Abstract
A conserved stem-loop motif of the constitutive decay element (CDE) in the 3′ UTR of mRNAs is recognized by the ROQ domain of Roquin, which mediates their degradation. Here we report two crystal structures of the Homo sapiens ROQ domain in complex with CDE RNA. The ROQ domain has an elongated shape, with three sub-domains. The 19-nt Hmgxb3 CDE is bound as a stem-loop to domain III. The 23-nt TNF RNA is bound as a duplex, to a separate site at the interface between domains I and II. Mutagenesis studies confirm that the ROQ domain has two separate RNA binding sites, one for stem-loop RNA (A site) and the other for dsRNA (B site). Mutation in either site perturbs the Roquin-mediated degradation of HMGXB3 and IL-6 mRNAs in human cells, demonstrating the importance of both sites for mRNA decay.
The cellular levels of mRNAs are regulated through many different mechanisms, which control their biosynthesis, stability and degradation. For example, microRNAs can recognize appropriate sequence motifs in the mRNAs and lead to their destruction and/or repression of their translation by the ribosome. Other sequence elements in the mRNAs can recruit protein factors to regulate their levels. A well-known example is the AU-rich element (ARE) found in the 3′ UTR of many cytokine, proto-oncogene and transcription factor mRNAs 1-3. The ARE mediates the rapid decay of these mRNAs by recruiting deadenylation and decapping enzymes as well as other proteins, in response to defined cellular signals. Therefore, the ARE functions primarily as a regulated decay element for these mRNAs.
Recent studies have identified a constitutive decay element (CDE) in the 3′ UTR of tumor necrosis factor α (TNF-α), inhibitor of κB (IκB), inducible T cell costimulator (ICOS), and other mRNAs 4,5. The CDE mediates the post-transcriptional, microRNA-independent repression of these mRNAs, irrespective of the cellular environment. A conserved stem-loop motif in the CDE is recognized by the ROQ domain of paralogous proteins Roquin and Roquin-2 (ref. 5), which recruit the deadenylation and decapping machineries to initiate mRNA degradation 5,6. Roquin has a central role in repressing autoimmunity, and mice deficient in both Roquin and Roquin-2 in T cells or carrying a single-point mutation in the ROQ domain of Roquin (M199R, sanroque mutation) have increased ICOS expression and exhibit a lupus-like autoimmune phenotype 6-11. On the other hand, overexpression of Roquin in T cells promotes the development of arthritis in a mouse model 12. Roquin knockout mice show perinatal lethality but not autoimmunity, suggesting that Roquin may have important developmental functions as well 9,13.
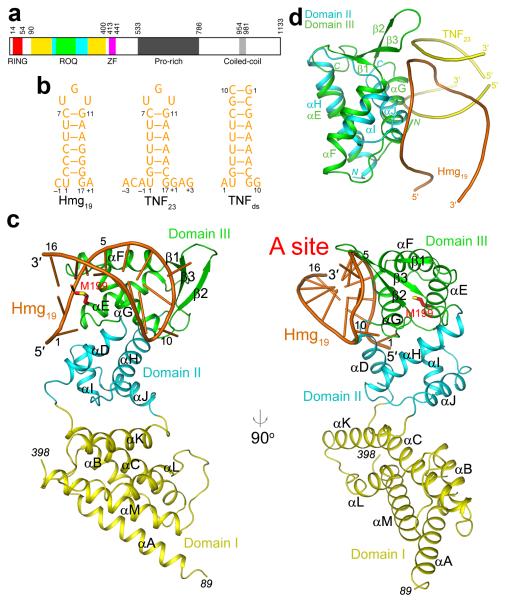
Human Roquin contains 1133 amino acid residues (125 kD, Fig. 1a). The N-terminal RING finger, ROQ and CCCH-type zinc finger (ZF) domains are well conserved among metazoans (Supplementary Fig. 1). The ROQ domain does not share recognizable sequence conservation with other proteins. It is required for binding the CDE, while the RING and ZF domains are dispensable for this interaction 5. The ROQ domain of Roquin-2 is 88% identical to that of Roquin (Supplementary Fig. 1) and Roquin-2 can complement Roquin’s function in repressing autoimmunity 9,10. The C-terminal segment of Roquin following the ZF is poorly conserved in sequence (Supplementary Fig. 1), but it recruits the deadenylation machinery and is required for CDE-mediated decay 5,6.
Figure 1. Structure of the ROQ domain of human Roquin in complex with the constitutive decay element (CDE) of Hmgxb3 mRNA.
(a). Domain organization of human Roquin. Domains I, II and III of the ROQ domain are colored yellow, cyan and green, respectively. ZF: zinc-finger. (b). Schematic drawing of three RNA molecules used in this study, Hmg19, TNF23, and TNFds. The numbering scheme of the nucleotides is also shown. (c). Two views of the structure of the ROQ domain of human Roquin in complex with Hmg19. The three sub-domains of the ROQ domain are colored in yellow, cyan and green, respectively, and the RNA in orange. The side chain of Met199 (site of the sanroque mutation) is shown in red for carbon atoms. (d). Overlay of the structures of domain III of ROQ domain (green) in complex with Hmg19 (orange) and domain II of ROQ domain (cyan) with TNF23 duplex (yellow). All the structure figures were produced with PyMOL (www.pymol.org).
To understand the molecular basis for CDE recognition by the ROQ domain, we have determined the crystal structures of this domain from human Roquin in complex with two different CDEs, from Hmgxb3 and TNF mRNAs. The ROQ domain is actually composed of three sub-domains, I, II and III, and the structural analysis reveals previously unrecognized similarity of domains II and III to the helix-turn-helix (HTH) and winged-helix (WH) motifs. The Hmgxb3 CDE stem-loop interacts with the WH motif of domain III, which primarily recognizes the 5′ arm and the loop of the RNA (A site). Two bases of the 3-nt loop are flipped out to make close contacts with the protein. Unexpectedly, the TNF RNA is bound as a duplex, due to its higher AU content in the stem and hence lower stability of the stem-loop conformation. This RNA is located in a separate binding site (B site), at the interface between domains I and II. Our mutagenesis, biochemical and cellular experiments confirm the importance of both RNA binding sites for the functions of Roquin in mediating mRNA decay.
