Chemical and physical control of biointerfaces is attractive because of its flexibility and effectiveness in cell and tissue regulation, as well as in diagnostic and therapeutic applications.[1–7] When introducing biochemical cues to cells, various cell activities (e.g., adhesion, spreading morphologies, proliferation) can be manipulated at their biointerfaces.[5–7] For example, in the development of stem cell therapies, surfaces modified with synthetic peptides can be used to support the self-renewal and differentiation of stem cells.[4] Notably, specific cell–substrate interactions observed on three-dimensional (3D) micro/nanostructures can provide the topographic cues regulating the cell spreading morphology of neurons,[8,9] thereby promoting the level of cell differentiation for stem cells,[10–12] enhancing the transfection efficiency of cells with targeted gene expression,[13–15] and improving the capturing efficiency of circulating tumor cells (CTCs) for noninvasive blood biopsies.[16–21] In addition to these biological applications, integrating additional electrical functionality into biointerfaces has recently attracted significant interest for the digital transformation of biological signals within bioelectronics. [22,23]
Bioelectronic interfaces (BEIs) are promising intermediate layers that can enhance communication between electronics and biological systems; they can be operated to couple the flows of electrons and ions in dual directions. Accordingly, BEIs have great potential for use in electrical signaling,[22,23] stimulation,[24–28] and electrically triggered-response toward the release and pumping of small molecules.[29–33] In the development of BEIs, organic conducting polymers [CPs; e.g., polypyrrole or poly(3,4-ethylenedioxythiophene) (PEDOT)] have been applied widely for their outstanding electrical transport properties, inherent biocompatibility, and high manufacturing flexibility. Indeed, CPs can be synthesized with a diverse array of chemical designs, such as the incorporation of various anionic dopants [e.g., poly(sodium styrene sulfonate) (PSS), tosylate (TOS)][34–36] and/or the presenting of various functional side chains,[20,37–39] thereby extending their applicability. For instance, TOS-doped PEDOT (PEDOT:TOS) materials are currently the most promising BEIs because of their high electrical stability and biocompatibility, allowing long-term cell culturing or implantation;[36] alternatively, carboxylic acid–grafted PEDOT (PEDOTAc) materials can be conjugated to a specific cell capturing agent for CTC assays.[20] Briefly, this bioconjugation process involves initial activation of the carboxylic acid groups, using N-hydroxysuccinimide (NHS) and 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC), and subsequent conjugation with streptavidin. The streptavidin-grafted PEDOT films are then incubated with biotinylated anti-EpCAM to introduce these capturing agents onto the surfaces. In previous studies toward the development of techniques for early cancer diagnosis, we demonstrated that the cell binding affinity of EpCAM (epithelial cell adhesion molecule) on vertically standing silicon (Si) nanowire substrates occurs with capture efficiencies for CTCs ranging from 40 to 70%.[16,18] In addition, we found that this NanoVelcro concept can also be applied to electrochemically deposited CP nanodot structures at the submicron scale.[20] Accordingly, it would be desirable to produce well-defined 3D CP structures and understand the topographical effects of EpCAM-grafted CPs on CTC assays. At present, 3D micro/nanostructures of CPs can be prepared using electrospinning,[40,41] stepwise electrochemical polymerization,[38] hard-template[42–44] and soft-template[45–47] approaches. Nevertheless, large-scale high-fidelity engineering of CP micro/nanostructures, in terms of both biochemical and topographical effects, remains a challenge. Based on our previously developed BEI concepts for spatial and temporal control of cells and drugs upon flat CP films,[27,28,33] we believe that integrating the 3D micro/nanostructures of CP films will lead to greater possibilities for the development of BEIs.
