII activates an alternative pathway such as STAT3/5 and poses a more direct means of inducing transcriptional changes (16). Therefore, the presence of multiple EGFR variants in a single tumor highlights the intratumoral heterogeneity of glioblastoma conferred by the plasticity of EGFR amplicons.
Heterogeneous expression of oncogenic EGFR mutants and cooperativity of EGFR with itself or other receptor tyrosine kinases have each been hypothesized to contribute to tumor growth and resistance to therapy (14, 16, 40). It is conceivable that heterogeneity and cooperativity mechanisms play a role in RTK diversification and will impact the efficacy of targeted therapies in cancer, and in particular, may explain the lack of response in glioblastoma. Further complicating this scenario is recent work demonstrating how EGFRvIII containing cells can evade EGFR inhibitors through amplicon loss and down-regulation of the oncogenic receptor (5). It is striking that small molecule inhibitors can be so successful in non-small cell lung cancers, but remain ineffective in glioblastomas, despite EGFR being functionally significant (2, 3, 46–49). This work suggests that combining multiple EGFR inhibitors that act through different mechanisms on the receptor could be required in patients with multiple variants in non-overlapping subclones and also potentially prevent the activation of a resistance pathway. Such concepts deserve further evaluation in preclinical models.
As cancer genomic research approaches the discovery of the plurality of somatic mutations, it becomes crucial to understand the distribution pattern of somatic mutations among individual tumor cells and clones and how these mutations work together to drive tumor fitness (50). Single-cell analysis has the potential to greatly contribute to our understanding of co-dependency of mutations and tumor heterogeneity. The input requirements of this approach also make it feasible to deliver great insights into biopsies, necrotic tissues or tumors with great immune infiltration in order to delineate the full spectrum of cell-specific oncogenic drivers. Once a thorough understanding of the tumor state can be integrated, it is then possible to select combinations of drugs to prevent resistance due to emergence of pre-existing subclonal populations of tumor cells not detected in bulk tumor sequencing.
METHODS
Isolation of single nuclei and preparation of bulk tumor DNA
Anonymized tumor and matching blood specimens were collected following informed, written consent and processed under protocols approved by the Institutional Review Boards of the Dana-Farber Cancer Institute and the Broad Institute (protocol 11–433). Two cases of de novo GBM, designated as brain tumor BT325 and BT340, were mechanically dissociated into pools of nuclei as previously described (24). A portion of the dissociated material was selected to represent bulk tumor. Genomic DNA was isolated from the pooled nuclei and matched blood using DNeasy columns (Qiagen).
Preparation of single-nucleus DNA libraries
Single nuclei (SN) were sorted directly into 96-well PCR plates containing 5 μl of phosphate buffered saline using BD FACS Aria IIu flow cytometer calibrated to deposit single-nuclei into each well. A DAPI-based DNA stain was used to select intact nuclei. Empty wells were included on every plate to control for background MDA amplification. See expanded methods for additional details.
Quality assessment of single-nucleus DNA libraries
We generated single-nucleus DNA libraries from ~100 single-nuclei with the aim of selecting the best libraries from each tumor for deeper sequencing and analysis. Quality assessment was done by multiplexed sequencing of SN libraries to an average depth of 0.01x per library. Two criteria were employed to select the best SNS libraries for deeper sequencing.
First we removed libraries with more than 20% read sequences not aligned to the human reference. We then evaluated the evenness of the remaining MDA libraries by two metrics. The first metric was adapted from the method recently described (21) to measure the evenness of genome-wide coverage by evaluating the distribution of reads in a DNA library across different genomic loci stratified by the sequencing coverage at each locus. The second metric, termed the pileup metric, estimates the percentage of sequenced reads that were accumulated in local regions at densities 10 times higher than the chromosomal average. The spacing between each read and the adjacent reads (left and right) was used to infer if the read is in a pileup region. Reads in pileup regions were then downsampled to the average density estimated from the flanking non-pileup regions. The downsampling was iterated until no pileup region exceeding a certain size (e.g., 100 reads) was remaining. The pileup metric was calculated from the percentage of reads removed during the downsampling procedure.
The two metrics showed a strong correlation between each other (Supplementary Fig. 2C): Both can be used to estimate of the evenness of coverage for SNS libraries in order to guide the optimization of SNS workflow or select better libraries for deeper sequencing. In addition, removing the pileup reads can smooth the sequence coverage and reveal true copy-number profiles for each SNS library, which can be utilized to identify (tumor) samples of interest.