Results
Overall structure of the ROQ domain
We determined the structure of the ROQ domain of human Roquin in complex with a 23-nt CDE of human TNF-α (TNF23) at 1.9 Å resolution (Table 1). TNF23 contains the 17-nt conserved stem-loop as well as 3-nt flanking sequences at its 5′ and 3′ ends (Fig. 1b) 5. We next determined the structure of the ROQ domain in complex with a 19-nt CDE of mouse Hmgxb3 (Hmg19) at 2.9 Å resolution, in a different crystal form. This CDE is more GC rich in the stem but has the same 3-nt loop as TNF23 (Fig. 1b). Efforts at crystallizing the ROQ domain alone have not been successful.
Table 1. Data collection and refinement statistics.
| ROQ-Hmg19 complex | ROQ-TNF23 complex | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 49.6, 170.0, 51.1 | 90.8, 93.1, 100.2 |
| α, β, γ (°) | 90, 97.0, 90 | 90, 90, 90 |
| Resolution (Å) | 40–2.9 (3.0–2.9) 1 | 40.0–1.9 (2.0–1.9) |
| Rmerge (%) | 9.4 (41.5) | 6.9 (43.9) |
| I / σI | 9.1 (1.7) | 19.4 (3.4) |
| Completeness (%) | 97 (88) | 100 (100) |
| Redundancy | 2.1 (1.9) | 3.8 (3.8) |
| Refinement | ||
| Resolution (Å) | 40–2.9 | 40–1.9 |
| No. reflections | 17,984 | 128,927 2 |
| Rwork / Rfree | 19.5 / 25.1 | 17.7 / 21.2 |
| No. atoms | ||
| Protein | 4701 | 4783 |
| Nucleic acid | 678 | 767 |
| Ligand/ion | 8 | 20 |
| Water | 2 | 885 |
| B factors (Å2) | ||
| Protein | 54.4 | 22.1 |
| Nucleic acid | 51.1 | 45.3 |
| Ligand/ion | 57.9 | 34.0 |
| Water | 41.9 | 34.0 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.011 |
| Bond angles (°) | 1.4 | 1.2 |
One crystal was used for each data collection.
Values in parentheses are for highest-resolution shell.
The Friedel pairs were kept as separate reflections during the refinement.
The structure of the ROQ domain is mostly helical, with 13 helices (αA-αM) and a small three-stranded β-sheet (β1-β3) (Fig. 1c). The structure has an elongated shape, with dimensions of approximately 25 Å × 40 Å × 75 Å, and can be divided into three sub-domains. Sub-domain I has six tightly packed anti-parallel helices, three from the N-terminal end of the ROQ domain (αA-αC) and three from the C-terminal end (αK-αM). The ROQ domain has generally been defined in the literature as containing residues 131-360. Our structure shows however that the domain actually covers residues 90-400 (Fig. 1a), and the extra ~40 residues at the N and C termini contribute helices αA and αM, respectively.
Sub-domain III is formed by residues in the middle of the ROQ domain (195-271) and contains three helices (αE-αG) and the three-stranded β-sheet (Fig. 1c). Unexpectedly, the backbone fold of this domain is similar to that of the winged-helix motif, especially the WH-B domain of the RING ubiquitin-protein ligase cullin 14-16 and related proteins 17. For example, the rms distance among equivalent Cα atoms of this domain and the WH-B domain of cullin 1 is 2.3 Å (Supplementary Note 1, Supplementary Fig. 2), although the amino acid sequence identity of the aligned residues is only 11%, based on an analysis with the program DaliLite (Z score of 9.2) 18. Another structural homolog of this domain is the WH motif of elongation factor SelB, which is responsible for selenocysteine incorporation during translation 19 (Supplementary Fig. 2). The rms distance is 2.7 Å and the sequence identity is 13% (Z score of 5.6).
Sub-domain II, with four helices (αD, αH-αJ), connects domains I and III, and there are no direct contacts between them (Fig. 1c). It has tight contacts with domain III, with ~900 Å2 surface area burial, whereas the interface with domain I is more limited (~200 Å2 surface area burial) and mostly hydrophilic in nature. To our surprise, helices αH-αJ form another helix-turn-helix (HTH) motif in the ROQ domain, and its structural homologs include other HTH and winged-helix proteins, such as the Zα domain of human adenosine deaminase (ADAR1, Supplementary Fig. 2) 20. The rms distance is 1.8 Å for 48 equivalent Cα atoms, and the sequence identity is 14% (Z score of 4.9).
In fact, the arrangement of helices αH-αJ in domain II is also similar to that of helices αE-αG in domain III, even though domain II lacks the three-stranded β-sheet (the ‘wing’) compared to domain III (Fig. 1d, Supplementary Note 1). The rms distance is 2.1 Å for 44 equivalent Cα atoms, and the sequence conservation is 16% (Z score 3.8, indicating a more remote similarity between the two domains). Helix αD of domain II is not a part of the HTH motif but has close interactions with helices αH and αI.
Binding mode of the Hmgxb3 CDE
We chose the Hmgxb3 CDE for our studies as it has four G-C base pairs in the stem and is therefore expected to be more stable as a stem-loop than the TNF CDE (Fig. 1b). The stem of Hmg19 contains six Watson-Crick base pairs and assumes a slightly distorted A-form RNA structure (Fig. 2a). A U1-G17 wobble at the base of the stem was proposed in an earlier report 5, but this base pair was not formed and in fact G17 is disordered (Fig. 2a). These two nucleotides are present in TNF and Hmgxb3 but are not conserved in the other CDEs (Supplementary Fig. 3).