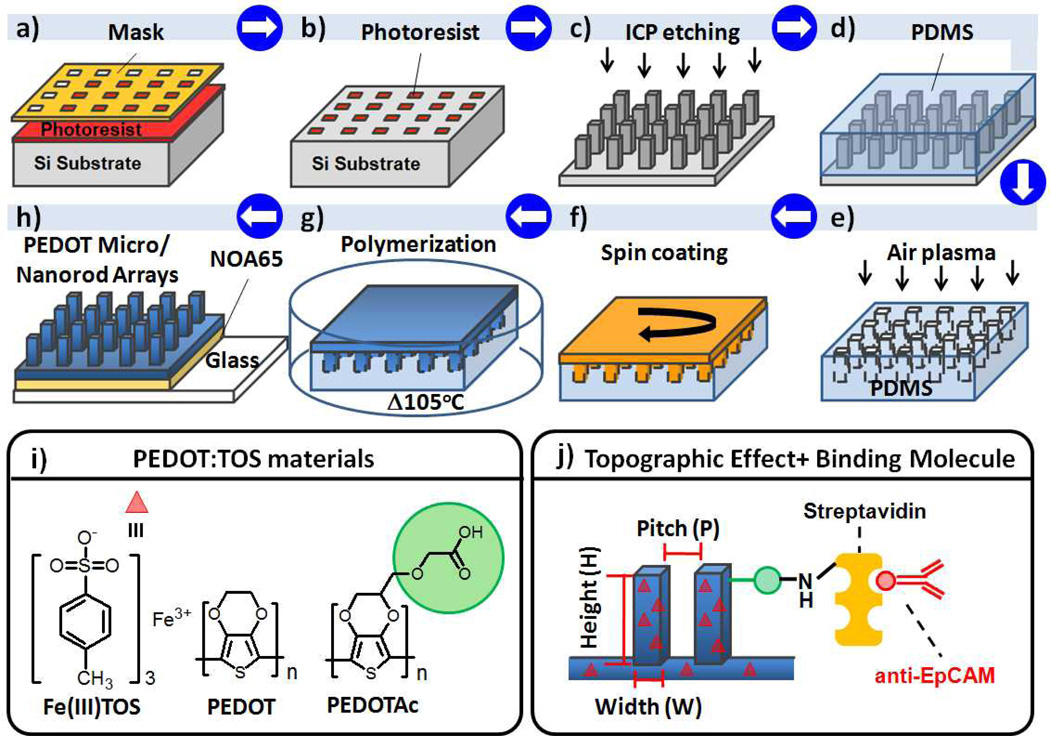
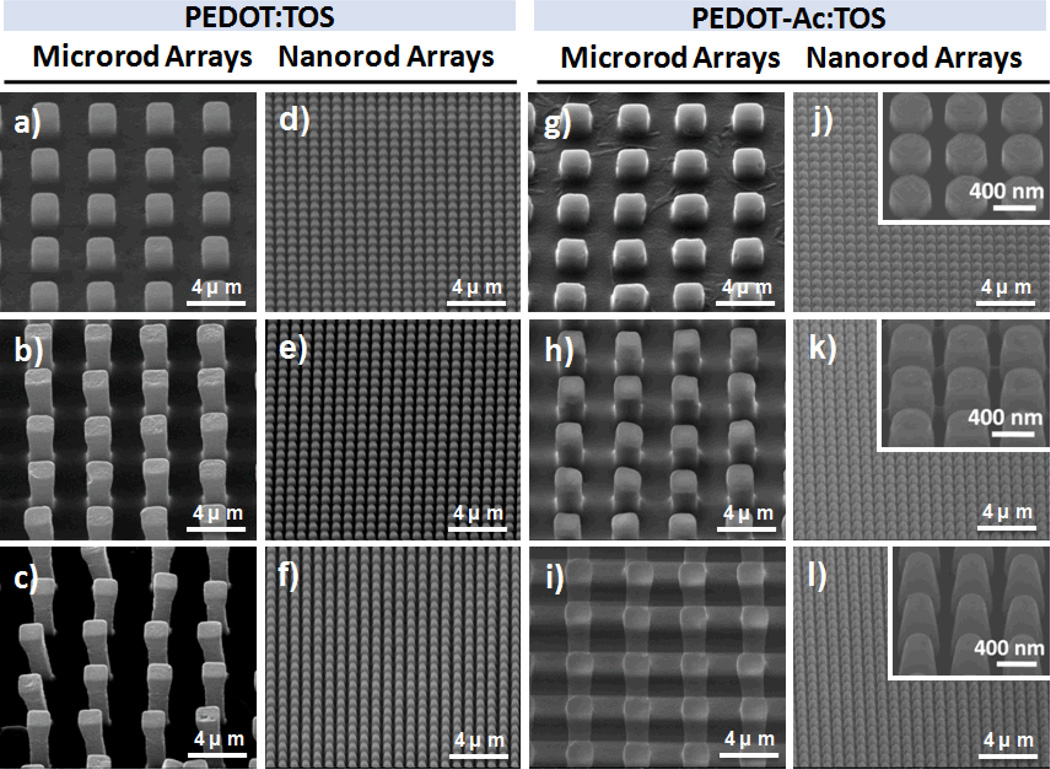
Herein, we report a method for fabricating large-scale PEDOT-based micro/nanorod arrays as 3D BEIs; it can be performed readily using a combination of chemical oxidative polymerization and modified poly(dimethylsiloxane) (PDMS) transfer printing techniques (Figure 1a–h). We generated the negative PDMS hole array replicas from the 1H,1H,2H,2H-perfluorooctyltrichlorosilane (FOTS)–coated Si micro/nanorod arrays, which could be patterned through I-line projection photolithography and inductively coupled plasma (ICP) reactive ion etching of exposed Si. By using a Si master featuring a diverse range of designed rod structures, we could prepare micro/nanostructures of PEDOT:TOS and PEDOTAc:TOS from the respective precursors through PDMS transfer printing with deliberate control as BEIs. However, the challenge was to develop a process for the fabrication of high-quality and large 3D PEDOT rod arrays with the rod size ranging from micrometer to nanometer. After several trials to obtain optimized condition, we found that the envisaged PEDOT precursor, containing EDOT derivatives with both iron (III) tosylate (as oxidizing agent) and imidazole (as inhibitor), underwent slow polymerization at 105 °C in a covered glass Petri dish. This slow polymerization had a positive effect on the quality of the PEDOT-based micro/nanorod array films (Figure 1g). We also noted that after spin-coating the PEDOT precursor onto the low-surface-energy (8.67 mJ mm−2) PDMS surface (Table S1, Supporting Information), droplets of the PEDOT precursor tended to aggregate on the PDMS surface during the slow polymerization process. Therefore, non-uniform PEDOT rod arrays were formed. To overcome this problem, we subjected the PDMS replicates to air plasma treatment (10 mtorr, 30 s) to partially change the PDMS surface energy (from 8.67 to 27.92 mJ mm−2), thereby promoting the PEDOT precursor to fill into the PDMS structures. After performing a demolding step using a biocompatible UV-curable polyurethane precursor (NOA65, Norland), we obtained highly uniform PEDOT rod arrays with excellent mechanical properties on the targeting glass substrate (Figure 1h; see also Figure S2a, Supporting Information). The surface energy of the PDMS replicates increased (up to 27.92 mJ mm−2) upon lengthening the air plasma treatment, which oxidized the PDMS surfaces to form a material more like SiO2;[48] this long-term (> 60 s) plasma treatment did, however, lead to a new interfacing problem—inefficient demolding of PEDOT micro/nanorod array films from the negative PDMS replicates. Figure 2 and Figure S1 and Table S1 (Supporting Information) reveal that positive Si masters featuring microstructural (diameter: 2 µm; height: 2, 4, or 6 µm; period: 2 µm) and nanostructural (diameter: 0.4 µm; height: 0.4, 0.8, or 1.2 µm; period: 0.8 µm) designs allowed us to efficiently produce PEDOT-based micro/nanorod arrays with either flat or various array structures. Indeed, Si master structures with similar geometric designs resulted in almost identical PEDOT micro/nanorod array structures (Figure 2).