Two-step strategy for somatic variant detection by SNS
Whole-genome amplification by MDA is known to generate artifacts including single-base errors and random chimera, which can be indistinguishable from true somatic mutations. Given these considerations, we adopted a two-step strategy to analyze somatic mutations by SNS of many tumor nuclei. First, we detect all clonal and subclonal somatic variants, including single-nucleotide variants and chromosomal rearrangements, from the pool of SNS complemented with bulk whole-genome sequencing. Second, we search for the detected variants within each SNS and in the bulk tumor to infer the clonality of these variants and construct the clonal hierarchy of these variants. See expanded methods for additional details.
Detection of somatic copy number alterations
Whole-genome amplification by MDA from single-nuclear DNA can generate multiple distortions to the original DNA copy-number profile, which pose a considerable challenge to the detection of true somatic copy-number events in SN libraries. We therefore did not utilize the read depth signal from SN libraries to detect copy-number breakpoints but only inferred copy-number breakpoints from read depths in the bulk-tumor against the germline reference using SegSeq (51). At 10x sequencing depth, the read depth signal from bulk tumor should be sufficient to detect somatic copy-number alterations of 10kb or longer at or above 15% clonal fraction, which is comparable to the smallest subclone we can identify from 50–60 single cells.
As the read depth signal often resulted in over-segmentation, we merged the copy-number breakpoints inferred from the bulk tumor read depth with the list of non-reciprocal chromosomal translocation breakpoints detected from the pool of bulk tumor and SN libraries. We then identified true chromosomal breaks as supported both by read depth difference across the breakpoint and by the presence of somatic rearrangement signatures on the higher copy-number side. This final set of chromosomal breakpoints was then used to segment the copy-number profile for the bulk tumor as well as for each single nucleus.
Haplotype-based detection of loss-of-heterozygosity
A unique feature of many tumor genomes is the loss of heterozygosity of large chromosomal regions as well as smaller homozygous deletions, leading to the elimination of one or both germline haplotypes (52, 53) (Supplemental Fig. 4A). For cells with loss-of-heterozygosity (LOH) in a region of the genome, the retained haplotype, or allelotype (54), can be directly determined from the sequencing data (Supplemental Fig. 5B). If whole-genome amplification from single-cells were complete and represented all alleles, then the haploid genotype of cells with LOH could be readily distinguished from the heterozygous genotype of diploid cells (Supplemental Fig. 5B, left panel). However, single-cell sequencing is inevitably incomplete and not all alleles are represented in the resulting data (Supplemental Fig. 5B, right panel). Single-base errors introduced during MDA can further confound the inference of true genotype.
We devised a clustering-based approach to dissect the cell populations harboring heterozygous and/or homozygous genotypes in each genomic region. First, the pairwise distance between single-cell genotypes is calculated from the fraction of germline heterozygous sites covered in both libraries with different genotypes (Supplemental. 5C); this distance is large if cells are discordant for heterozygosity/homozygosity, and small if they are both homozygous (concordant). We then perform clustering analysis of cellular populations to identify cells with loss of heterozygosity (LOH) in a given region, which form a very tight cluster (Supplemental Fig. 5C, right panel). Supplemental Figure 5D showed the two clusters formed by 60 SNS libraries from BT325 identified by LOH in Chromosome 10, one cluster displaying a unique haploid genotype (LOH), and a second cluster that were heterozygous.
By pooling the genotype data of LOH cells in Cluster 1, we can determine both the retained haplotype “R” and the deleted haplotype “D” and assign individual or combined “RD” haplotypes to each nucleus at all covered germline heterozygous sites (Supplemental Fig. 5E). This analysis showed that Cluster 1 contains BT325 nuclei with only allele “R” while Cluster 2 contains an admixture of alleles “R” and “D” on chromosome 10 (Supplemental Fig. 5F). The ratio of the deleted to retained haplotypes is approximately 0 in LOH nuclei, and close to 1 in diploid nuclei. Interestingly, the D:R ratio is near 0.5 in another population of nuclei (Supplemental Fig. 5G).
We inferred that the population with D:R ratio of 0.5 represents libraries derived from a 1:1 mixture of tumor and normal nuclei. The normal/tumor mixture was later confirmed by the presence of clonal drivers at subclonal quantities in these samples (Supplemental Fig. 7). Therefore, the haplotype analysis also provides an accurate method to identify and characterize cellular mixtures in presumably single-nuclear samples.
Fluorescent in situ hybridization on FFPE tissue
FISH was performed on 5 micron formalin-fixed paraffin-embedded (FFPE) tissues, which were baked at 60°C for at least two hours, then de-paraffinized and digested using methods described previously (55). See supplemental experimental procedures for probes used in this study. Tissue sections and probes were co-denatured, hybridized at least 16 hrs at 37°C in a darkened humid chamber, washed in 2X SSC at 70°C for 10 min, rinsed in room temperature 2X SSC, and counterstained with DAPI (4′,6-diamidino-2-phenylindole, Abbott Molecular/Vysis, Inc.). Slides were imaged using an Olympus BX51 fluorescence microscope. Individual images were captured using an Applied Imaging system running CytoVision Genus version 3.9 and all aberrations detected by FISH were reviewed and confirmed by a cytogeneticist (AHL).