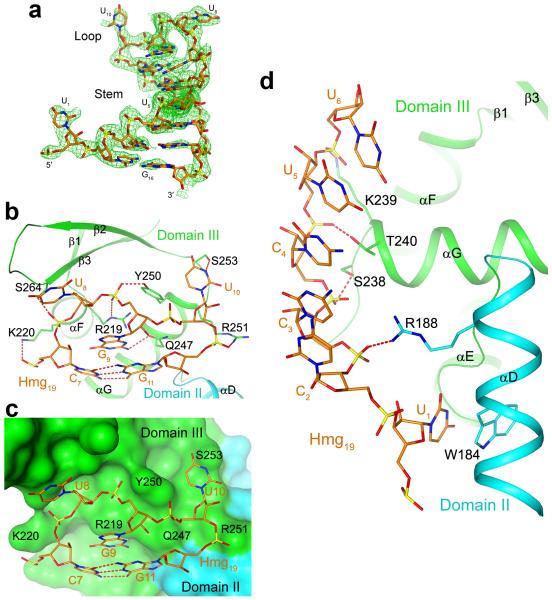
Figure 2. Interactions between the Hmg19 CDE and the ROQ domain.
(a). Composite omit Fo-Fc electron density map at 2.9 Å resolution for the Hmg19 RNA, contoured at 3σ. (b). Interactions of the 3-nt UGU loop and the top base pair of the stem with domain III of the ROQ domain. Hydrogen bonds and ionic interactions are shown as red dashed lines. (c). Molecular surface of the ROQ domain near the binding site for the 3-nt UGU loop, showing the close contacts between domain III of the ROQ domain and the tri-loop. (d). Interactions between the 5′ arm and U1 of Hmg19 with domains III and II of the ROQ domain.
The majority of the interactions between Hmg19 and the ROQ domain are mediated through the 3-nt loop and the 5′ arm of the stem, which contact primarily domain III (helices αF and αG of the HTH motif and the β2-β3 loop of the wing) of the ROQ domain (Figs. 2b-d). In contrast, the 3′ arm of the stem has few contacts with the protein and weaker electron density (Fig. 2a).
We will refer to this binding site for the Hmg19 RNA as the A site (Fig. 1c). It is located in an electropositive surface patch in the ROQ domain (Supplementary Fig. 4). The protein-RNA interface involves hydrogen-bonding and ionic interactions with the backbone phosphate groups of the RNA, and π-stacking interactions for a few of the bases (Supplementary Note 2). Both pyrimidine bases of the U8G9U10 tri-loop are flipped out and have close contacts with the protein (Figs. 2b, 2c). The base of G9 is stacked with the last base pair of the stem as well as the guanidinium group of Arg219 (αF). The base of U1 is flipped ~180° from the helical pattern of the stem (Fig. 2a) and is π-stacked with the side chain of Trp184, in helix αD of domain II (Fig. 2d).
Amino acids in this interface are highly conserved among the ROQ domains (Supplementary Fig. 1). However, none of the nucleotides of Hmg19 appears to be recognized specifically by the A site. On the other hand, this site recognizes the shape of the 3-nt loop (Fig. 2c) and is unlikely to accommodate other types of loops. This provides a degree a selectivity in the interactions between the ROQ domain and Hmg19, and explains the fact that all known CDEs have a tri-loop with pyrimidines at the first and third positions and purine at the second position 5.
Binding mode of the TNF23 RNA
Unexpectedly, our crystallographic analysis revealed that the TNF23 RNA is not in a stem-loop conformation in the crystal. Instead, we found the two TNF23 molecules in an anti-parallel, double helical structure, formed by the stem regions of the two RNAs (Supplementary Fig. 5). The 3-nt loop gives rise to an unpaired region in the middle of this duplex, which has weaker electron density, suggesting that this region is somewhat flexible. In fact, one nucleotide in this region (U10) is disordered in the crystal (Supplementary Fig. 5). Our further experiments showed that the formation of this duplex was due to the lower stability of the stem-loop conformation (low GC content) and the high concentration that was needed for crystallization (Supplementary Note 3). Under physiological conditions, the TNF CDE exists as a stem-loop and is expected to be recognized by Roquin through the A site in domain III.
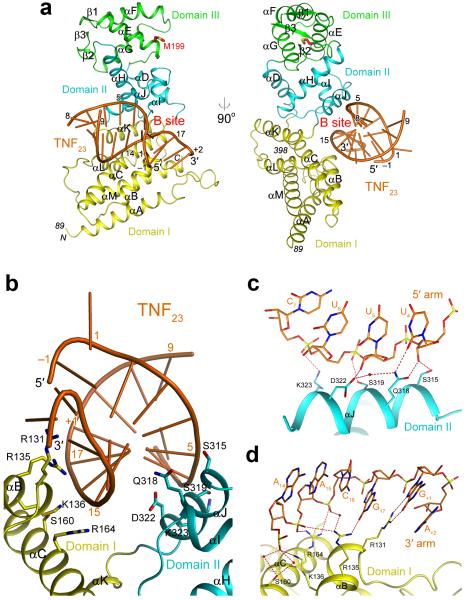
The TNF23 duplex contains six Watson-Crick base pairs from each of the stem regions of the TNF23 RNA (Supplementary Fig. 5). The two ROQ domains in the asymmetric unit have contacts with the base-paired regions of the TNF23 RNA but have no interactions with the nucleotides in the loop region in the middle of the duplex (Supplementary Fig. 5). The RNA is positioned in an electropositive surface depression at the interface of domains I and II (Supplementary Fig. 4), and we will refer to this binding site as the B site (Fig. 3a).