Figure 1.
a–h) Schematic representation of the integrated fabrication of a PEDOT-based micro/nanorod array film. a, b) I-line projection photolithography on a Si substrate. c) Formation of Si micro/nanorod array master after ICP. d) Preparation of PDMS on Si micro/nanorod arrays modified with the FOTS vapor. e) Negative PDMS micro/nanohole array replicate treated with air plasma (10 mtorr; 30 s). f) PEDOT precursor spin-coated on the PDMS replicate. g) Optimized chemical oxidative polymerization of PEDOT film in a covered glass Petri dish. h) Transfer of the PEDOT micro/nanorod array film to the glass substrate using a UV-curable prepolymer (NOA65, Norland) as the adhesive. i) Chemical structures of the PEDOT precursors. j) Schematic representation of the bioconjugation of epithelial cellular adhesion molecule antibody (anti-EpCAM) to TOS-doped PEDOT micro/nanorod arrays and films.
Figure 2.
SEM images of PEDOT-based micro/nanorod array films. a–f) PEDOT-X with designed structures (X) of a) 2, b) 4, c) 6, d) 0.4, e) 0.8, and f) 1.2. g–l) PEDOTAc-X with designed structures (X) of g) 2, h) 4, i) 6, j) 0.4, k) 0.8, and l) 1.2. (Inset: higher-magnification 40° tilted-view of PEDOTAc-X).
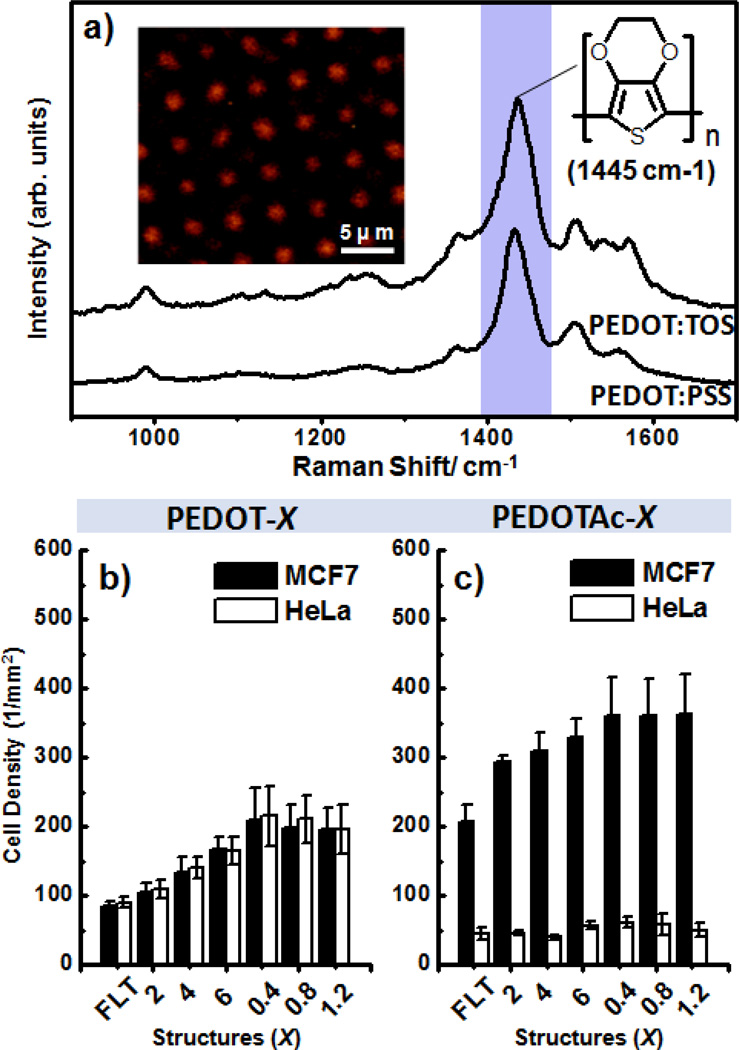
Previous investigations of the biological applications of micro/nanostructural features have revealed that biochemical and dimensional control of topographic structures is extremely important to ensure high CTC capture efficiency because it is necessary to match the structures of the cellular surface components (e.g., microvilli) and the underlying substrates.[49] Therefore, we selected CTCs as a cell model to explore the correlation between topographic effects and capture performance on chips, with the aim to possibly using PEDOT-based micro/nanorod arrays for CTC diagnostics. Toward developing PEDOT-based BEIs, we first needed to confirm that the PEDOT:TOS materials in our design geometry exhibit good redox chemical and electrical conducting properties. Through comparison with a commercial PEDOT:PSS material (Clevios P VP Al 4083), we performed Raman characterization of our PEDOT:TOS microrod arrays with excitation at 514.5 nm; we observed similar resonant structures for the PEDOT chains, with benzoid and quinoid structure bands appearing in the range from 1400 to 1500 cm−1 (Figure 3a).[50] By mapping a 2D Raman image according to the intensity of the signal at 1445 cm−1, we observed the PEDOT:TOS microrod array structures (inset to Figure 3a). Upon applying an alternating chemical potential to the PEDOT:TOS materials, the polymers underwent reversible switching between their oxidation and reduction states [oxidation state: treated with sodium persulfate (Na2SO8); reduction state: treated with hydrazine (N2H4)], appearing with dark blue and purple colors, respectively (Figures S2a–b, Supporting Information). The benefit of using this process for the fabrication of 3D PEDOT-based BEIs is that it allows the formation of higher-quality film, with tunable biochemical surfaces and tunable topographical morphologies, relative to those prepared using other approaches; these films also exhibit electrical conductivities (300–400 S cm−1) suitable for integration of this 3D PEDOT-based BEI platform in bioelectronic systems.