BAC probes in Fluorescent in situ hybridization
BAC probes were all obtained from CHORI BACPAC Resources Center, Oakland, CA, USA and included: clone RP11-815K24 (corresponding to EGFR, at 7p11.2), clone RP11-121K16 (corresponding to 6q13) and clone RP11-482I10 (mapping to 9p21.3). Commercially available probes (all obtained from Abbott Molecular) included CEP6 (6p11.1-q11), CEP7 (7p11.1-q11.1), and CEP9 (9p11-q11). Final homebrew probe concentrations were 100 nag/ul. Final concentrations for commercial reference probes followed manufacturer’s directions.
Agilent SureFISH
Specific detection of the EGFRvIII and EGFR wild-type alleles was performed using custom-designed pooled oligonucleotide probe sets (SureFISH) which tiled exons 2–7 (red probe, retained in wild type EGFR) and exons 9–28 (green probe, used to enumerate copies of both EGFR wild type and EGFRvIII). Hybridization was performed according to the manufacturers protocol (Agilent Technologies) on FFPE sections (4um) of BT325.
Expression Constructs
Wild-type EGFR and EGFR vIII p-BABE-puro vectors were prepared as previously described (38). QuikChange site-directed mutagenesis (Stratagene) was used for generating EGFR vII using wild-type EGFR as a template. All plasmids were confirmed by sequencing.
Cell culture and generation of cell lines by viral transduction
Ba/F3 cells were obtained, thawed and maintained as previously described (38). Proliferation assays were performed as previously described (37). The BaF3 cells were confirmed to be the parental line through a cell viability test following removal of IL-3.
Mouse neural stem cells were derived from ganglionic eminences of E14.5 p16Ink4a:p19Arf null embryos were obtained and genotyped by qPCR of genomic DNA as described (13, 39). Cells were transduced by EGFR wild-type, EGFR vII or EGFR vIII p-BABE retrovirus and selected with puromycin after 3 days for 2 passages. Transduction of the EGFR variant was confirmed by immuno blot. For subcutaneous xenograft studies, EGFRvII or EGFRvIII transduced mouse neural stem cells (106 cells) were injected in the left flank of nude mice with EGFRwt injected on the right as control. Five mice were injected for each EGFR variant. All animal experiments were performed in accordance with Dana-Farber Cancer Institute animal facility regulations and policy under the protocol #09-016.
Drug sensitivity assay
For growth inhibition assays, Ba/F3 cells (10,000 cells) were plated in 180 μL media in 96-well flat-bottom plates. Twenty-four hours after plating, drugs prepared in 20 μL cell culture medium were added to cells. All drugs were purchased from LC Laboratories. The concentrations of erlotinib and lapatinib used for the assay ranged from 0.1 nM to 10 μM. The concentrations of afatinib and neratinib ranged from 0.001 to 100 nM. The cells were incubated for another 72 hrs and the viable cell numbers were measured using CellTiter Glo (Promega).
Immunoblotting and antibodies
Cultures were serum-starved for 3 hours prior to EGF stimulation and harvesting. Epidermal growth factor (EGF, PeproTech) stimulations were performed using 25 ng/ml for 10 minutes. Anti-EGFR, phospho-EGFR (pY1068), AKT, phospho-AKT (pS473), ERK1/2 and phospho-ERK1/2 (pT202/Y204) antibodies were purchased from Cell Signaling Technology. Anti-vinculin was from Sigma-Aldrich.
Supplementary Material
SIGNIFICANCE.
We developed a novel single-cell sequencing methodology capable of identifying unique, non-overlapping subclonal alterations from archived frozen clinical specimens. Using GBM as an example, we validated our method to successfully define tumor cell subpopulations containing distinct genetic and treatment resistance profiles and potentially mutually cooperative combinations of alterations in EGFR and other genes.
Acknowledgments
Financial Support: This work was supported by the Bridge Project, a collaboration between The David H. Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center (DF/HCC) and the National Brain Tumor Society to (J.C.L, K.L.L and M.M.). This work was supported in part by grants to K.L.L. from NCI R01CA170592, P01CA142536 and the Sontag Foundation and to M.M. from NCI RO1CA116020.