Figure 3. Structure of the ROQ domain of human Roquin in complex with the CDE of TNF-α mRNA.
(a). Two views of the structure of the ROQ domain of human Roquin in complex with the TNF23 RNA. Only half of the TNF23 RNA duplex is shown. (b). Overview of the interface between TNF23 RNA (orange) and the ROQ domain (yellow and cyan). (c). Interactions between the 5′ arm of TNF23 and domain II of the ROQ domain. Water molecules are shown as red spheres. (d). Interactions between the 3′ arm of TNF23 and domain I of the ROQ domain
Both arms of the base-paired region interact with the ROQ domain (Fig. 3b, Supplementary Note 4, Supplementary Fig. 6). For the 5′ arm, nucleotides U4, U5 and U6, near the middle of the base-paired region, have hydrogen-bonding interactions with residues in helix αJ of domain II (Fig. 3c). In addition, the interactions with the base of U4 are consistent with pyrimidines, as they involve the carbonyl oxygen on the C2 atom that is shared between U and C. For the 3′ arm, nucleotides A14, A15 and C16 near the end of the duplex contact residues in helices αB and αC of domain I (Fig. 3d). These residues that interact with the RNA are highly conserved among the ROQ domains (Supplementary Fig. 1). However, neither arm of the RNA appears to make strong contacts with the ROQ domain. Therefore, simultaneous binding to both arms may be required for association with the B site, indicating that it is likely for dsRNA. Moreover, the exact sequence of the RNA is not specifically recognized in this binding site either.
Helix αJ in domain II that mediates the interactions with TNF23 is the second helix of the HTH motif. However, the binding mode of TNF23 to this domain is rather different from that of nucleic acids to other HTH motifs (Supplementary Fig. 2). Moreover, the position of TNF23 relative to domain II is also strikingly different from that of Hmg19 relative to domain III (Fig. 1d). The helical axes of the two RNAs are at a nearly 90° angle, most likely due to the involvement of domain I in the binding of TNF23.
Mutations in the interfaces abolish RNA binding
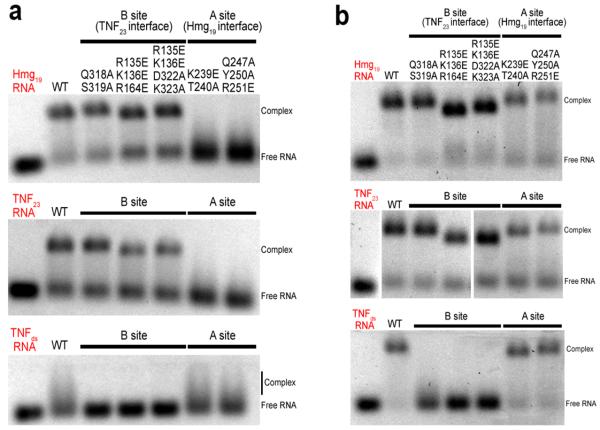
We next introduced mutations in the ROQ domain residues that are in the interfaces with RNA and assessed their effects on binding by the electrophoretic mobility shift assay (EMSA). Mutations in the interface with Hmg19 (A site) included K239E T240A (Fig. 2d) and Q247A Y250A R251E (Fig. 2b), and those in the interface with TNF23 (B site) included Q318A S319A of domain II (Fig. 3c), R135E K136E R164E of domain I (Fig. 3d), and R135E K136E D322A K323A of both domains. The mutants produced similar elution profiles from cation exchange chromatography as that for the wild-type ROQ domain, suggesting that the mutations did not disrupt the overall structure of the domain.
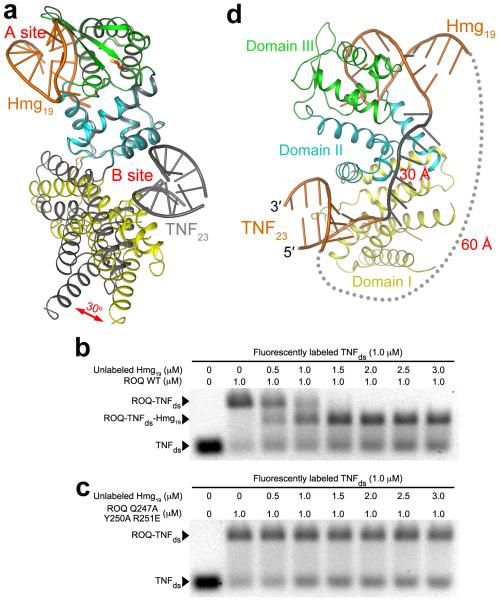
The Hmg19 and TNF23 RNAs were incubated with the wild-type and mutant ROQ domain proteins in the presence of 200 mM NaCl. The EMSA results showed that mutations in the A site blocked binding to both Hmg19 and TNF23, while mutations in the B site had only small effects on binding of the two RNAs (Fig. 4a). These results confirm the structural observations and suggest that both Hmg19 and TNF23 predominantly adopt the stem-loop conformation under these experimental conditions (1 μM RNA concentration).
Figure 4. Biochemical characterizations of the ROQ domain-RNA interactions.
(a). Electrophoretic mobility shift assay (EMSA) results for wild-type and mutant ROQ domains with Hmg19, TNF23 and TNFds RNAs. The NaCl concentration was 200 mM. (b). EMSA results at 50 mM NaCl concentration. Please see Supplementary Fig. 7 for the uncropped images of the middle panel.