Figure 3.
a) Raman spectra of PEDOT:TOS and PEDOT:PSS films recorded at an excitation laser wavelength at 514.5 nm; inset: Raman image plotted with respect to the intensity of the peak of the PEDOT thiophene rings at 1445 cm−1. b, c) Capture yields of various anti-EpCAM–conjugated PEDOT:TOS films [a) PEDOT-X; b) PEDOTAc-X; X is the designed structure of Si master] on MCF7 (■) and HeLa (□) cells. MCF7 cells, EpCAM-(+); HeLa cells, EpCAM-(−).
To prove the concept of developing PEDOT-based 3D bioelectronic interfaces for CTC assays, we first examined the size- and/or chemical-dependent cell capture (Figures 3b and 3c) on films of PEDOT:TOS (PEDOT-X) and EpCAM-grafted PEDOTAc:TOS (PEDOTAc-X), where X refers to a PEDOT film that is flat or has a designed microstructure (diameter: 2 µm; height: 2, 4, or 6 µm; period: 2 µm) or nanostructure (diameter: 0.4 µm; height: 0.4, 0.8, or 1.2 µm; period: 0.8 µm). When we individually incubated the MCF7 breast cancer cells (EpCAM-positive cell line) and HeLa cervical cancer cells (EpCAM-negative cell line) onto PEDOT-based bioelectronic interfaces for 1 h, the nonspecific cell capture performance of PEDOT resulted in similar cell-capture yields of MCF7 and HeLa cells on all of our structures (Figure 3b); the cell-capture density did, however, experience a size effect, decreasing in the order (X; 0.4 < 0.8 < 1.2 > 6 > 4 > 2 > FLT), but existing in the range 75–200 cells mm−2. In contrast, we observed relatively higher cell-capture yields of MCF7 cells and lower nonspecific background of HeLa cells on all of the PEDOTAc structures; that is, PEDOTAc appears to be more sensitive and specific than PEDOT for the capture of EpCAM-positive cells (Figure 3c). The optimal time required to achieve call capture were obtained in Figure S2c (Supporting Information), which summarized the correlation between incubation time and the number of captured cells on PEDOTAc-0.4. For the EpCAM positive MCF7 cells, the maximal cell-capture numbers were achieved at an incubation time of 50 min. Whereas relatively low cell numbers were observed for EpCAM-negative HeLa cells on the 3D PEDOTAc-0.4, we selected an incubation period of 1-h for capturing EpCAM-positive cells. During the 1-h incubation time for cell capturing, the cell-capture density of MCF7 cells on the PEDOTAc nanostructures was more than seven times than that of HeLa cells, with better capture efficiency than those of the corresponding microstructures and flat structures; the cell-capture densities on the PEDOTAc-X structures also exhibited a size effect, decreasing in the same order as that of the PEDOT-X systems. Although all of our PEDOTAc nanostructures exhibited greater than 350 cells mm−2, the differences in the cell-capture yields of PEDOTAc-6 and PEDOTAc-0.4 were as low as approximately 25 cells mm−2 in our 3D PEDOT-based micro/nanorod array system. To investigate the