We thank Dr. Juliann Chmielecki for generating the EGFRvII expression construct and Drs. Scott Carter and Marcin Imielinski for helpful discussions. We are thankful to the BWH Tissue Biorepository Core (Dr. William Richards, Director) and the Department of Pathology faculty and staff of the DFCI Tissue Bank for their help and dedication. This work was supported by the Bridge Project, a collaboration between The David H. Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center (DF/HCC) and the National Brain Tumor Society to (J.C.L, K.L.L and M.M.). This work was supported in part by grants to K.L.L. from NCI R01CA170592, P01CA142536 and the Sontag Foundation and to M.M. from NCI RO1CA116020.
Abbreviation List
- EGFR
Epidermal Growth Factor Receptor
- EGFRvII
EGFR variant II
- EGFRvIII
EGFR variant III
- GBM
Glioblastoma
Footnotes
Conflict of Interest: M.M. is a founder and equity holder of Foundation Medicine, a for-profit company that provides next-generation sequencing diagnostic services.
Competing Interests
M.M. is a founder and equity holder of Foundation Medicine, a for-profit company that provides next-generation sequencing diagnostic services.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–7. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–6. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–29. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanborn JZ, Salama SR, Grifford M, Brennan CW, Mikkelsen T, Jhanwar S, et al. Double minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res. 2013;73:6036–45. doi: 10.1158/0008-5472.CAN-13-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng S, Fu J, Vegesna R, Mao Y, Heathcock LE, Torres-Garcia W, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013 doi: 10.1101/gad.213686.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–77. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 14.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–45. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho J, Pastorino S, Zeng Q, Xu X, Johnson W, Vandenberg S, et al. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer Res. 2011;71:7587–96. doi: 10.1158/0008-5472.CAN-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA, et al. EGFR Phosphorylates Tumor-Derived EGFRvIII Driving STAT3/5 and Progression in Glioblastoma. Cancer Cell. 2013;24:438–49. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishikawa R, Sugiyama T, Narita Y, Furnari F, Cavenee WK, Matsutani M. Immunohistochemical analysis of the mutant epidermal growth factor, deltaEGFR, in glioblastoma. Brain Tumor Pathol. 2004;21:53–6. doi: 10.1007/BF02484510. [DOI] [PubMed] [Google Scholar]
- 18.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–7. [PubMed] [Google Scholar]
- 21.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voet T, Kumar P, Van Loo P, Cooke SL, Marshall J, Lin ML, et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Aa N, Cheng J, Mateiu L, Esteki MZ, Kumar P, Dimitriadou E, et al. Genome-wide copy number profiling of single cells in S-phase reveals DNA-replication domains. Nucleic Acids Res. 2013;41:e66. doi: 10.1093/nar/gks1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–85. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–95. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potter NE, Ermini L, Papaemmanuil E, Cazzaniga G, Vijayaraghavan G, Titley I, et al. Single cell mutational profiling and clonal phylogeny in cancer. Genome Res. 2013 doi: 10.1101/gr.159913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 31.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphrey PA, Gangarosa LM, Wong AJ, Archer GE, Lund-Johansen M, Bjerkvig R, et al. Deletion-mutant epidermal growth factor receptor in human gliomas: effects of type II mutation on receptor function. Biochem Biophys Res Commun. 1991;178:1413–20. doi: 10.1016/0006-291x(91)91051-d. [DOI] [PubMed] [Google Scholar]
- 36.Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89:2965–9. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Greulich H, Janne PA, Sellers WR, Meyerson M, Griffin JD. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–74. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- 38.Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2006;103:7817–22. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 40.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 41.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–14. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–6. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 46.Yeh P, Chen H, Andrews J, Naser R, Pao W, Horn L. DNA-Mutation Inventory to Refine and Enhance Cancer Treatment (DIRECT): A Catalog of Clinically Relevant Cancer Mutations to Enable Genome-Directed Anticancer Therapy. Clin Cancer Res. 2013;19:1894–901. doi: 10.1158/1078-0432.CCR-12-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med. 2004;351:2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 48.Lee JC, Vivanco I, Beroukhim R, Huang JH, Feng WL, DeBiasi RM, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL, et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–71. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang DY, Getz G, Jaffe DB, O’Kelly MJ, Zhao X, Carter SL, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–21. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 53.Kern SE, Fearon ER, Tersmette KW, Enterline JP, Leppert M, Nakamura Y, et al. Clinical and pathological associations with allelic loss in colorectal carcinoma [corrected] Jama. 1989;261:3099–103. doi: 10.1001/jama.261.21.3099. [DOI] [PubMed] [Google Scholar]
- 54.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, et al. Allelotype of colorectal carcinomas. Science. 1989;244:207–11. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 55.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–51. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.