To confirm that the B site is truly for binding dsRNA, we created another RNA molecule by replacing the 3-nt loop of TNF23 with two GC base pairs (TNFds, Fig. 1b). The two separate chains were mixed and annealed to form the dsRNA, and mutations in the B site completely blocked binding to this RNA (Fig. 4a). Binding of wild-type ROQ domain as well as the mutants in the A site to this RNA was observed, but the complex migrated as a smear on the gel, likely due to its poor stability. We then decreased the NaCl concentration to 50 mM and observed a clear band for the complex in the EMSA experiment, while mutations in the B site still abolished the interactions with the TNFds RNA (Fig. 4b). The lower ionic strength also stabilized the complex with the stem-loop RNA, such that the effects of mutations in the A site were reduced. This is consistent with the structural observations where the interface is mostly ionic and hydrophilic in nature (Fig. 2b). Finally, the ssRNA species of TNFds did not cause a gel shift (data not shown), proving that the B site is a dsRNA binding site.
Two separate RNA binding sites in the ROQ domain
The structures of the Hmg19 and TNF23 complexes demonstrate that the ROQ domain has two separate RNA binding sites, with no overlaps between them (Fig. 5a). The binding mode of Hmg19 RNA in the A site clearly indicates that a dsRNA cannot be accommodated there, as the loop makes intimate contacts with the ROQ domain (Fig. 2c). Our mutagenesis and EMSA results confirm that the A site in domain III is for binding stem-loop RNA, while the B site at the interface of domains I and II is for binding dsRNA.
Figure 5. Two separate RNA binding sites in the ROQ domain.
(a). Overlay of the structures of the Hmg19 complex (in color) and TNF23 complex (gray) of the ROQ domain, based on domains II and III. A 30° change in the orientation of domain I is indicated with the red arrow. (b). EMSA results showing the formation of the ROQ-TNFds-Hmg19 ternary complex. Wild-type ROQ domain was pre-incubated with labeled TNFds, and then with increasing concentrations of unlabeled Hmg19. The NaCl concentration was 50 mM. (c). EMSA results with mutations in the A site. The mutations blocked the formation of the ternary complex. (d). A model showing possible linkages between the RNA molecules in the B site and A site. A single-stranded RNA (gray, 7 nts) was modeled to connect the 3′ end of the RNA in the B site to the 5′ end of the stem-loop RNA in the A site (~30 Å distance). A linker between the 3′ end of the RNA in the A site to the 5′ end of the RNA in the B site would need to span ~60 Å (gray dots).
The position of domain I relative to domains II and III in the Hmg19 complex is substantially different from that in the TNF23 complex, corresponding to a rotation of ~30° (Fig. 5a). This conformational change reduces the size of the groove between domains I and II, such that the B site in the Hmg19 complex cannot accommodate dsRNA due to steric clashes. However, this change is likely due to the limited interface between domains I and II, and the consequent flexibility of domain I, rather than being caused by Hmg19 binding.
To obtain direct experimental evidence that the ROQ domain can bind both stem-loop RNA and dsRNA at the same time, we pre-incubated wild-type ROQ domain with fluorescently labeled TNFds RNA, and then introduced increasing concentrations of unlabeled Hmg19 RNA. The EMSA result clearly showed a faster migrating species in the presence of Hmg19, corresponding to the formation of the ROQ-TNFds-Hmg19 ternary complex (Fig. 5b). In comparison, mutations in the A site that blocked Hmg19 binding also abolished the formation of the ternary complex (Fig. 5c). We also reversed the sequence of incubation of the two RNAs and obtained the same results (data not shown). Therefore, the A and B sites of the ROQ domain can function independently of each other and bind RNA individually (as observed in the crystal structures) as well as simultaneously.
Both RNA binding sites are important for mRNA decay
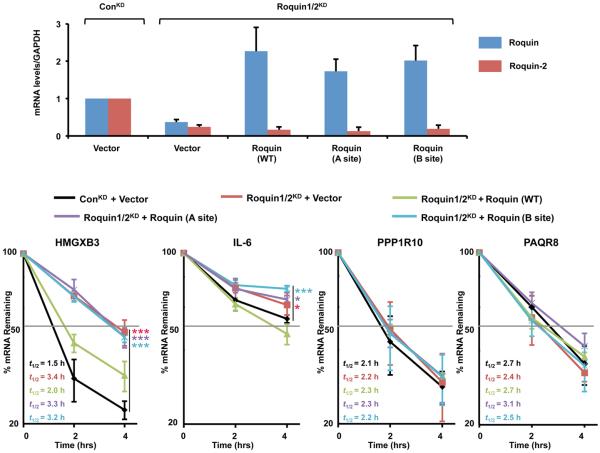
To assess the functional significance of the two RNA binding sites in promoting decay of mRNAs targeted by Roquin, we simultaneously knocked down human Roquin and Roquin-2 in 293T cells and complemented with exogenous wild-type and mutant Roquin (Fig. 6a), which were confirmed to be stable and expressed at comparable protein levels (MZ and MK, data not shown). We tested the stability of HMGXB3 and PPP1R10 mRNAs, which were shown to be responsive to Roquin in mouse NIH3T3 cells 5, as well as the pleiotropic cytokine IL-6 mRNA which may possess a CDE (Supplementary Fig. 3). Reduction of endogenous Roquin and Roquin-2, by 70% and 90% respectively, led to the stabilization of the human HMGXB3 and IL-6 mRNAs that was reversed upon complementation with shRNA-resistant wild-type Roquin (Fig. 6b). In contrast, despite comparable levels of expression (Fig. 6a), neither the A site nor the B site mutant Roquin complementation could restore the decay of these mRNAs, demonstrating that both sites are necessary for efficient Roquin-mediated decay of target mRNAs in cells. The effect of Roquin knockdown on IL-6 stability is smaller, consistent with the presence of other mechanisms that regulate this mRNA 21. As expected, an mRNA lacking a predicted CDE, PAQR8, was not responsive to changes in Roquin levels. Surprisingly, the PPP1R10 mRNA was also nonresponsive to a reduction in Roquin levels or complementation with Roquin in human cells, indicating some functional differences between the human and mouse cells for this mRNA.