working mechanism for the capture of CTCs on the PEDOTAc micro/nanostructures, we recorded SEM images of MCF7 cells on PEDOTAc-6 and PEDOTAc-0.4 to examine any differences in their cell morphologies. As revealed in Figure S2d (Supporting Information), each captured MCF7 cells on PEDOTAc-6 exhibited a round cell body with extended filopodia-like protrusion, which generated mechanical tension around the underlying microrod structures and bent them. Therefore, some of rods were easily distorted due to the SEM sample preparation process during drying because it was quite often observed for cell sitting on PEDOTAc-6. In addition to size matching between the cellular filopodia and the PEDOT nanostructures, we also observed extended lamellipodia (sheet-like protrusion) of MCF7 cells on PEDOTAc-0.4, revealing that the increased cell-capture yield of 25 cells mm−2 was due partially to more cell–substrate interactions between the pseudopodia and the PEDOTAc nanostructures (Figure S2e, Supporting Information).
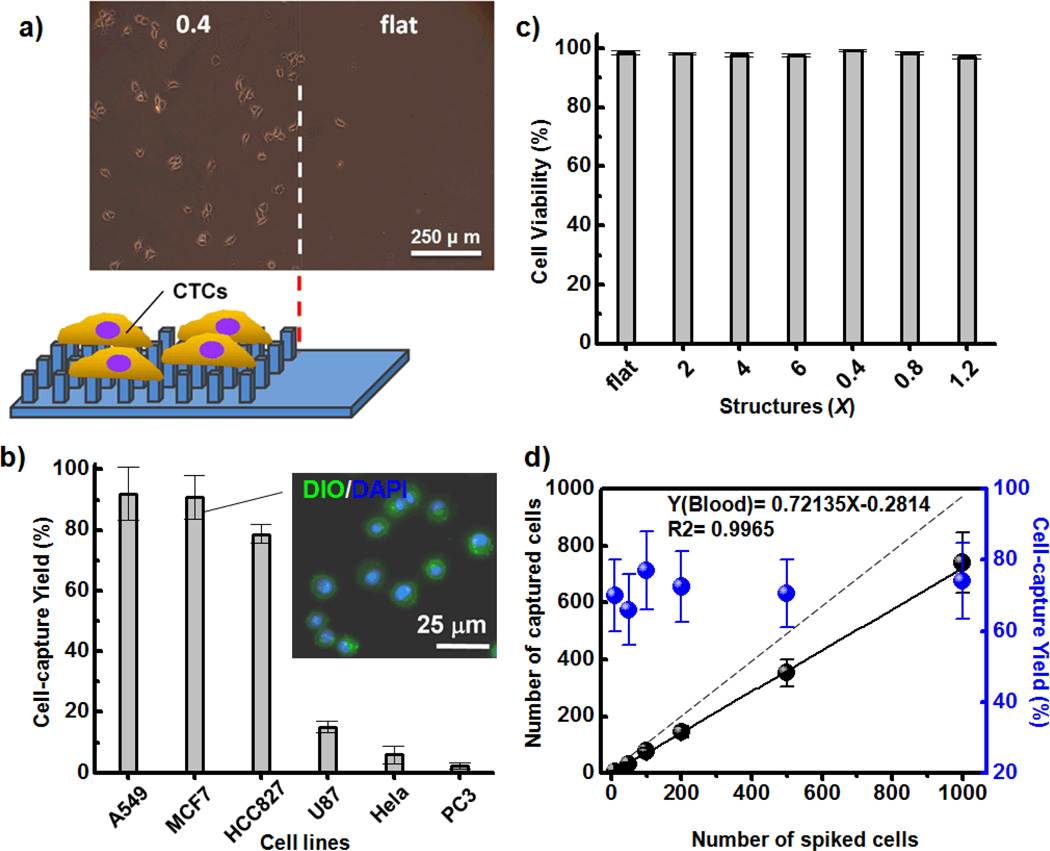
Because of the specific biochemical and topographic interactions between cells and substrates and their high degrees of transparency, we could use an inverted optical microscope to monitor the cell-capture efficiency of our PEDOT-based micro/nanorod arrays. Indeed, the bright field images of MCF7 cells captured on patterned PEDOTAc-0.4 revealed greater cell-capture efficiency than that of MCF7 cells on PEDOTAc-FLT (Figure 4a). Furthermore, a fluorescence Live/Dead staining assay [calcein AM (green) for live cells; EtH-1 (red) for dead cells] of the captured MCF7 cells revealed the high cell viability on our PEDOTAc films, with the cell-capture yield of PEDOT-0.4 being higher than that of PEDOTAc-6 (Figure S3, Supporting Information). In addition to MCF7 and HeLa cancer cell lines, we also tested the cell-capture efficiency of PEDOTAc-0.4 toward other EpCAM-positive (A549, HCC827) and EpCAM-negative (U87, PC3) cell lines (Figure 4b). Our results reveal that the topographical effect of EpCAM-grafted PEDOT-based BEIs can improve the cell isolation performance toward targeted cells while mitigating nonspecific binding of non-demanded cells. The inset of Figure 4b displays a fluoresce image of captured MCF7 cells; the spherical conformation, with slightly extended lamellipodia and filopodia morphologies, is consistent with that observed using SEM (Figures S2d and S2e, Supporting Information). Moreover, we used a cell viability analyzer (Vi-CELL™ XR, Beckman Coulter) to investigate the influence of the various geometrical designs of PEDOTAc on the cell viability. The viability of the MCF7 cells captured on all of the PEDOTAc structures was approximately 97%, confirming negligible damage on our PEDOT-based micro/nanorod array systems (Figure 4c). To examine the dynamic range and clinical utility of the optimized PEDOT-based nanorod array system, we used it to capture artificial MCF7 cells from healthy blood at a series of densities from 10 to 1000 cell mL−1. Notably, the cell-capture yield at each different spiked cell density was greater than 70%; thus, our PEDOTAc platform exhibits efficient performance for clinical samples (Figure 4d).
Figure 4.
a) Microscopy image of MCF7 cells captured on (left) patterned PEDOTAc-0.4 and (right) the flat substrate. b) Cell-capture efficiencies from suspensions of breast (MCF), lung (A549, HCC827), cervical (HeLa), prostate (PC3), and brain (U87) cell lines; inset: two-color fluorescence image based on DiO membrane (green) and DAPI nuclear staining, used to identify the captured morphology of MCF7 cells on PEDOTAc-0.4. c) Quantitative evaluation of cell viability of MCF7 cells captured on various PEDOTAc structures; error bar represents the standard deviation from three repeats. d) Capture efficiencies of various contents of MCF7 cells in whole blood; dashed line: ideal cell-capture yield when the various spiked cell densities were greater than 100%.
In conclusion, we develop a facile solution-processing approach for producing 3D PEDOT-based micro/nanorod array films, which can be further surface-grafted with capture agents for directed specific recognition to study the cell–substrate interactions on BEIs. This BEI platform features the advantageous characteristics: (1) diverse dimensional structures (tunable from the microscale to the nanoscale), (2) varied surface chemical properties (tunable from nonspecific to specific), (3) high electrical conductivity, and (4) reversible chemical redox switching, and (5) high optical transparency. Furthermore, through systematic studies of PEDOT and PEDOTAc systems, we explore the effects of both chemistry and topography on the CTC-capture performance. The PEDOTAc-0.4 surface exhibits the optimal cell-capture efficiency; it could be used to isolate CTCs with minimal contamination from surrounding nontargeted cells (e.g., EpCAM-negative cells, white blood cells) and negligible disruption of the CTCs’ viability and functions. It is conceivable that PEDOT-based micro/nanorod array films function as a critical therapeutic intervention for monitoring tumor progression and metathesis, providing valuable insight into the use of electronics for tissue engineering and regenerative medicine.
Supplementary Material
Acknowledgments
The research endeavors at Academia Sinica were supported by the National Science Council (NSC) of Taiwan (contracts NSC 100-2113-M-001-027-MY3 and NSC 101-2120-M-001-011) and by the Academia Sinica Research Project on Nano Science and Technology; those at RIKEN were supported by RIKEN and by Grants-In-Aid for Young Scientists (No. 22681016 and 23710138) from JSPS/MEXT; those at UCLA were supported by a Creativity Award from the Prostate Cancer Foundation and by research grants (R33 CA157396 and R33 CA174562) from the National Institute of Health; and those at Ming Chi University of Technology were supported by the NSC of Taiwan (contracts NSC 103-2218-E-131-002).
Contributor Information
Yu-Sheng Hsiao, Department of Materials Engineering, Ming Chi University of Technology, 84 Gunjuan Road, Taishan, New Taipei City 243 (Taiwan); Research Center for Applied Sciences, Academia Sinica, Taipei 11529 (Taiwan).