Figure 6. Functional study of Roquin in human 293T cells.
(a). mRNA levels as measured by quantitative real-time PCR. Roquin and Roquin-2 mRNA levels in 293T cells are shown after control knockdown (ConKD), Roquin/Roquin-2 double knockdown (Roquin1/2KD) as indicated above the figure, and complementation by add back with shRNA-resistant Roquin (WT), and Roquin (A site) and Roquin (B site) mutants. Complementation with the vector plasmid lacking the Roquin cDNA is denoted (Vector). Error bars, s.d. (n=3 cell cultures) (b). Plot showing the stability of the HMGXB3, IL-6 and PPP1R10 mRNAs. Degradation of the mRNAs was measured by quantitative real-time PCR in the same 293T cells as in panel a. Level of mRNA remaining following actinomycin D-directed transcriptional silencing at the indicated times is shown. Error bars, s.d. (n=3 cell cultures). Half-lives (t1/2) are denoted in the figure and presented relative to GAPDH mRNA and derived from three independent experiments. Comparison of the various complementation data relative to the control are indicated by the bars, and p-values from comparison of the decay rates are presented with asterisks (* p<0.05, ** p<0.01, *** p<0.001, from two-tailed extra sum-of-squares F test).
Discussion
Our structural studies reveal that the ROQ domain actually consists of three sub-domains. Moreover, although the ROQ domain does not share recognizable sequence homology with other proteins, the structures show that it contains two HTH motifs in tandem, a WH motif in domain III and an HTH motif in domain II. These motifs are known to bind DNA and RNA as well as mediate protein-protein interactions 22. Our mutagenesis and biochemical studies show that the WH motif in domain III is crucial for binding stem-loop RNA (the A site), while the HTH motif in domain II contribute to binding dsRNA (the B site). The tight interface between domains II and III suggests that they may function together as a unit. Domain I of the ROQ domain, with a unique arrangement of helices, is also involved in dsRNA binding. Therefore, all three domains of the ROQ domain mediate or contribute to RNA binding.
The presence of two RNA binding sites in the ROQ domain is consistent with and explains the current data on Roquin. Immunoprecipitation experiments with Roquin have identified a large number of mRNAs, many of which do not contain the conserved CDE stem-loop 5. Within the ICOS 3′ UTR, Roquin also recognizes sequences in addition to the CDE 6,23. Therefore, Roquin may bind sequences other than the CDE stem-loop, and the B site may have an important role in recognizing such RNAs.
The distance from the 3′ end of the dsRNA in the B site to the 5′ end of the stem-loop in the A site is ~30 Å, which can be bridged by a linker of at least 7 nts (Fig. 5d). On the other hand, the distance from the 3′ end of the stem-loop in the A site to the 5′ end of the dsRNA in the B site is ~60 Å, and a longer linker will be needed to connect these two ends (Supplementary Fig. 3). Alternatively, the two binding sites in the ROQ domain may function independently and bind separate RNA molecules.
Our studies show that domain III has a crucial role in recognizing the CDE stem-loop. The functional importance of this domain is further supported by the fact that it also contains the M199R mutation that produces the sanroque phenotype in mice 7. The Met199 residue is located at the N-terminal end of helix αE, but it is not in the interface with RNA (Fig. 1c). The side chain is partially exposed to the surface (Supplementary Fig. 6), and the mutation to Arg can be accommodated without significantly perturbing the structure. The structural observations are consistent with the data that the M199R mutant has similar binding affinity for the CDE as the wild-type protein 5, and that the mutation did not affect the stability of Roquin 7 but partially reduced its ability to repress ICOS expression 8. The role of the M199R mutation in the sanroque phenotype is likely due to a different mechanism, for example disrupting interactions with a protein partner as the Arg side chain introduced by the mutation is expected to change the surface shape and electrostatic properties of that region of domain III.
Overall, our structural and biochemical studies have defined the molecular basis for the recognition of the CDE stem-loop by the ROQ domain, and unexpectedly revealed a separate site for dsRNA recognition by this domain. Our functional studies showed that both binding sites are important for Roquin-mediated decay of target mRNAs. The structures also provide a foundation for understanding the functions of Roquin and the CDE in mediating mRNA decay as well as other cellular processes.
Online Methods
Protein expression and purification
Residues 89-407 of human Roquin (containing the ROQ domain) were sub-cloned into the pET26b vector (Novagen), and the resulting recombinant protein carried a C-terminal hexa-His tag. E. coli BL21 Star (DE3) cells transformed with this plasmid were induced with 0.5 mM IPTG and grown at 37 °C for 18 h. The soluble protein was purified through nickel-agarose affinity (Qiagen), cation-exchange (SP Sepharose Fast Flow, GE Healthcare) and gel-filtration (Sephacryl S-300, GE Healthcare) chromatography. The purified protein was concentrated to 20 mg/ml and stored at −80 °C in a solution containing 20 mM Tris (pH 8.5), 250 mM NaCl, 5 mM DTT and 5% (v/v) glycerol. The His tag was not cleaved for crystallization.
The selenomethionyl protein sample was produced in E. coli B834 (DE3) cells, and the bacteria were grown in defined LeMaster media supplemented with selenomethionine 24. The purification procedure was the same as that for the native protein.