Shyh-Chyang Luo, Responsive Organic Materials Laboratory, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198 (Japan); Department of Materials Science and Engineering, National Cheng Kung University, Tainan (Taiwan).
Shuang Hou, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Bo Zhu, Responsive Organic Materials Laboratory, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198 (Japan); State Key Laboratory for Modification of Chemical Fibers and Polymer Materials, College of Materials Science and Engineering, Donghua University, Shanghai (China).
Jun Sekine, Responsive Organic Materials Laboratory, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198 (Japan).
Chiung-Wen Kuo, Research Center for Applied Sciences, Academia Sinica, Taipei 11529 (Taiwan).
Di-Yen Chueh, Research Center for Applied Sciences, Academia Sinica, Taipei 11529 (Taiwan).
Hsiaohua Yu, Responsive Organic Materials Laboratory, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198 (Japan).
Hsian-Rong Tseng, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Peilin Chen, Research Center for Applied Sciences, Academia Sinica, Taipei 11529 (Taiwan).
References
- 1.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Benoit DS, Schwartz MP, Durney AR, Anseth KS. Nat. Mater. 2008;7:816. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melkoumian Z, Weber JL, Weber DM, Fadeev AG, Zhou Y, Dolley-Sonneville P, Yang J, Qiu L, Priest CA, Shogbon C, Martin AW, Nelson J, West P, Beltzer JP, Pal S, Brandenberger R. Nat. Biotechnol. 2010;28:606. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- 5.Cranford SW, de Boer J, van Blitterswijk C, Buehler MJ. Adv. Mater. 2013;25:802. doi: 10.1002/adma.201202553. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, Papoutsakis ET. Adv. Healthc. Mater. 2013;2:25. doi: 10.1002/adhm.201200169. [DOI] [PubMed] [Google Scholar]
- 7.Leal-Egaña A, Díaz-Cuenca A, Boccaccini AR. Adv. Mater. 2013;25:4049. doi: 10.1002/adma.201301227. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari A, Cecchini M, Serresi M, Faraci P, Pisignano D, Beltram F. Biomaterials. 2010;31:4682. doi: 10.1016/j.biomaterials.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari A, Cecchini M, Dhawan A, Micera S, Tonazzini I, Stabile R, Pisignano D, Beltram F. Nano Lett. 2011;11:505. doi: 10.1021/nl103349s. [DOI] [PubMed] [Google Scholar]
- 10.Fu J, Wang Y-K, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nat. Methods. 2010;7:733. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen A, Lieu DK, Freschauf L, Lew V, Sharma H, Wang J, Nguyen D, Karakikes I, Hajjar RJ, Gopinathan A, Botvinick E, Fowlkes CC, Li RA, Khine M. Adv. Mater. 2011;23:5785. doi: 10.1002/adma.201103463. [DOI] [PubMed] [Google Scholar]
- 12.Kim TG, Shin H, Lim DW. Adv. Funct. Mater. 2012;22:2446. [Google Scholar]
- 13.McKnight TE, Melechko AV, Hensley DK, Mann DGJ, Griffin GD, Simpson ML. Nano Lett. 2004;4:1213. [Google Scholar]
- 14.Peckys DB, Melechko AV, Simpson ML, McKnight TE. Nanotechnology. 2009;20:145304. doi: 10.1088/0957-4484/20/14/145304. [DOI] [PubMed] [Google Scholar]
- 15.Shiu J-Y, Kuo C-W, Whang W-T, Chen P. Lab Chip. 2010;10:556. doi: 10.1039/c000798f. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Wang H, Jiao J, Chen K-J, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angew. Chem. Int. Ed. 2009;121:9132. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekine J, Luo S-C, Wang S, Zhu B, Tseng H-R, Yu H-h. Adv. Mater. 2011;23:4788. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK-F, Tseng HR. Angew. Chem. Int. Ed. 2011;50:3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ. Adv. Mater. 2012;24:2756. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 20.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo S-C, Tseng H-R, Yu H-h. Adv. Mater. 2013;25:1547. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR. Angew. Chem. Int. Ed. 2013;52:3379. doi: 10.1002/anie.201208452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SY, Kim BN, Zakhidov AA, Taylor PG, Lee JK, Ober CK, Lindau M, Malliaras GG. Adv. Mater. 2011;23:H184. doi: 10.1002/adma.201100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khodagholy D, Doublet T, Gurfinkel M, Quilichini P, Ismailova E, Leleux P, Herve T, Sanaur S, Bernard C, Malliaras GG. Adv. Mater. 2011;23:H268. doi: 10.1002/adma.201102378. [DOI] [PubMed] [Google Scholar]