The RNA samples were purchased from Integrated DNA Technologies (IDT). The TNF23 and Hmg19 RNAs were diluted to 300 and 50 μM concentration, respectively, in a buffer containing 20 mM Tris (pH 8.5), 250 mM NaCl and 4 mM MgCl2 and the solution was heated at 98 °C for 5 min and then slowly cooled to room temperature. To form the ROQ-TNF23 complex, 60 nmol of the protein and 72 nmol of the RNA were mixed in a solution (total volume 3 ml) containing 20 mM Tris (pH 7.9), 150 mM NaCl and 5 mM MgCl2. The Hmg19 complex was prepared with 13 nmol of ROQ domain and 15 nmol of RNA in the same buffer (total volume 1.5 ml). The mixtures were incubated on ice for 30 min prior to gel filtration chromatography. Fractions corresponding to the complex were collected and concentrated to 7.5 mg/ml (TNF23 complex) or 6.4 mg/ml (Hmg19 complex) in a buffer containing 20 mM Tris (pH 8.5), 250 mM NaCl, 5 mM DTT and 5% (v/v) glycerol.
Protein crystallization
Crystals of the ROQ-TNF23 complex were obtained with the sitting-drop vapor-diffusion method at 20 °C. The reservoir solution contained 100 mM sodium citrate (pH 5.7), 1.5 M LiCl, and 18-21% (w/v) PEG 6000. 1 μl of the protein-RNA complex solution at 1.5 mg/ml concentration (diluted with the storage buffer without glycerol) was mixed with 1 μl of the reservoir solution. Crystals grew to full size in one week. They belong to space group P212121, with unit cell parameters of a=90.8 Å, b=93.1 Å, c=100.2 Å. There are two molecules of the ROQ domain and a TNF23 duplex in the asymmetric unit.
Crystals of the ROQ-Hmg19 complex were obtained with the hanging-drop vapor-diffusion method at 20 °C. The reservoir solution contained 100 mM Tris (pH 8.2), 0.3 M calcium acetate and 12-14% (w/v) PEG 8000. 1 μl of the protein-RNA complex solution at 3 mg/ml concentration was mixed with 1 μl of the reservoir solution. Crystals grew to full size in two weeks. They belong to space group P21, with unit cell parameters of a=49.6 Å, b=170.0 Å, c=51.1 Å, and β=97.0°. There are two copies of the ROQ-Hmg19 complex in the asymmetric unit.
The crystals were cryo-protected by the reservoir solution supplemented to 35% (w/v) PEG6000 (TNF23 complex) or PEG8000 (Hmg19 complex) and flash-frozen in liquid nitrogen for data collection at 100K.
Data collection and structure determination
The structure of the ROQ-TNF23 complex was determined by the selenomethionyl anomalous diffraction method 25. A single-wavelength anomalous diffraction (SAD) data set to 1.9 Å resolution was collected on a selenomethione-substituted crystal using an ADSC Quantum-315 CCD at beamline X29A of the National Synchrotron Light Source (NSLS). The diffraction data were processed and scaled with the HKL package 26. The data processing statistics are summarized in Table 1.
The Se atoms were located with the program SHELXD 27. The reflections were then phased with the program SOLVE and the phases were improved with the program RESOLVE 28. Most of the protein residues were automatically built by RESOLVE, and the model was completed by manual building with the program Coot 29. The structure refinement was performed using the programs CNS 30 and Phenix 31.
A native data set to 2.9 Å resolution on the ROQ-Hmg19 complex was collected at the X25 beamline of the NSLS, on a Pilatus 6M detector. The structure was determined by the molecular replacement method, with the program COMO 32. The three regions of the ROQ domain were searched independently in the calculation.
ROQ domain mutants
ROQ domain mutants were made with the QuikChange kit (Stratagene) and verified through sequencing. The mutant proteins were purified by nickel affinity and cation exchange chromatography.
Electrophoretic mobility shift assay (EMSA)
The Hmg19, TNF23 and TNFds 5′ arm RNAs with 6-FAM fluorescent label at the 5′ end and the unlabeled TNFds 3′ arm were purchased from IDT. The RNAs were diluted to 5 μM and heated at 98 °C for 5 min and then slow-cooled to room temperature. The protein-RNA mixtures were prepared in a buffer containing 20 mM Tris (pH 8.2), 5 mM MgCl2, and 50 or 200 mM NaCl. The samples were loaded to 0.8% native agarose gel. The electrophoresis was performed in 1× TAE buffer (pH 8.0) at 4 °C, and then the RNA was visualized on a UV illuminator.
Plasmid construction
The FLAG-tagged pTK-IREShyg-FLAG plasmid was constructed in three steps. First, the polylinker of pIRESpuro3 (Clontech) was replaced with the FLAG-tag containing polylinker in pcDNA3 (ref. 33) using Sac I and Not I restriction sites to generate pIRES-FLAG-puro3 vector. Second, the puromycin resistant gene in pIRES-FLAG-puro3 was removed by Sma I and Xba I digestion and replaced with the hygromycin B resistant gene derived from pTRE2hyg (Addgene) using Stu I and Xba I to generate pIREShyg-FLAG vector. In the last step, IREShyg-FLAG cassette from the pIREShyg-FLAG vector was PCR amplified with the addition of adaptors containing NheI and NotI restriction sites at 5′ and 3′ end, respectively. Then, the Renilla luciferase cDNA within pRL-TK (Promega) was replaced by the NheI-NotI flanked IREShyg-FLAG cassette to generate the pTK-IREShyg-FLAG plasmid. The full-length human Roquin cDNA was PCR amplified using primers 5′ GAATTCGATATCCATGCCTGTACAAGCTCC 3′ and 5′ GAATTCTGCGGCCGCTTAAGGAGCAGAACTGG 3′ containing the EcoRV and NotI restriction endonuclease sites respectively and inserted into the same restriction sites of the pTK-IREShyg-FLAG plasmid in frame and downstream of the Flag-tag to generate pTK-IREShyg-FLAG-Roquin. shRNA-resistant Roquin was generated by PCR mutagenesis using primers 5′ AACGAGTAGTTAATTCACAATACGGCACACAGCCACA 3′ and 5′ TGTGCCGTATTGTGAATTAACTACTCGTTCTTGGAGGTAGGG 3′ to alter the shRNA target sequence to generate pRoquin (WT). shRNA-resistant mutants, pRoquin (A site) and pRoquin (B site), were constructed by replacing the wild-type ROQ domain region within pRoquin (WT) with the corresponding A site mutant (Q247A Y250A R251E) and B site mutant (R135E K136E D322A K323A) using BlpI/BstEII and BlpI/MfeI restriction sites, respectively. All constructs were confirmed by sequencing.
Generation of lentiviral particles
All shRNA plasmids and packaging plasmids were purchased from Sigma. 293T cells were transfected by pLKO.1-shRoquin (Sigma #TRCN0000144045) / pLKO1-shRoquin-2 (Sigma # TRCN0000294353), and pCMV-VSVG and psPAX2 with Lipofectamin2000 (Life technology) to generate viral particles. Culture medium was harvested two days post transfection and frozen at −80°C for storage.
Determination of mRNA half-lives
293T cells infected by control shRNA expressing lentivirus (ConKD) or both Roquin and Roquin-2 specific shRNA lentivirus (Roquin1/2KD) were transiently transfected with pTK-IREShyg-FLAG vector, pRoquin (WT), pRoquin (A site) or pRoquin (B site). Cells were treated with actinomycin D (5 mg/L, Sigma) 48 hours post transfection/infection, cells harvested and RNA isolated (TRIzol Reagent; Life Technology) at the indicated time intervals. RNAs were then reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega) and oligo(dT) primer according to the manufacturer’s instructions. Values were quantified by real-time PCR using SYBR green PCR core reagent (Applied Biosystems) and the abundance of specific mRNAs were quantified using the standard-curve method according to the recommendations of the manufacturer. mRNA levels were normalized to the GAPDH mRNA and plotted against time. The primers used for real-time PCR are listed in Supplementary Table 1. Real-time PCR was carried out with an ABI Prism 7900HT sequence detection system. Assuming equation of first order decay rate was fitted to the data points by linear regression: y=100 × e(b × t), where y stands for the percentage of mRNA remaining and the time. mRNA half-lives were determine by t1/2=(−ln2)/b. p-values for the decay rate b in Figure 6b were derived using extra sum-of-squares F test calculated by PRISM® software.
Supplementary Material
Acknowledgements
We thank X. Jiao (Rutgers University) for constructing the pTK-IREShyg and pIREShyg-FLAG plasmids; N. Whalen, R. Jackimowicz and H. Robinson for access to the X29A beamline at the NSLS. The in-house instrument for X-ray diffraction screening was purchased with a US National Institutes of Health (NIH) grant S10OD012018 (LT). This research is supported by NIH grants GM077175 (LT) and GM067005 (MK).
Footnotes
Accession codes
The atomic coordinates and the observed structure factors have been deposited in the Protein Data Bank, with accession codes 4QIK (TNF23 complex) and 4QIL (Hmg19 complex).
Author contributions
DT carried out protein expression, purification and crystallization, X-ray diffraction data collection and processing, structure determination and refinement, mutagenesis and EMSA experiments. MZ carried out the mRNA stability experiments. LT and MK supervised the experiments, analyzed the data and wrote the manuscript. All authors commented on the manuscript.
References
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Beisang D, Bohjanen PR. Perspectives on the ARE as it turns 25 years old. Wiley Interdiscip. Rev. RNA. 2012;3:719–731. doi: 10.1002/wrna.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks SA, Blackshear PJ. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanism of action. Biochim. Biophys. Acta. 2013;1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoecklin G, Lu M, Rattenbacher B, Moroni C. A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway. Mol. Cell. Biol. 2003;23:3506–3515. doi: 10.1128/MCB.23.10.3506-3515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leppek K, et al. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–891. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Glasmacher E, et al. Roquin binds inducible constimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat. Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- 7.Vinuesa CG, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 8.Yu D, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 9.Vogel KU, et al. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Nat. Immunol. 2013;38:655–668. doi: 10.1016/j.immuni.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Pratama A, et al. Roquin-2 shares functions with its paralog roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38:669–680. doi: 10.1016/j.immuni.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Heissmeyer V, Vogel KU. Molecular control of Tfh-cell differentiation by Roquin family proteins. Immunol. Rev. 2013;253:273–289. doi: 10.1111/imr.12056. [DOI] [PubMed] [Google Scholar]
- 12.Ji YR, et al. Enforced expression of roquin protein in T cells exacerbates the incidence and severity of experimental arthritis. J. Biol. Chem. 2012;287:42269–42277. doi: 10.1074/jbc.M112.374835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertossi A, et al. Loss of Roquin induces early death and immune deregulation but not autoimmunity. J. Exp. Med. 2011;208:1749–1756. doi: 10.1084/jem.20110578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-FboxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 15.Goldenberg SJ, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott DC, et al. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soler N, Fourmy D, Yoshizawa S. Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB. J. Mol. Biol. 2007;370:728–741. doi: 10.1016/j.jmb.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Za domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 22.Teichmann M, Dumay-Odelot H, Fribourg S. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription. 2012;3:2–7. doi: 10.4161/trns.3.1.18917. [DOI] [PubMed] [Google Scholar]
- 23.Athanasopoulos V, et al. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J. 2010;277:2109–2127. doi: 10.1111/j.1742-4658.2010.07628.x. [DOI] [PubMed] [Google Scholar]
References
- 24.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Cryst. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Meth. Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan KD. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: building a new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 32.Jogl G, Tao X, Xu Y, Tong L. COMO: A program for combined molecular replacement. Acta Cryst. 2001;D57:1127–1134. doi: 10.1107/s0907444901006783. [DOI] [PubMed] [Google Scholar]
- 33.Jiao X, Wang Z, Kiledjian M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol. Cell. 2006;24:713–722. doi: 10.1016/j.molcel.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.