- 24.Xie J, Macewan MR, Willerth SM, Li X, Moran DW, Sakiyama-Elbert SE, Xia Y. Adv. Funct. Mater. 2009;19:2312. doi: 10.1002/adfm.200801904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quigley AF, Razal JM, Thompson BC, Moulton SE, Kita M, Kennedy EL, Clark GM, Wallace GG, Kapsa RMI. Adv. Mater. 2009;21:4393. doi: 10.1002/adma.200901165. [DOI] [PubMed] [Google Scholar]
- 26.Abidian MR, Corey JM, Kipke DR, Martin DC. Small. 2010;6:421. doi: 10.1002/smll.200901868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao Y-S, Lin C-C, Hsieh H-J, Tsai S-M, Kuo C-W, Chu C-W, Chen P. Lab Chip. 2011;11:3674. doi: 10.1039/c1lc20675c. [DOI] [PubMed] [Google Scholar]
- 28.Lu C-H, Hsiao Y-S, Kuo C-W, Chen P. Biochim. Biophys. Acta. 2013;1830:4321. doi: 10.1016/j.bbagen.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 29.Abidian MR, Martin DC. Adv. Funct. Mater. 2009;19:573. [Google Scholar]
- 30.Simon DT, Kurup S, Larsson KC, Hori R, Tybrandt K, Goiny M, Jager EWH, Berggren M, Canlon B, Richter-Dahlfors A. Nat. Mater. 2009;8:742. doi: 10.1038/nmat2494. [DOI] [PubMed] [Google Scholar]
- 31.Thompson BC, Richardson RT, Moulton SE, Evans AJ, O’Leary S, Clark GM, Wallace GG. J. Control. Release. 2010;141:161. doi: 10.1016/j.jconrel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Stavrinidou E, Leleux P, Rajaona H, Khodagholy D, Rivnay J, Lindau M, Sanaur S, Malliaras GG. Adv. Mater. 2013;25:4488. doi: 10.1002/adma.201301240. [DOI] [PubMed] [Google Scholar]
- 33.Hsiao Y-S, Kuo C-W, Chen P. Adv. Funct. Mater. 2013;37:4648. [Google Scholar]
- 34.Svennersten K, Larsson KC, Berggren M, Richter-Dahlfors A. Biochim. Biophys. Acta. 2011;1810:276. doi: 10.1016/j.bbagen.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Martin DC, Wu J, Shaw CM, King Z, Spanninga SA, Richardson-Burns S, Hendricks J, Yang J. Polym. Rev. 2010;50:340. [Google Scholar]
- 36.Green RA, Hassarati RT, Bouchinet L, Lee CS, Cheong GL, Yu JF, Dodds CW, Suaning GJ, Poole-Warren LA, Lovell NH. Biomaterials. 2012;33:5875. doi: 10.1016/j.biomaterials.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 37.Luo S-C, Mohamed Ali E, Tansil NC, Yu H-h, Gao S, Kantchev EAB, Ying JY. Langmuir. 2008;24:8071. doi: 10.1021/la800333g. [DOI] [PubMed] [Google Scholar]
- 38.Luo S-C, Liour SS, Yu H-h. Chem. Commun. 2010;46:4731. doi: 10.1039/c002321c. [DOI] [PubMed] [Google Scholar]
- 39.Luo S-C, Zhu B, Nakao A, Nakatomi R, Yu H-h. Adv. Eng. Mater. 2011;13:B423. [Google Scholar]
- 40.Laforgue A, Robitaille L. Macromolecules. 2010;43:4194. [Google Scholar]
- 41.Feng Z-Q, Wu J, Cho W, Leach MK, Franz EW, Naim YI, Gu Z-Z, Corey JM, Martin DC. Polymer. 2013;54:702. doi: 10.1016/j.polymer.2012.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao R, Cho SI, Liu R, Lee SB. J. Am. Chem. Soc. 2007;129:4483. doi: 10.1021/ja068924v. [DOI] [PubMed] [Google Scholar]
- 43.Han MG, Foulger SH. Chem. Commun. 2005:3092. doi: 10.1039/b504727g. [DOI] [PubMed] [Google Scholar]
- 44.Lin H-A, Luo S-C, Zhu B, Chen C, Yamashita Y, Yu H-h. Adv. Funct. Mater. 2013;23:3212. [Google Scholar]
- 45.Han MG, Foulger SH. Small. 2006;2:1164. doi: 10.1002/smll.200600135. [DOI] [PubMed] [Google Scholar]
- 46.Yoon H, Hong JY, Jang J. Small. 2007;3:1774. doi: 10.1002/smll.200700245. [DOI] [PubMed] [Google Scholar]
- 47.Luo S-C, Sekine J, Zhu B, Zhao H, Nakao A, Yu H-h. ACS Nano. 2012;6:3018. doi: 10.1021/nn300737e. [DOI] [PubMed] [Google Scholar]
- 48.Fuard D, Tzvetkova-Chevolleau T, Decossas S, Tracqui P, Schiavone P. Microelectron. Eng. 2008;85:1289. [Google Scholar]
- 49.Mattila PK, Lappalainen P. Nat. Rev. Mol. Cell Biol. 2008;9:446. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 50.Hsiao Y-S, Whang W-T, Chen C-P, Chen Y-C. J. Mater. Chem. 2008;18:5948. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.